Oral Absorbent AST-120 Is Associated with Compositional and Functional Adaptations of Gut Microbiota and Modification of Serum Short and Medium-Chain Fatty Acids in Advanced CKD Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Patient Recruitment
2.2. Gut-Producing Metabolites
2.3. Fecal 16S rRNA Gene Sequencing and Functional Prediction of Bacterial Gene
2.4. Statistical Analysis
3. Results
3.1. Subject Characteristics
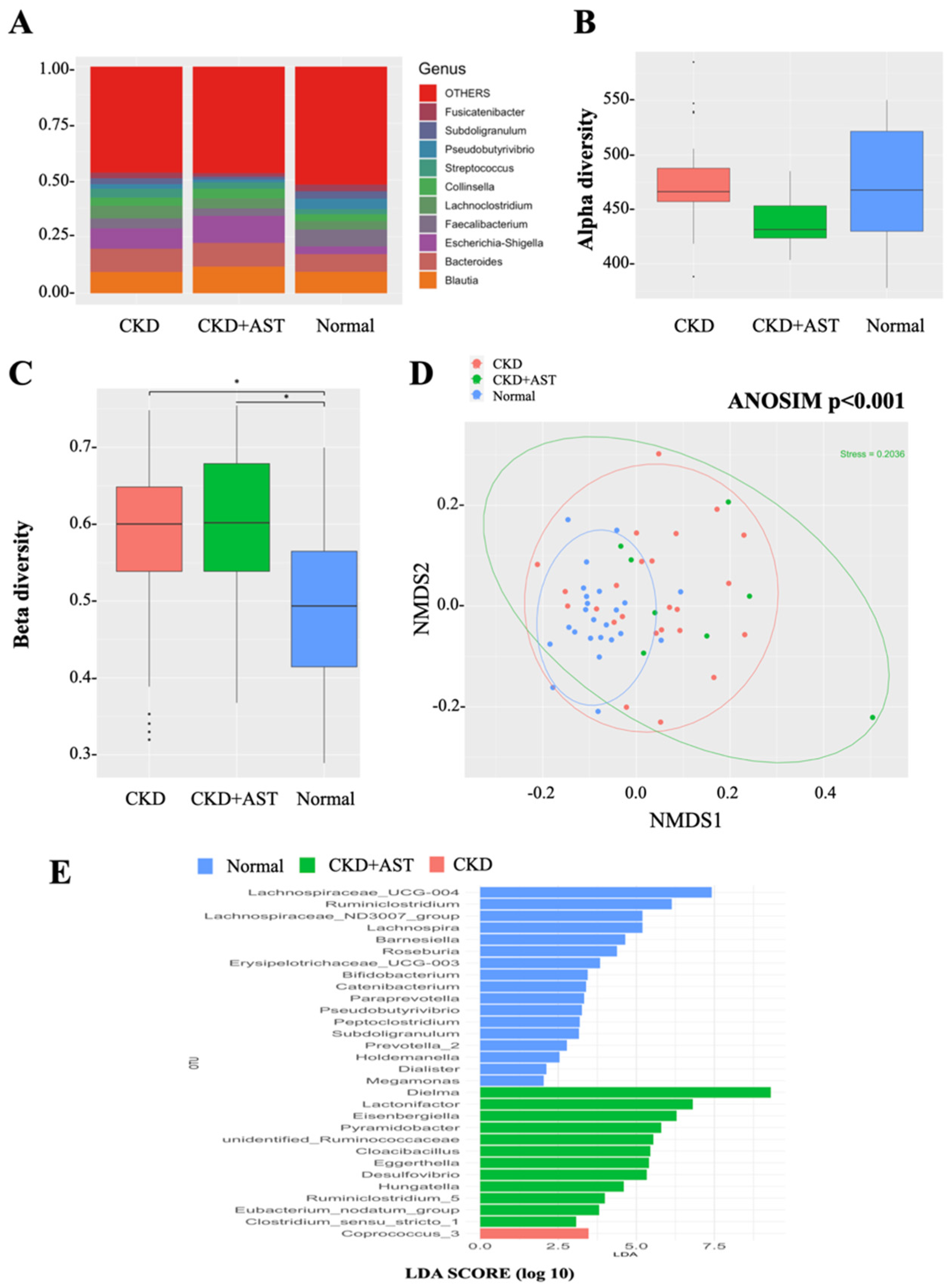
3.2. Modification of Gut Microbial Architecture in Advanced CKD Patients Receiving AST-120
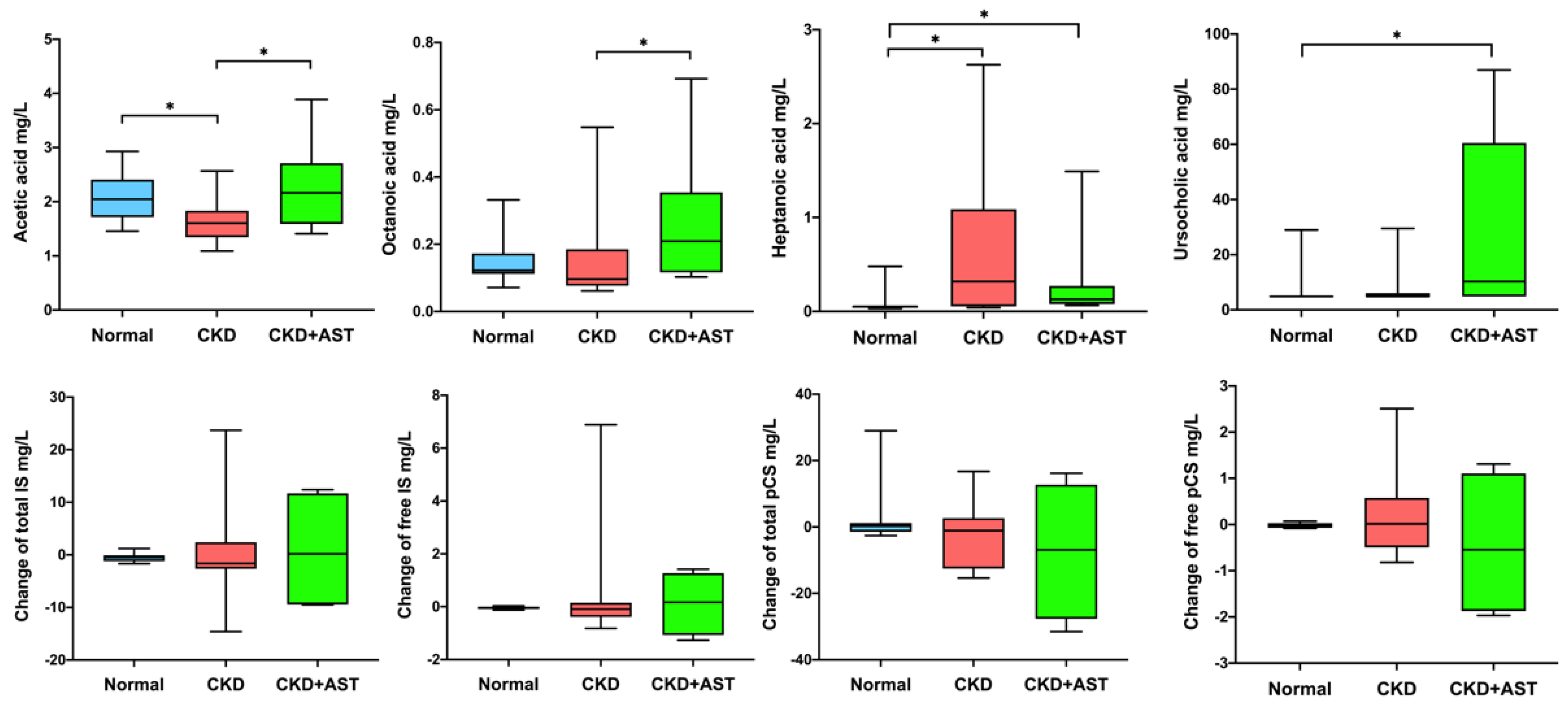
3.3. Description of Variation of Metabolites in Advanced CKD Patients Receiving AST-120
3.4. Prediction of Functional Capability of Genes of Gut Microbiota Associated with AST-120
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Felizardo, R.J.F.; Watanabe, I.K.M.; Dardi, P.; Rossoni, L.V.; Câmara, N.O.S. The interplay among gut microbiota, hypertension and kidney diseases: The role of short-chain fatty acids. Pharmacol. Res. 2019, 141, 366–377. [Google Scholar] [CrossRef]
- Wang, X.; Yang, S.; Li, S.; Zhao, L.; Hao, Y.; Qin, J.; Zhang, L.; Zhang, C.; Bian, W.; Zuo, L.; et al. Aberrant gut microbiota alters host metabolome and impacts renal failure in humans and rodents. Gut 2020, 69, 2131–2142. [Google Scholar] [CrossRef] [PubMed]
- Laville, S.M.; Massy, Z.A.; Kamel, S.; Chillon, J.M.; Choukroun, G.; Liabeuf, S. Intestinal Chelators, Sorbants, and Gut-Derived Uremic Toxins. Toxins 2021, 13, 91. [Google Scholar] [CrossRef] [PubMed]
- Vanholder, R.; Schepers, E.; Pletinck, A.; Nagler, E.V.; Glorieux, G. The uremic toxicity of indoxyl sulfate and p-cresyl sulfate: A systematic review. J. Am. Soc. Nephrol. 2014, 25, 1897–1907. [Google Scholar] [CrossRef] [PubMed]
- Wu, I.W.; Hsu, K.H.; Lee, C.C.; Sun, C.Y.; Hsu, H.J.; Tsai, C.J.; Tzen, C.Y.; Wang, Y.C.; Lin, C.Y.; Wu, M.S. p-Cresyl sulphate and indoxyl sulphate predict progression of chronic kidney disease. Nephrol. Dial. Transpl. 2011, 26, 938–947. [Google Scholar] [CrossRef]
- Wu, I.W.; Hsu, K.H.; Hsu, H.J.; Lee, C.C.; Sun, C.Y.; Tsai, C.J.; Wu, M.S. Serum free p-cresyl sulfate levels predict cardiovascular and all-cause mortality in elderly hemodialysis patients—A prospective cohort study. Nephrol. Dial. Transpl. 2012, 27, 1169–1175. [Google Scholar] [CrossRef]
- Cha, R.H.; Kang, S.W.; Park, C.W.; Cha, D.R.; Na, K.Y.; Kim, S.G.; Yoon, S.A.; Kim, S.; Han, S.Y.; Park, J.H.; et al. Sustained uremic toxin control improves renal and cardiovascular outcomes in patients with advanced renal dysfunction: Post-hoc analysis of the Kremezin Study against renal disease progression in Korea. Kidney Res. Clin. Pract. 2017, 36, 68–78. [Google Scholar] [CrossRef]
- Schulman, G.; Berl, T.; Beck, G.J.; Remuzzi, G.; Ritz, E.; Shimizu, M.; Shobu, Y.; Kikuchi, M. The effects of AST-120 on chronic kidney disease progression in the United States of America: A post hoc subgroup analysis of randomized controlled trials. BMC Nephrol. 2016, 17, 141. [Google Scholar] [CrossRef]
- Asai, M.; Kumakura, S.; Kikuchi, M. Review of the efficacy of AST-120 (KREMEZIN(®)) on renal function in chronic kidney disease patients. Ren. Fail. 2019, 41, 47–56. [Google Scholar] [CrossRef]
- Ueda, H.; Shibahara, N.; Takagi, S.; Inoue, T.; Katsuoka, Y. AST-120 treatment in pre-dialysis period affects the prognosis in patients on hemodialysis. Ren. Fail. 2008, 30, 856–860. [Google Scholar] [CrossRef]
- Wu, I.W.; Hsu, K.H.; Sun, C.Y.; Tsai, C.J.; Wu, M.S.; Lee, C.C. Oral adsorbent AST-120 potentiates the effect of erythropoietin-stimulating agents on Stage 5 chronic kidney disease patients: A randomized crossover study. Nephrol. Dial. Transpl. 2014, 29, 1719–1727. [Google Scholar] [CrossRef] [PubMed]
- Sato, E.; Hosomi, K.; Sekimoto, A.; Mishima, E.; Oe, Y.; Saigusa, D.; Ito, S.; Abe, T.; Sato, H.; Kunisawa, J.; et al. Effects of the oral adsorbent AST-120 on fecal p-cresol and indole levels and on the gut microbiota composition. Biochem. Biophys. Res. Commun. 2020, 525, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Yoshifuji, A.; Wakino, S.; Irie, J.; Matsui, A.; Hasegawa, K.; Tokuyama, H.; Hayashi, K.; Itoh, H. Oral adsorbent AST-120 ameliorates gut environment and protects against the progression of renal impairment in CKD rats. Clin. Exp. Nephrol. 2018, 22, 1069–1078. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, N.D.; Yuan, J.; Khazaeli, M.; Masuda, Y.; Ichii, H.; Liu, S. Oral activated charcoal adsorbent (AST-120) ameliorates chronic kidney disease-induced intestinal epithelial barrier disruption. Am. J. Nephrol. 2013, 37, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Al Khodor, S.; Shatat, I.F. Gut microbiome and kidney disease: A bidirectional relationship. Pediatr. Nephrol. 2017, 32, 921–931. [Google Scholar] [CrossRef]
- Wu, I.W.; Lin, C.Y.; Chang, L.C.; Lee, C.C.; Chiu, C.Y.; Hsu, H.J.; Sun, C.Y.; Chen, Y.C.; Kuo, Y.L.; Yang, C.W.; et al. Gut Microbiota as Diagnostic Tools for Mirroring Disease Progression and Circulating Nephrotoxin Levels in Chronic Kidney Disease: Discovery and Validation Study. Int. J. Biol. Sci. 2020, 16, 420–434. [Google Scholar] [CrossRef]
- Wu, I.W.; Gao, S.S.; Chou, H.C.; Yang, H.Y.; Chang, L.C.; Kuo, Y.L.; Dinh, M.C.V.; Chung, W.H.; Yang, C.W.; Lai, H.C.; et al. Integrative metagenomic and metabolomic analyses reveal severity-specific signatures of gut microbiota in chronic kidney disease. Theranostics 2020, 10, 5398–5411. [Google Scholar] [CrossRef]
- Wu, I.W.; Lee, C.C.; Hsu, H.J.; Sun, C.Y.; Chen, Y.C.; Yang, K.J.; Yang, C.W.; Chung, W.H.; Lai, H.C.; Chang, L.C.; et al. Compositional and Functional Adaptations of Intestinal Microbiota and Related Metabolites in CKD Patients Receiving Dietary Protein Restriction. Nutrients 2020, 12, 2799. [Google Scholar] [CrossRef]
- Lin, C.N.; Wu, I.W.; Huang, Y.F.; Peng, S.Y.; Huang, Y.C.; Ning, H.C. Measuring serum total and free indoxyl sulfate and p-cresyl sulfate in chronic kidney disease using UPLC-MS/MS. J. Food Drug Anal. 2019, 27, 502–509. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, V.M.; Chen, I.M.; Palaniappan, K.; Chu, K.; Szeto, E.; Grechkin, Y.; Ratner, A.; Jacob, B.; Huang, J.; Williams, P.; et al. IMG: The Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 2012, 40, D115–D122. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Sumida, K.; Lau, W.L.; Kalantar-Zadeh, K.; Kovesdy, C.P. Novel intestinal dialysis interventions and microbiome modulation to control uremia. Curr. Opin. Nephrol. Hypertens. 2022, 31, 82–91. [Google Scholar] [CrossRef]
- Schulman, G.; Berl, T.; Beck, G.J.; Remuzzi, G.; Ritz, E.; Arita, K.; Kato, A.; Shimizu, M. Randomized Placebo-Controlled EPPIC Trials of AST-120 in CKD. J. Am. Soc. Nephrol. 2015, 26, 1732–1746. [Google Scholar] [CrossRef]
- Cha, R.H.; Kang, S.W.; Park, C.W.; Cha, D.R.; Na, K.Y.; Kim, S.G.; Yoon, S.A.; Han, S.Y.; Chang, J.H.; Park, S.K.; et al. A Randomized, Controlled Trial of Oral Intestinal Sorbent AST-120 on Renal Function Deterioration in Patients with Advanced Renal Dysfunction. Clin. J. Am. Soc. Nephrol. 2016, 11, 559–567. [Google Scholar] [CrossRef]
- Kaakoush, N.O. Insights into the Role of Erysipelotrichaceae in the Human Host. Front. Cell. Infect. Microbiol. 2015, 5, 84. [Google Scholar] [CrossRef]
- Mo, Y.; Sun, H.; Zhang, L.; Geng, W.; Wang, L.; Zou, C.; Wu, Y.; Ji, C.; Liu, X.; Lu, Z. Microbiome-Metabolomics Analysis Reveals the Protection Mechanism of α-Ketoacid on Adenine-Induced Chronic Kidney Disease in Rats. Front. Pharmacol. 2021, 12, 657827. [Google Scholar] [CrossRef]
- Hiraga, Y.; Kubota, T.; Katoh, M.; Horai, Y.; Suzuki, H.; Yamashita, Y.; Hirata, R.; Moroi, M. AST-120 Treatment Alters the Gut Microbiota Composition and Suppresses Hepatic Triglyceride Levels in Obese Mice. Endocr. Res. 2021, 46, 178–185. [Google Scholar] [CrossRef]
- Hirakawa, H.; Takita, A.; Uchida, M.; Kaneko, Y.; Kakishima, Y.; Tanimoto, K.; Kamitani, W.; Tomita, H. Adsorption of Phenazines Produced by Pseudomonas aeruginosa Using AST-120 Decreases Pyocyanin-Associated Cytotoxicity. Antibiotics 2021, 10, 434. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Uchida, M.; Kurabayashi, K.; Nishijima, F.; Takita, A.; Tomita, H. In vitro activity of AST-120 that suppresses indole signaling in Escherichia coli, which attenuates drug tolerance and virulence. PLoS ONE 2020, 15, e0232461. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.; Piceno, Y.M.; DeSantis, T.Z.; Pahl, M.; Andersen, G.L.; Vaziri, N.D. Expansion of urease- and uricase-containing, indole- and p-cresol-forming and contraction of short-chain fatty acid-producing intestinal microbiota in ESRD. Am. J. Nephrol. 2014, 39, 230–237. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Schönfeld, P.; Wojtczak, L. Short- and medium-chain fatty acids in energy metabolism: The cellular perspective. J. Lipid Res. 2016, 57, 943–954. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [CrossRef]
- Miao, W.; Wu, X.; Wang, K.; Wang, W.; Wang, Y.; Li, Z.; Liu, J.; Li, L.; Peng, L. Sodium Butyrate Promotes Reassembly of Tight Junctions in Caco-2 Monolayers Involving Inhibition of MLCK/MLC2 Pathway and Phosphorylation of PKCβ2. Int. J. Mol. Sci. 2016, 17, 1696. [Google Scholar] [CrossRef]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef]
- Frampton, J.; Murphy, K.G.; Frost, G.; Chambers, E.S. Short-chain fatty acids as potential regulators of skeletal muscle metabolism and function. Nat. Metab. 2020, 2, 840–848. [Google Scholar] [CrossRef]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef]
- Kelly, C.J.; Zheng, L.; Campbell, E.L.; Saeedi, B.; Scholz, C.C.; Bayless, A.J.; Wilson, K.E.; Glover, L.E.; Kominsky, D.J.; Magnuson, A.; et al. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe 2015, 17, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.C.; Tomino, Y.; Lu, K.C. Impacts of Indoxyl Sulfate and p-Cresol Sulfate on Chronic Kidney Disease and Mitigating Effects of AST-120. Toxins 2018, 10, 367. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, S.; Hashimoto, R.; Tezuka, T.; Sakuma, M.; Abe, S.; Ishikawa, T.; Taguchi, I.; Inoue, T. Antioxidative effect of an oral adsorbent, AST-120, and long-term outcomes in chronic kidney disease patients with cardiovascular disease. Hypertens. Res. 2020, 43, 1128–1131. [Google Scholar] [CrossRef]

| All Patients, n = 56 | Normal, n = 24 | CKD, n = 24 | CKD+AST, n = 8 | p | |
|---|---|---|---|---|---|
| Age, mean (SD) | 66.25 ± 7.65 | 64.04 ± 6.54 | 68.04 ± 7.31 | 67.5 ± 10.67 | 0.691 |
| Male, n (%) | 24 (42.9) | 12 (50) | 10 (41.7) | 2 (25) | 0.676 |
| Diabetes, n (%) | 23 (41.1) | 9 (37.5) | 12 (50) | 2 (25) | 0.412 |
| Hypertension, n (%) | 44 (78.6) | 15 (62.5) | 23 (95.8) | 6 (75) | 0.147 |
| Gout, n (%) | 6 (10.7) | 0 (0) | 4 (16.7) | 2 (25) | 0.169 |
| Hyperlipidemia, n (%) | 25 (44.6) | 11 (45.8) | 12 (50) | 2 (25) | 0.133 |
| Systolic pressure, mm Hg | 134.20 ± 16.52 | 128.42 ± 17.02 | 138.71 ± 14.59 | 138.57 ± 17.13 | 0.95 |
| Body mass index kg/m2 | 25.72 ± 4.03 | 25.87 ± 4.09 | 25.4 ± 4.1 | 26.23 ± 4.08 | 0.482 |
| Blood urea nitrogen, mg/dL | 41.73 ± 30.14 | 14.42 ± 3.62 | 62.96 ± 22.99 | 59.96 ± 29.57 | 0.615 |
| Serum creatinine, mg/dL | 3.08 ± 2.61 | 0.78 ± 0.19 | 4.51 ± 1.95 | 5.68 ± 2.83 | 0.133 |
| Estimated GFR, mL/min/m2 | 52.16 ± 61.07 | 102.7 ± 63.94 | 13.94 ± 7.09 | 15.24 ± 19.48 | 0.335 |
| Hemoglobin, g/dL | 11.15 ± 2.43 | 13.5 ± 1.18 | 9.35 ± 1.37 | 9.54 ± 1.71 | 0.934 |
| Serum albumin, mg/dL | 4.3 ± 0.53 | 4.55 ± 0.24 | 4.06 ± 0.67 | 4.28 ± 0.34 | 0.904 |
| Serum calcium, mg/dL | 8.94 ± 0.67 | 9.29 ± 0.36 | 8.7 ± 0.77 | 8.64 ± 0.62 | 0.454 |
| Serum phosphate, mg/dL | 4.29 ± 1.03 | 3.79 ± 0.54 | 4.66 ± 1.09 | 4.65 ± 1.41 | 0.6 |
| Serum potassium, mEq/L | 4.29 ± 0.58 | 4.1 ± 0.35 | 4.41 ± 0.52 | 4.51 ± 0.97 | 0.315 |
| Serum uric acid, mg/dL | 5.98 ± 1.81 | 5.64 ± 1.48 | 6.48± 2.07 | 5.55 ± 1.79 | 0.452 |
| Fasting sugar, mg/dL | 116.4 ± 36.98 | 119.42 ± 31.28 | 116.58 ± 45.98 | 105.43 ± 16.72 | 0.58 |
| Total cholesterol, mg/dL | 186.25 ± 40.5 | 192.71 ± 25.05 | 191.04 ± 46.66 | 152.5 ± 49.55 | 0.22 |
| hs-CRP, mg/L | 1.46 (2.56) | 0.89 (1.21) | 2.4 (7.29) | 0.8 (2.08) | 0.457 |
| Urine protein-creatinine ratio, mg/g | 1331.69 (2237.75) | 81.5 (122.21) | 2144 (1582.46) | 1279.5 (701) | 0.404 |
| CKD stage, n (%) | 0.15 | ||||
| 4 | 8 (14.3) | 0 | 7 (29.2) | 1 (12.5) | |
| 5 | 24 (42.9) | 0 | 17 (70.8) | 7 (87.5) |
| Gut Microbiota | RA, Non-CKD | RA, CKD+AST | RA, CKD | p * | p (CKD5+AST vs. CKD) # | p (CKD+AST vs. Non-CKD) # | p (CKD vs. Non-CKD) # | |
|---|---|---|---|---|---|---|---|---|
| Family | Genus | |||||||
| Desulfovibrionaceae | Bilophila ↑ | 0.001321 | 0.002437 | 0.001151 | 0.01777 | 0.0023 | 0.0222 | 0.1221 |
| Desulfovibrionaceae | Desulfovibrio ↑ | 0.00131 | 0.006965 | 0.002721 | 0.00005694 | 0.008106 | 0.00001035 | 0.004386 |
| Synergistaceae | Cloacibacillus ↑ | 0.00005216 | 0.001904 | 0.0001465 | 0.0003161 | 0.004336 | 0.00003474 | 0.02781 |
| Synergistaceae | Pyramidobacter ↑ | 0.00009877 | 0.002647 | 0.0003385 | 0.002609 | 0.001757 | 0.0003311 | 0.246 |
| Clostridiaceae | Hungatella ↑ | 0.0004262 | 0.002574 | 0.001379 | 0.0004461 | 0.01319 | 0.00006986 | 0.01236 |
| Clostridiaceae | Clostridium_sensu_stricto_1 ↑ | 0.00682 | 0.0283 | 0.01751 | 0.0009109 | 0.04831 | 0.0002663 | 0.005404 |
| Acidaminococcaceae | Phascolarctobacterium ↑ | 0.001441 | 0.002211 | 0.0007402 | 0.02947 | 0.004157 | 0.03693 | 0.1143 |
| Eggerthellaceae | Eggerthella ↑ | 0.0007291 | 0.001958 | 0.0009766 | 0.001362 | 0.00127 | 0.0001586 | 0.2051 |
| Eubacteriaceae | Eubacterium_nodatum ↑ | 0.00006437 | 0.0006392 | 0.0004018 | 0.00009063 | 0.01742 | 0.00002331 | 0.002764 |
| Ruminococcaceae | Ruminococcus_2 ↑ | 0.01591 | 0.03468 | 0.01968 | 0.02802 | 0.01359 | 0.003985 | 0.2649 |
| Erysipelotrichaceae | Erysipelotrichaceae_UCG-003 ↓ | 0.01026 | 0.001492 | 0.007197 | 0.0001766 | 0.000575 | 0.0000165 | 0.1013 |
| Erysipelotrichaceae | Catenibacterium ↓ | 0.01586 | 0.001242 | 0.00566 | 0.0002244 | 0.01655 | 0.0000453 | 0.005829 |
| Lachnospiraceae | Coprococcus_3 ↓ | 0.005866 | 0.00245 | 0.01383 | 0.006751 | 0.0009957 | 0.002919 | 0.3179 |
| Lachnospiraceae | Lachnospira ↓ | 0.002425 | 0.0005027 | 0.001978 | 0.000772 | 0.02734 | 0.0001599 | 0.008844 |
| Prevotellaceae | Prevotella_9 ↓ | 0.04353 | 0.001145 | 0.009813 | 0.0002952 | 0.01108 | 0.00004583 | 0.01081 |
| Porphyromonadaceae | Barnesiella ↓ | 0.003292 | 0.0004162 | 0.001276 | 0.00299 | 0.03622 | 0.0005491 | 0.01896 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, C.-K.; Su, S.-C.; Chang, L.-C.; Yang, K.-J.; Lee, C.-C.; Hsu, H.-J.; Chen, Y.-T.; Sun, C.-Y.; Wu, I.-W. Oral Absorbent AST-120 Is Associated with Compositional and Functional Adaptations of Gut Microbiota and Modification of Serum Short and Medium-Chain Fatty Acids in Advanced CKD Patients. Biomedicines 2022, 10, 2234. https://doi.org/10.3390/biomedicines10092234
Hsu C-K, Su S-C, Chang L-C, Yang K-J, Lee C-C, Hsu H-J, Chen Y-T, Sun C-Y, Wu I-W. Oral Absorbent AST-120 Is Associated with Compositional and Functional Adaptations of Gut Microbiota and Modification of Serum Short and Medium-Chain Fatty Acids in Advanced CKD Patients. Biomedicines. 2022; 10(9):2234. https://doi.org/10.3390/biomedicines10092234
Chicago/Turabian StyleHsu, Cheng-Kai, Shih-Chi Su, Lun-Ching Chang, Kai-Jie Yang, Chin-Chan Lee, Heng-Jung Hsu, Yih-Ting Chen, Chiao-Yin Sun, and I-Wen Wu. 2022. "Oral Absorbent AST-120 Is Associated with Compositional and Functional Adaptations of Gut Microbiota and Modification of Serum Short and Medium-Chain Fatty Acids in Advanced CKD Patients" Biomedicines 10, no. 9: 2234. https://doi.org/10.3390/biomedicines10092234
APA StyleHsu, C.-K., Su, S.-C., Chang, L.-C., Yang, K.-J., Lee, C.-C., Hsu, H.-J., Chen, Y.-T., Sun, C.-Y., & Wu, I.-W. (2022). Oral Absorbent AST-120 Is Associated with Compositional and Functional Adaptations of Gut Microbiota and Modification of Serum Short and Medium-Chain Fatty Acids in Advanced CKD Patients. Biomedicines, 10(9), 2234. https://doi.org/10.3390/biomedicines10092234







