Two Loci Contribute to Age-Related Hearing Loss Resistance in the Japanese Wild-Derived Inbred MSM/Ms Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Hearing Tests
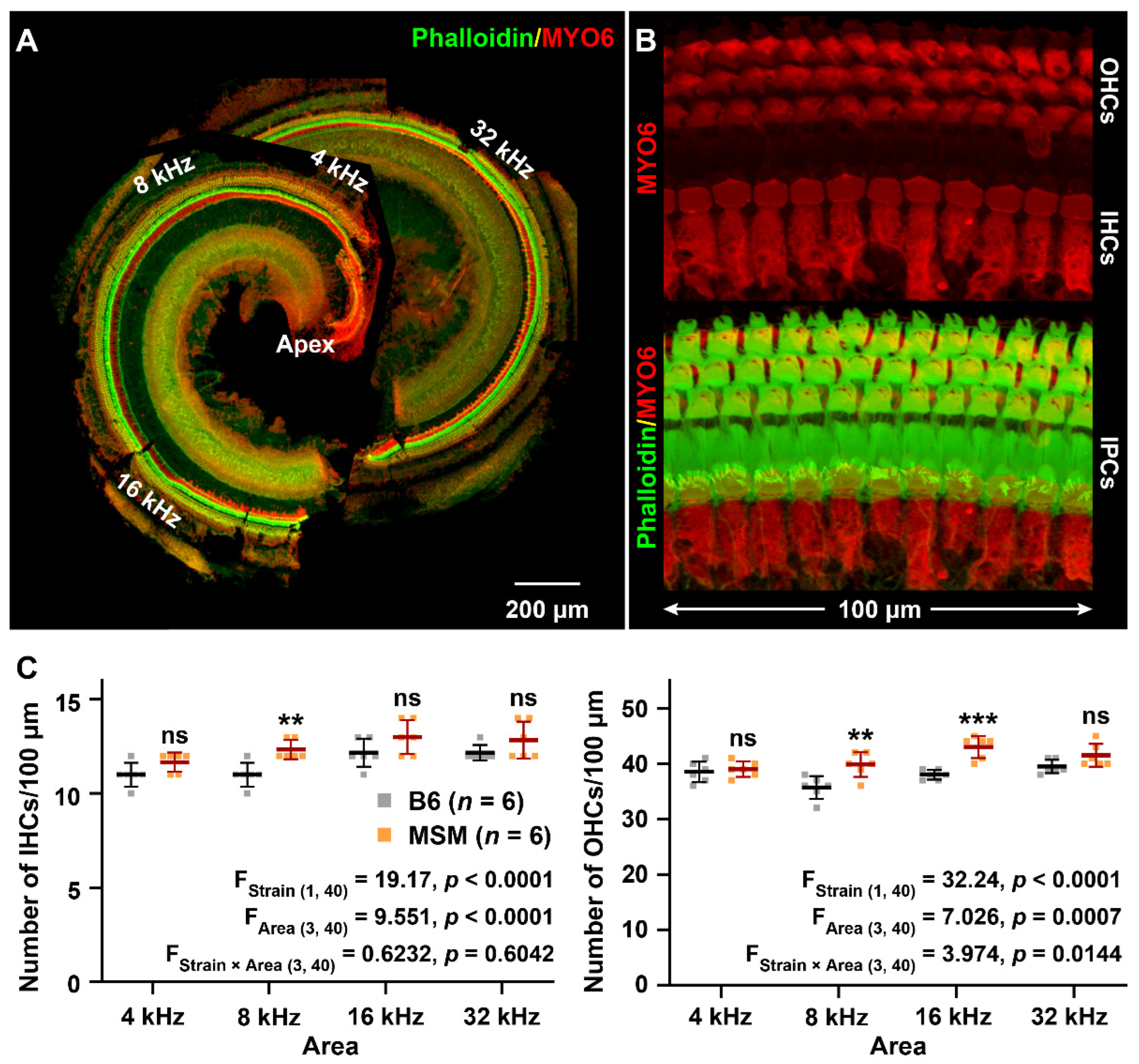
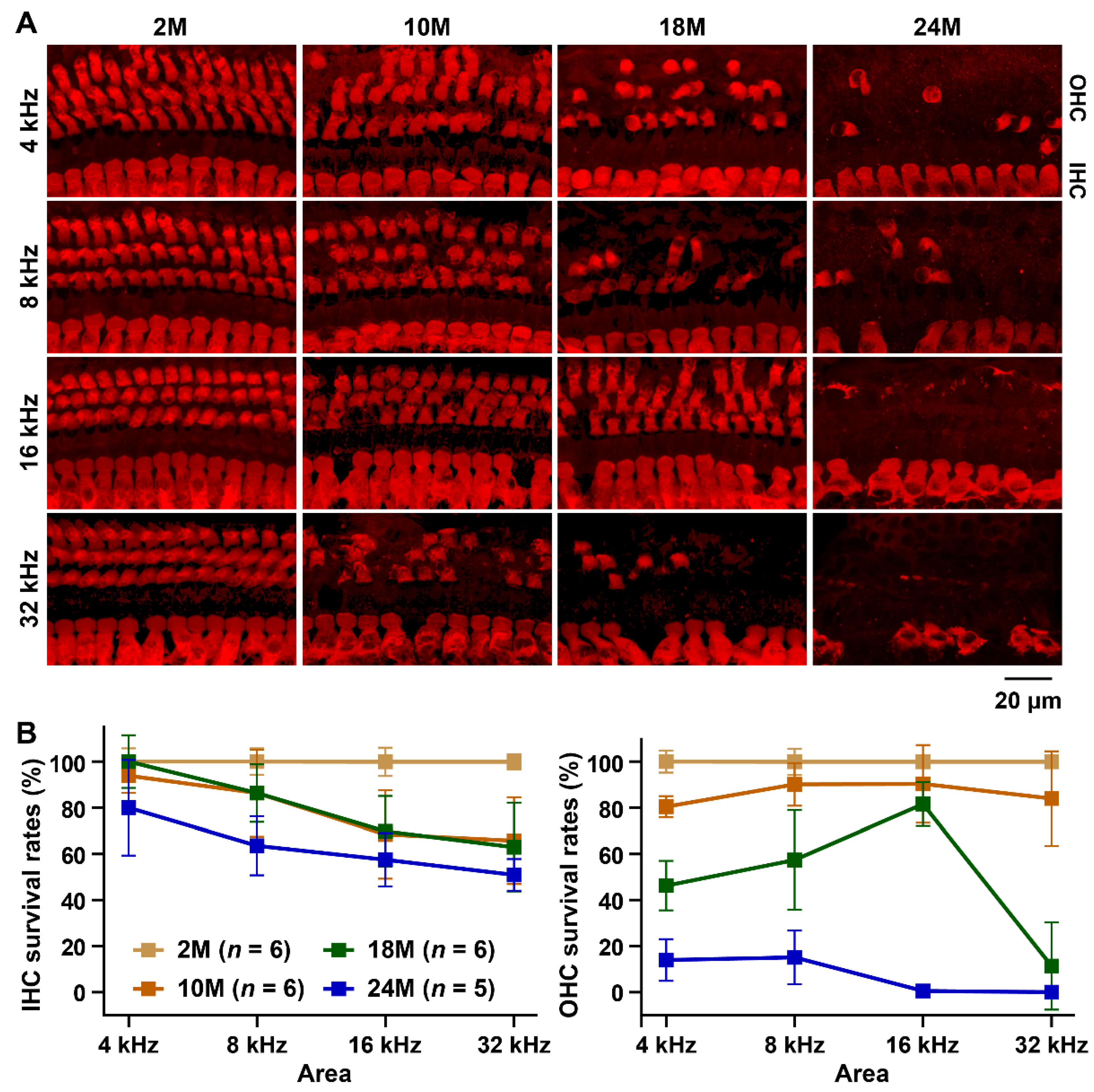
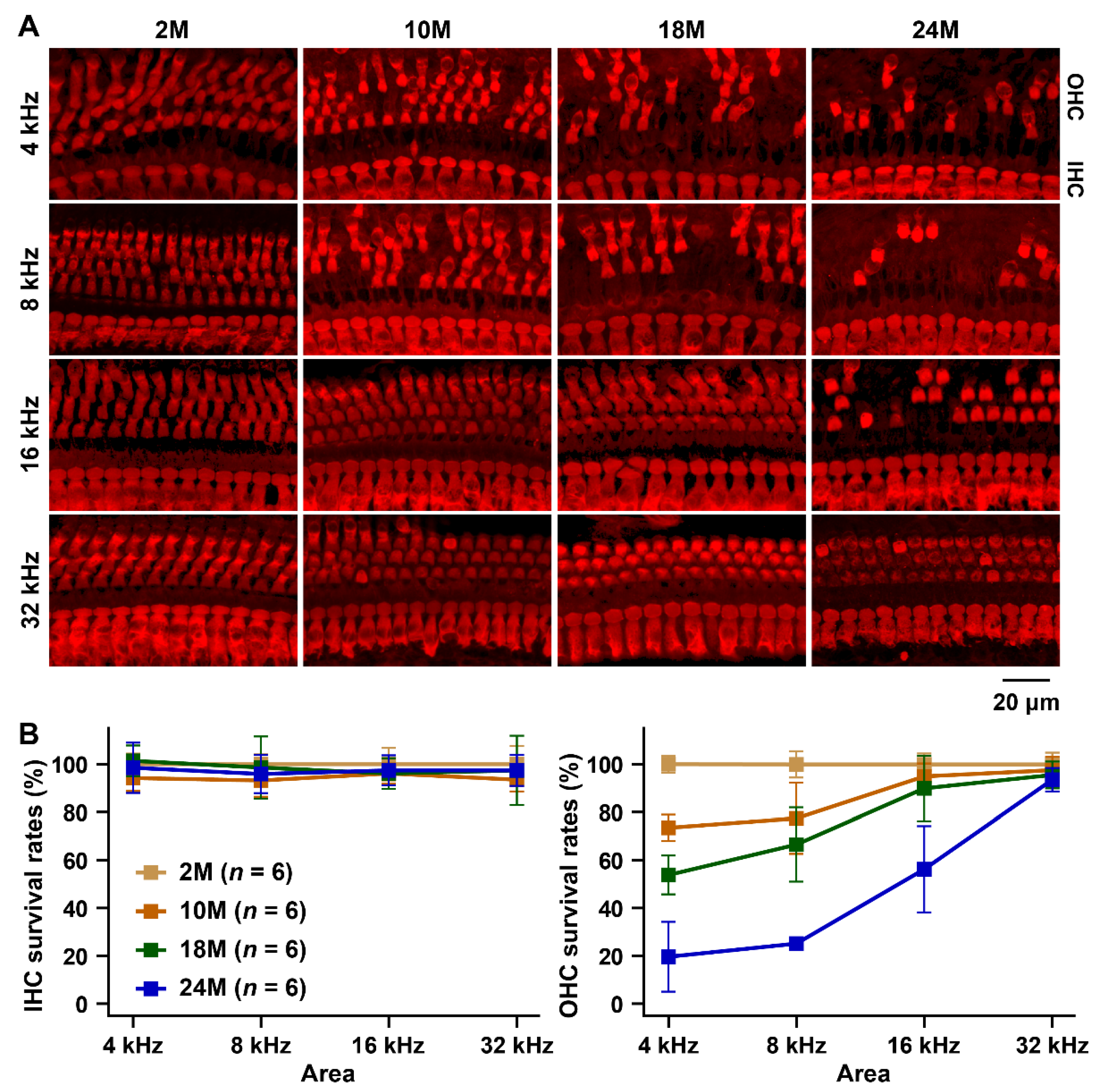
2.3. Hair Cell Counts
2.4. Scanning Electron Microscopy
2.5. Linkage Analysis
2.6. Statistical Analysis
3. Results
3.1. Characterization of the ARHL Associated Phenotypes in MSM Strain
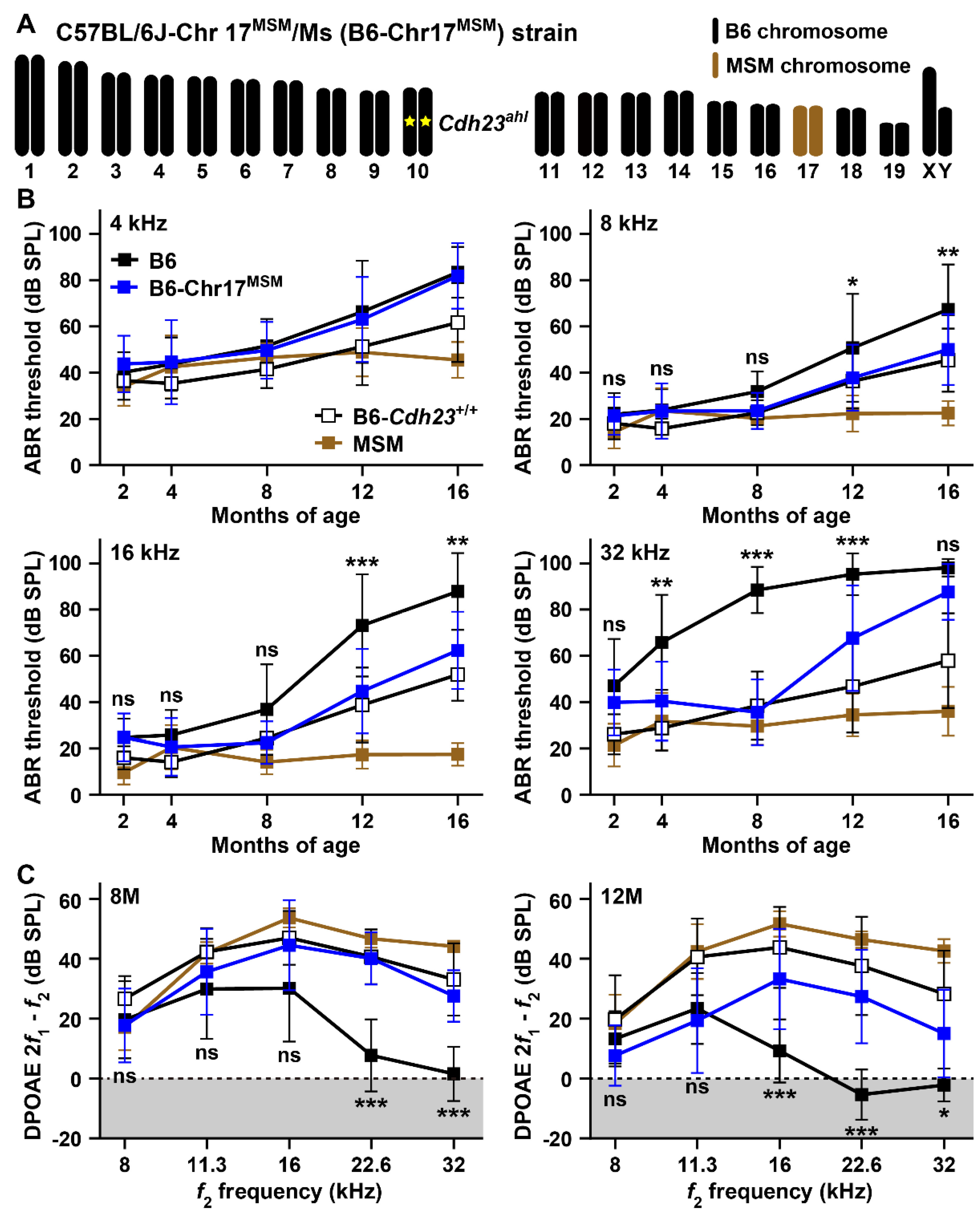
3.2. Hearing Assessments in B6 Strain by Substitution to MSM-Derived Chromosome 17
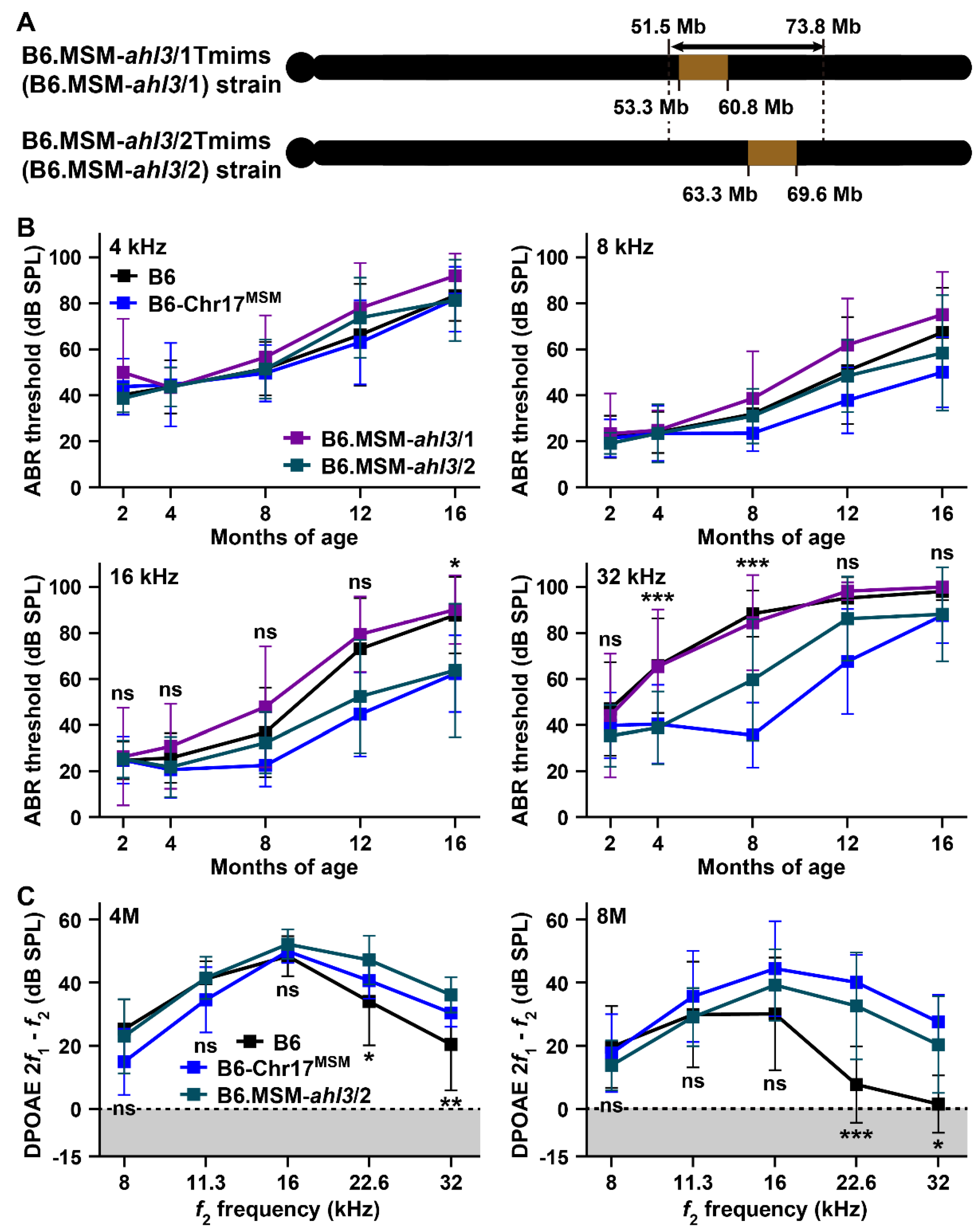
3.3. Hearing Assessments in B6 Strain by Transferring of MSM-Derived ahl3 Genomic Regions
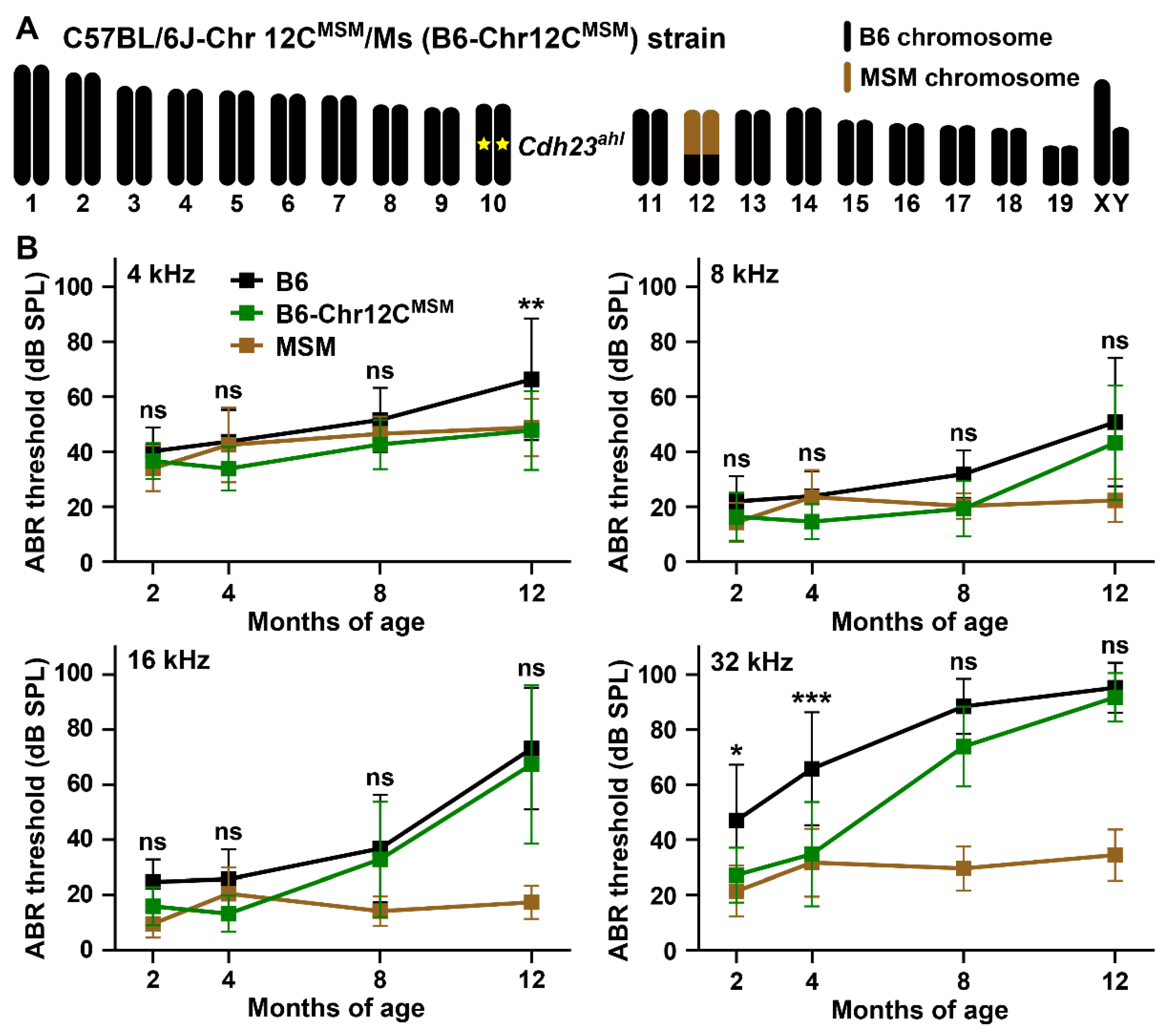
3.4. ARHL Resistance in B6 Strain by Transferring MSM-Derived Centromeric Region on Chromosome 12
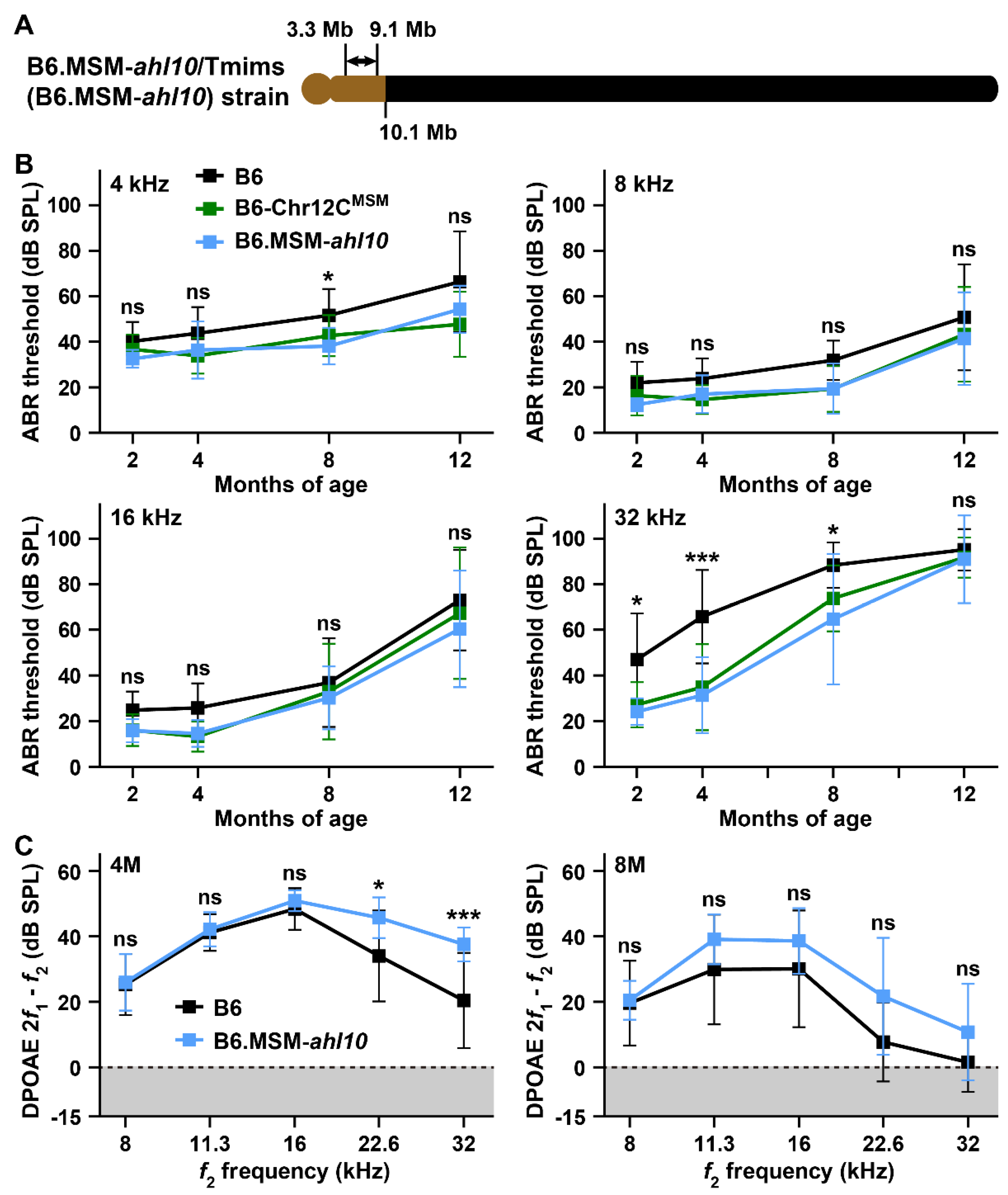
3.5. Identification of the ahl10 Locus on Mouse Chromosome 12
3.6. Exploration of Candidate Genes in ahl3 and ahl10 Regions
4. Discussion
4.1. Pathological Factors That Prevent ARHL Onset in MSM Strain
4.2. ARHL Resistance Effect in B6 Strain by Transferring MSM-Derived Chromosome 17 and ahl3 Genomic Regions
4.3. ARHL Resistance Effect in B6 Strain by Transfer of MSM-Derived Chromosome 12C and ahl10 Genomic Regions
4.4. Prediction of Candidate Genes for ARHL Resistance by ahl3 and ahl10 loci
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kikkawa, Y.; Seki, Y.; Okumura, K.; Ohshiba, Y.; Miyasaka, Y.; Suzuki, S.; Ozaki, M.; Matsuoka, K.; Noguchi, Y.; Yonekawa, H. Advantages of a Mouse Model for Human Hearing Impairment. Exp. Anim. 2012, 61, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Ohlemiller, K.K.; Jones, S.M.; Johnson, K.R. Application of Mouse Models to Research in Hearing and Balance. J. Assoc. Res. Otolaryngol. 2016, 17, 493–523. [Google Scholar] [CrossRef]
- Beck, J.A.; Lloyd, S.; Hafezparast, M.; Lennon-Pierce, M.; Eppig, J.T.; Festing, M.F.; Fisher, E.M. Genealogies of Mouse Inbred Strains. Nat. Genet. 2000, 24, 23–25. [Google Scholar] [CrossRef] [PubMed]
- Keithley, E.M.; Canto, C.; Zheng, Q.Y.; Fischel-Ghodsian, N.; Johnson, K.R. Age-Related Hearing Loss and the Ahl Locus in Mice. Hear. Res. 2004, 188, 21–28. [Google Scholar] [CrossRef]
- Zheng, Q.Y.; Johnson, K.R.; Erway, L.C. Assessment of Hearing in 80 Inbred Strains of Mice by ABR Threshold Analyses. Hear. Res. 1999, 130, 94–107. [Google Scholar] [CrossRef]
- Yasuda, S.P.; Seki, Y.; Suzuki, S.; Ohshiba, Y.; Hou, X.; Matsuoka, K.; Wada, K.; Shitara, H.; Miyasaka, Y.; Kikkawa, Y. c.753A>G Genome Editing of a Cdh23ahl Allele Delays Age-Related Hearing Loss and Degeneration of Cochlear Hair Cells in C57BL/6J Mice. Hear. Res. 2020, 389, 107926. [Google Scholar] [CrossRef]
- Suzuki, S.; Ishikawa, M.; Ueda, T.; Ohshiba, Y.; Miyasaka, Y.; Okumura, K.; Yokohama, M.; Taya, C.; Matsuoka, K.; Kikkawa, Y. Quantitative Trait Loci on Chromosome 5 for Susceptibility to Frequency-Specific Effects on Hearing in DBA/2J Mice. Exp. Anim. 2015, 64, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.R.; Longo-Guess, C.; Gagnon, L.H.; Yu, H.; Zheng, Q.Y. A Locus on Distal Chromosome 11 (ahl8) and Its Interaction with Cdh23ahl Underlie the Early Onset, Age-Related Hearing Loss of DBA/2J Mice. Genomics 2008, 92, 219–225. [Google Scholar] [CrossRef]
- Noben-Trauth, K.; Zheng, Q.Y.; Johnson, K.R. Association of Cadherin 23 with Polygenic Inheritance and Genetic Modification of Sensorineural Hearing Loss. Nat. Genet. 2003, 35, 21–23. [Google Scholar] [CrossRef]
- Miyasaka, Y.; Shitara, H.; Suzuki, S.; Yoshimoto, S.; Seki, Y.; Ohshiba, Y.; Okumura, K.; Taya, C.; Tokano, H.; Kitamura, K.; et al. Heterozygous Mutation of Ush1g/Sans in Mice Causes Early-Onset Progressive Hearing Loss, Which Is Recovered by Reconstituting the Strain-Specific Mutation in Cdh23. Hum. Mol. Genet. 2016, 25, 2045–2059. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.-B.; Longo-Guess, C.M.; Gagnon, L.H.; Saylor, K.W.; Dumont, R.A.; Spinelli, K.J.; Pagana, J.M.; Wilmarth, P.A.; David, L.L.; Gillespie, P.G.; et al. The R109H Variant of Fascin-2, A Developmentally Regulated Actin Crosslinker in Hair-Cell Stereocilia, Underlies Early-Onset Hearing Loss of DBA/2J Mice. J. Neurosci. 2010, 30, 9683–9694. [Google Scholar] [CrossRef] [PubMed]
- Perrin, B.J.; Strandjord, D.M.; Narayanan, P.; Henderson, D.M.; Johnson, K.R.; Ervasti, J.M. β-Actin and Fascin-2 Cooperate to Maintain Stereocilia Length. J. Neurosci. 2013, 33, 8114–8121. [Google Scholar] [CrossRef]
- Johnson, K.R.; Longo-Guess, C.M.; Gagnon, L.H. A QTL on Chr 5 Modifies Hearing Loss Associated with the Fascin-2 Variant of DBA/2J Mice. Mamm. Genome 2015, 26, 338–347. [Google Scholar] [CrossRef]
- Takada, T.; Mita, A.; Maeno, A.; Sakai, T.; Shitara, H.; Kikkawa, Y.; Moriwaki, K.; Yonekawa, H.; Shiroishi, T. Mouse Inter-Subspecific Consomic Strains for Genetic Dissection of Quantitative Complex Traits. Genome Res. 2008, 18, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Takada, T.; Ebata, T.; Noguchi, H.; Keane, T.M.; Adams, D.J.; Narita, T.; Shin-I, T.; Fujisawa, H.; Toyoda, A.; Abe, K.; et al. The Ancestor of Extant Japanese Fancy Mice Contributed to the Mosaic Genomes of Classical Inbred Strains. Genome Res. 2013, 23, 1329–1338. [Google Scholar] [CrossRef] [PubMed]
- Moriwaki, K.; Miyashita, N.; Mita, A.; Gotoh, H.; Tsuchiya, K.; Kato, H.; Mekada, K.; Noro, C.; Oota, S.; Yoshiki, A.; et al. Unique Inbred Strain MSM/Ms Established from the Japanese Wild Mouse. Exp. Anim. 2009, 58, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Ohno, T.; Tsuchiya, T.; Horio, F. Characterization of Diabetes-Related Traits in MSM and JF1 Mice on High-Fat Diet. J. Nutr. Biochem. 2004, 15, 614–621. [Google Scholar] [CrossRef]
- Nakanishi, M.; Tazawa, H.; Tsuchiya, N.; Sugimura, T.; Tanaka, T.; Nakagama, H. Mouse Strain Differences in Inflammatory Responses of Colonic Mucosa Induced by Dextran Sulfate Sodium Cause Differential Susceptibility to PhIP-Induced Large Bowel Carcinogenesis. Cancer Sci. 2007, 98, 1157–1163. [Google Scholar] [CrossRef] [PubMed]
- Okumura, K.; Saito, M.; Isogai, E.; Tokunaga, Y.; Hasegawa, Y.; Araki, K.; Wakabayashi, Y. Functional Polymorphism in Pak1-3′ Untranslated Region Alters Skin Tumor Susceptibility by Alternative Polyadenylation. J. Investig. Dermatol. 2022, 142, 2323–2333.e12. [Google Scholar] [CrossRef]
- Okumura, K.; Saito, M.; Isogai, E.; Wakabayashi, Y. The Japanese Wild-Derived Inbred Mouse Strain, MSM/Ms in Cancer Research. Cancers 2021, 13, 1026. [Google Scholar] [CrossRef]
- Nemoto, M.; Morita, Y.; Mishima, Y.; Takahashi, S.; Nomura, T.; Ushiki, T.; Shiroishi, T.; Kikkawa, Y.; Yonekawa, H.; Kominami, R. Ahl3, A Third Locus on Mouse Chromosome 17 Affecting Age-Related Hearing Loss. Biochem. Biophys. Res. Commun. 2004, 324, 1283–1288. [Google Scholar] [CrossRef]
- Henry, K.R. Sex- and Age-Related Elevation of Cochlear Nerve Envelope Response (CNER) and Auditory Brainstem Response (ABR) Thresholds in C57BL/6 Mice. Hear. Res. 2002, 170, 107–115. [Google Scholar] [CrossRef]
- Mock, B.E.; Vijayakumar, S.; Pierce, J.; Jones, T.A.; Jones, S.M. Differential effects of Cdh23753A on auditory and vestibular functional aging in C57BL/6J mice. Neurobiol. Aging 2016, 43, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Seki, Y.; Shitara, H.; Ishii, R.; Ouchi, T.; Yasuda, S.P.; Kikkawa, Y. Myosin VI Haploinsufficiency Reduced Hearing Ability in Mice. Neuroscience 2021, 478, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Miyasaka, Y.; Suzuki, S.; Ohshiba, Y.; Watanabe, K.; Sagara, Y.; Yasuda, S.P.; Matsuoka, K.; Shitara, H.; Yonekawa, H.; Kominami, R.; et al. Compound Heterozygosity of the Functionally Null Cdh23v-ngt and Hypomorphic Cdh23ahl Alleles Leads to Early-onset Progressive Hearing Loss in Mice. Exp. Anim. 2013, 62, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Sergeyenko, Y.; Lall, K.; Liberman, M.C.; Kujawa, S.G. Age-Related Cochlear Synaptopathy: An Early-Onset Contributor to Auditory Functional Decline. J. Neurosci. 2013, 33, 13686–13694. [Google Scholar] [CrossRef] [PubMed]
- Takada, T.; Fukuta, K.; Usuda, D.; Kushida, T.; Kondo, S.; Kawamoto, S.; Yoshiki, A.; Obata, Y.; Fujiyama, A.; Toyoda, A.; et al. MoG+: A Database of Genomic Variations across Three Mouse Subspecies for Biomedical Research. Mamm. Genome 2022, 33, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the Effects of Coding Non-Synonymous Variants on Protein Function Using the SIFT Algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A Method and Server for Predicting Damaging Missense Mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Gojobori, T.; Ikeo, K.; Katayama, Y.; Kawabata, T.; Kinjo, A.R.; Kinoshita, K.; Kwon, Y.; Migita, O.; Mizutani, H.; Muraoka, M.; et al. VaProS: A Database-Integration Approach for Protein/genome Information Retrieval. J. Struct. Funct. Genom. 2016, 17, 69–81. [Google Scholar] [CrossRef]
- Kolla, L.; Kelly, M.; Mann, Z.F.; Anaya-Rocha, A.; Ellis, K.; Lemons, A.; Palermo, A.T.; So, K.S.; Mays, J.C.; Orvis, J.; et al. Characterization of the Development of the Mouse Cochlear Epithelium at the Single Cell Level. Nat. Commun. 2020, 11, 2389. [Google Scholar] [CrossRef]
- Liu, H.; Chen, L.; Giffen, K.P.; Stringham, S.T.; Li, Y.; Judge, P.D.; Beisel, K.W.; He, D.Z.Z. Cell-Specific Transcriptome Analysis Shows That Adult Pillar and Deiters’ Cells Express Genes Encoding Machinery for Specializations of Cochlear Hair Cells. Front. Mol. Neurosci. 2018, 11, 356. [Google Scholar] [CrossRef] [PubMed]
- Orvis, J.; Gottfried, B.; Kancherla, J.; Adkins, R.S.; Song, Y.; Dror, A.A.; Olley, D.; Rose, K.; Chrysostomou, E.; Kelly, M.C.; et al. gEAR: Gene Expression Analysis Resource Portal for Community-Driven, Multi-Omic Data Exploration. Nat. Methods 2021, 18, 843–844. [Google Scholar] [CrossRef]
- Wells, H.R.R.; Freidin, M.B.; Zainul Abidin, F.N.; Payton, A.; Dawes, P.; Munro, K.J.; Morton, C.C.; Moore, D.R.; Dawson, S.J.; Williams, F.M.K. GWAS Identifies 44 Independent Associated Genomic Loci for Self-Reported Adult Hearing Difficulty in UK Biobank. Am. J. Hum. Genet. 2019, 105, 788–802. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.A.; Van Laer, L.; Huentelman, M.J.; Sheth, S.S.; Van Eyken, E.; Corneveaux, J.J.; Tembe, W.D.; Halperin, R.F.; Thorburn, A.Q.; Thys, S.; et al. GRM7 Variants Confer Susceptibility to Age-Related Hearing Impairment. Hum. Mol. Genet. 2009, 18, 785–796. [Google Scholar] [CrossRef] [PubMed]
- Girotto, G.; Pirastu, N.; Sorice, R.; Biino, G.; Campbell, H.; d’Adamo, A.P.; Hastie, N.D.; Nutile, T.; Polasek, O.; Portas, L.; et al. Hearing Function and Thresholds: A Genome-Wide Association Study in European Isolated Populations Identifies New Loci and Pathways. J. Med. Genet. 2011, 48, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Van Laer, L.; Huyghe, J.R.; Hannula, S.; Van Eyken, E.; Stephan, D.A.; Mäki-Torkko, E.; Aikio, P.; Fransen, E.; Lysholm-Bernacchi, A.; Sorri, M.; et al. A Genome-Wide Association Study for Age-Related Hearing Impairment in the Saami. Eur. J. Hum. Genet. 2010, 18, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Vuckovic, D.; Dawson, S.; Scheffer, D.I.; Rantanen, T.; Morgan, A.; Di Stazio, M.; Vozzi, D.; Nutile, T.; Concas, M.P.; Biino, G.; et al. Genome-Wide Association Analysis on Normal Hearing Function Identifies PCDH20 and SLC28A3 as Candidates for Hearing Function and Loss. Hum. Mol. Genet. 2015, 24, 5655–5664. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, T.J.; Keats, B.J.; Yoshikawa, N.; Schaefer, C.; Risch, N.; Lustig, L.R. A Large Genome-Wide Association Study of Age-Related Hearing Impairment Using Electronic Health Records. PLoS Genet. 2016, 12, e1006371. [Google Scholar] [CrossRef]
- Nagtegaal, A.P.; Broer, L.; Zilhao, N.R.; Jakobsdottir, J.; Bishop, C.E.; Brumat, M.; Christiansen, M.W.; Cocca, M.; Gao, Y.; Heard-Costa, N.L.; et al. Genome-Wide Association Meta-Analysis Identifies Five Novel Loci for Age-Related Hearing Impairment. Sci. Rep. 2019, 9, 15192. [Google Scholar] [CrossRef]
- Ivarsdottir, E.V.; Holm, H.; Benonisdottir, S.; Olafsdottir, T.; Sveinbjornsson, G.; Thorleifsson, G.; Eggertsson, H.P.; Halldorsson, G.H.; Hjorleifsson, K.E.; Melsted, P.; et al. The Genetic Architecture of Age-Related Hearing Impairment Revealed by Genome-Wide Association Analysis. Commun. Biol. 2021, 4, 706. [Google Scholar] [CrossRef] [PubMed]
- Skuladottir, A.T.; Bjornsdottir, G.; Nawaz, M.S.; Petersen, H.; Rognvaldsson, S.; Moore, K.H.S.; Olafsson, P.I.; Magnusson, S.H.; Bjornsdottir, A.; Sveinsson, O.A.; et al. A Genome-Wide Meta-Analysis Uncovers Six Sequence Variants Conferring Risk of Vertigo. Commun. Biol. 2021, 4, 1148. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Johansson, Å.; Rask-Andersen, H.; Rask-Andersen, M. A Combined Genome-Wide Association and Molecular Study of Age-Related Hearing Loss in H. Sapiens. BMC Med. 2021, 19, 302. [Google Scholar] [CrossRef]
- Kalra, G.; Milon, B.; Casella, A.M.; Herb, B.R.; Humphries, E.; Song, Y.; Rose, K.P.; Hertzano, R.; Ament, S.A. Biological Insights from Multi-Omic Analysis of 31 Genomic Risk Loci for Adult Hearing Difficulty. PLoS Genet. 2020, 16, e1009025. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.Y.; Kui, L.; Xu, F.; Zheng, T.; Li, B.; McCarty, M.; Sun, Z.; Zhang, A.; Liu, L.; Starlard-Davenport, A.; et al. An Age-Related Hearing Protection Locus on Chromosome 16 of BXD Strain Mice. Neural Plast. 2020, 2020, 8889264. [Google Scholar] [CrossRef]
- Jaramillo, F.; Markin, V.S.; Hudspeth, A.J. Auditory Illusions and the Single Hair Cell. Nature 1993, 364, 527–529. [Google Scholar] [CrossRef] [PubMed]
- Liberman, M.C.; Zuo, J.; Guinan, J.J., Jr. Otoacoustic Emissions without Somatic Motility: Can Stereocilia Mechanics Drive the Mammalian Cochlea? J. Acoust. Soc. Am. 2004, 116, 1649–1655. [Google Scholar] [CrossRef] [PubMed]
- Hudspeth, A.J. Integrating the Active Process of Hair Cells with Cochlear Function. Nat. Rev. Neurosci. 2014, 15, 600–614. [Google Scholar] [CrossRef]
- Lavinsky, J.; Crow, A.L.; Pan, C.; Wang, J.; Aaron, K.A.; Ho, M.K.; Li, Q.; Salehide, P.; Myint, A.; Monges-Hernadez, M.; et al. Genome-Wide Association Study Identifies Nox3 as a Critical Gene for Susceptibility to Noise-Induced Hearing Loss. PLoS Genet. 2015, 11, e1005094. [Google Scholar] [CrossRef]
- Street, V.A.; Kujawa, S.G.; Manichaikul, A.; Broman, K.W.; Kallman, J.C.; Shilling, D.J.; Iwata, A.J.; Robinson, L.C.; Robbins, C.A.; Li, J.; et al. Resistance to Noise-Induced Hearing Loss in 129S6 and MOLF Mice: Identification of Independent, Overlapping, and Interacting Chromosomal Regions. J. Assoc. Res. Otolaryngol. 2014, 15, 721–738. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mashimo, T.; Erven, A.E.; Spiden, S.L.; Guénet, J.-L.; Steel, K.P. Two Quantitative Trait Loci Affecting Progressive Hearing Loss in 101/H Mice. Mamm. Genome 2006, 17, 841–850. [Google Scholar] [CrossRef]
- Oka, A.; Takada, T.; Fujisawa, H.; Shiroishi, T. Evolutionarily Diverged Regulation of X-Chromosomal Genes as a Primal Event in Mouse Reproductive Isolation. PLoS Genet. 2014, 10, e1004301. [Google Scholar] [CrossRef] [PubMed]
- Ohlemiller, K.K.; Kiener, A.L.; Gagnon, P.M. QTL Mapping of Endocochlear Potential Differences between C57BL/6J and BALB/cJ Mice. J. Assoc. Res. Otolaryngol. 2016, 17, 173–194. [Google Scholar] [CrossRef] [PubMed]
- Hardy, S.; Julien, S.G.; Tremblay, M.L. Impact of Oncogenic Protein Tyrosine Phosphatases in Cancer. Anticancer Agents Med. Chem. 2012, 12, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Eisenberger, T.; Di Donato, N.; Decker, C.; Delle Vedove, A.; Neuhaus, C.; Nürnberg, G.; Toliat, M.; Nürnberg, P.; Mürbe, D.; Bolz, H.J. A C-Terminal Nonsense Mutation Links PTPRQ with Autosomal-Dominant Hearing Loss, DFNA73. Genet. Med. 2018, 20, 614–621. [Google Scholar] [CrossRef]
- Schraders, M.; Oostrik, J.; Huygen, P.L.M.; Strom, T.M.; van Wijk, E.; Kunst, H.P.M.; Hoefsloot, L.H.; Cremers, C.W.R.J.; Admiraal, R.J.C.; Kremer, H. Mutations in PTPRQ Are a Cause of Autosomal-Recessive Nonsyndromic Hearing Impairment DFNB84 and Associated with Vestibular Dysfunction. Am. J. Hum. Genet. 2010, 86, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Goodyear, R.J.; Legan, P.K.; Wright, M.B.; Marcotti, W.; Oganesian, A.; Coats, S.A.; Booth, C.J.; Kros, C.J.; Seifert, R.A.; Bowen-Pope, D.F.; et al. A Receptor-like Inositol Lipid Phosphatase Is Required for the Maturation of Developing Cochlear Hair Bundles. J. Neurosci. 2003, 23, 9208–9219. [Google Scholar] [CrossRef] [PubMed]
- Ricci, A.J.; Fettiplace, R. The Effects of Calcium Buffering and Cyclic AMP on Mechano-Electrical Transduction in Turtle Auditory Hair Cells. J. Physiol. 1997, 501, 111–124. [Google Scholar] [CrossRef]
- Kimitsuki, T.; Matsuda, K.; Komune, S. Calcium Action on the Membrane Currents Possessing the Properties of Mechano-Electric Transducer Currents in Inner Hair Cells of the Guinea-Pig Cochlea. Int. J. Neurosci. 2006, 116, 1327–1335. [Google Scholar] [CrossRef]
- Santos-Cortez, R.L.P.; Lee, K.; Giese, A.P.; Ansar, M.; Amin-Ud-Din, M.; Rehn, K.; Wang, X.; Aziz, A.; Chiu, I.; Hussain Ali, R.; et al. Adenylate Cyclase 1 (ADCY1) Mutations Cause Recessive Hearing Impairment in Humans and Defects in Hair Cell Function and Hearing in Zebrafish. Hum. Mol. Genet. 2014, 23, 3289–3298. [Google Scholar] [CrossRef] [PubMed]
- Devasani, K.; Yao, Y. Expression and Functions of Adenylyl Cyclases in the CNS. Fluids Barriers CNS 2022, 19, 23. [Google Scholar] [CrossRef]
- Currall, B.B.; Chen, M.; Sallari, R.C.; Cotter, M.; Wong, K.E.; Robertson, N.G.; Penney, K.L.; Lunardi, A.; Reschke, M.; Hickox, A.E.; et al. Loss of LDAH Associated with Prostate Cancer and Hearing Loss. Hum. Mol. Genet. 2018, 27, 4194–4203. [Google Scholar] [CrossRef]
- Flachs, P.; Mihola, O.; Simeček, P.; Gregorová, S.; Schimenti, J.C.; Matsui, Y.; Baudat, F.; de Massy, B.; Piálek, J.; Forejt, J.; et al. Interallelic and Intergenic Incompatibilities of the Prdm9 (Hst1) Gene in Mouse Hybrid Sterility. PLoS Genet. 2012, 8, e1003044. [Google Scholar] [CrossRef]
- Arends, D.; Kärst, S.; Heise, S.; Korkuc, P.; Hesse, D.; Brockmann, G.A. Transmission Distortion and Genetic Incompatibilities between Alleles in a Multigenerational Mouse Advanced Intercross Line. Genetics 2022, 220, iyab192. [Google Scholar] [CrossRef]



| Congenic Region | Protein | Amino acid Substitution | Possible Impact | ||||
|---|---|---|---|---|---|---|---|
| Position | B6 | MSM | SIFT | PROVEAN | PolyPhen-2 | ||
| ahl3/2 | FBXL17 | 182 | Pro | Ser | Deleterious | Neutral | Benign |
| TMEM232 | 285 | Thr | Asn | NA * | Deleterious | Possibly damaging | |
| TXNDC2 | 133 | Glu | Gly | Tolerated | Deleterious | Benign | |
| 203 | Val | Ile | Tolerated | Neutral | Probably damaging | ||
| ANKRD12 | 910 | Asn | Ser | Deleterious | Neutral | Benign | |
| DDX11 | 657 | Val | Met | Tolerated | Deleterious | Benign | |
| MTCL1 | 848 | Arg | His | Tolerated | Deleterious | Benign | |
| 1254 | Ile | Leu | Deleterious | Neutral | Benign | ||
| PTPRM | 263 | Cys | Arg | Tolerated | Deleterious | Probably damaging | |
| LAMA1 | 656 | Asp | Asn | Tolerated | Deleterious | Benign | |
| 1122 | Gly | Ser | Tolerated | Deleterious | Benign | ||
| 1131 | Ala | Val | Deleterious | Deleterious | Possibly damaging | ||
| 1994 | Met | Thr | Tolerated | Deleterious | Possibly damaging | ||
| 2120 | Val | Ala | Tolerated | Deleterious | Probably damaging | ||
| 3033 | Ile | Val | Tolerated | Neutral | Possibly damaging | ||
| ARHGAP28 | 256 | Ala | Pro | Deleterious | Deleterious | Benign | |
| TMEM200C | 249 | Ser | Pro | Tolerated | Deleterious | Benign | |
| 344 | His | Asn | Deleterious | Neutral | Benign | ||
| 633 | Lys | Glu | Deleterious | Deleterious | Probably damaging | ||
| EPB41L3 | 803 | Ile | Val | Deleterious | Neutral | Benign | |
| ahl10 | ASXL2 | 940 | Ile | Thr | Tolerated | Deleterious | Possibly damaging |
| 1180 | Gln | Lys | Tolerated | Neutral | Probably damaging | ||
| 1191 | Ser | Phe | Tolerated | Deleterious | Probably damaging | ||
| ADCY3 | 113 | Phe | Val | Deleterious | Deleterious | Benign | |
| CENPO | 177 | Arg | Ser | Tolerated | Neutral | Probably damaging | |
| GM17541 | 46 | Leu | Ser | Tolerated | Deleterious | Benign | |
| 94 | Gly | Glu | Deleterious | Neutral | Benign | ||
| FAM228A | 113 | Gln | His | Tolerated | Deleterious | Benign | |
| 267 | Arg | Gly | Tolerated | Neutral | Probably damaging | ||
| WDCP | 474 | Pro | Ala | Tolerated | Deleterious | Benign | |
| ATAD2B | 702 | Thr | Ile | Deleterious | Neutral | Benign | |
| APOB | 260 | Thr | Lys | Deleterious | Deleterious | Probably damaging | |
| 459 | Thr | Lys | Deleterious | Neutral | Probably damaging | ||
| MATN3 | 333 | Ala | Thr | Tolerated | Deleterious | Benign | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yasuda, S.P.; Miyasaka, Y.; Hou, X.; Obara, Y.; Shitara, H.; Seki, Y.; Matsuoka, K.; Takahashi, A.; Wakai, E.; Hibino, H.; et al. Two Loci Contribute to Age-Related Hearing Loss Resistance in the Japanese Wild-Derived Inbred MSM/Ms Mice. Biomedicines 2022, 10, 2221. https://doi.org/10.3390/biomedicines10092221
Yasuda SP, Miyasaka Y, Hou X, Obara Y, Shitara H, Seki Y, Matsuoka K, Takahashi A, Wakai E, Hibino H, et al. Two Loci Contribute to Age-Related Hearing Loss Resistance in the Japanese Wild-Derived Inbred MSM/Ms Mice. Biomedicines. 2022; 10(9):2221. https://doi.org/10.3390/biomedicines10092221
Chicago/Turabian StyleYasuda, Shumpei P., Yuki Miyasaka, Xuehan Hou, Yo Obara, Hiroshi Shitara, Yuta Seki, Kunie Matsuoka, Ai Takahashi, Eri Wakai, Hiroshi Hibino, and et al. 2022. "Two Loci Contribute to Age-Related Hearing Loss Resistance in the Japanese Wild-Derived Inbred MSM/Ms Mice" Biomedicines 10, no. 9: 2221. https://doi.org/10.3390/biomedicines10092221
APA StyleYasuda, S. P., Miyasaka, Y., Hou, X., Obara, Y., Shitara, H., Seki, Y., Matsuoka, K., Takahashi, A., Wakai, E., Hibino, H., Takada, T., Shiroishi, T., Kominami, R., & Kikkawa, Y. (2022). Two Loci Contribute to Age-Related Hearing Loss Resistance in the Japanese Wild-Derived Inbred MSM/Ms Mice. Biomedicines, 10(9), 2221. https://doi.org/10.3390/biomedicines10092221





