A Quantitative Digital Analysis of Tissue Immune Components Reveals an Immunosuppressive and Anergic Immune Response with Relevant Prognostic Significance in Glioblastoma
Abstract
:1. Introduction
2. Methods
2.1. Inclusion Criteria
2.2. Histopathological Study, Digital Image Analysis, and Semiquantitative Evaluation
2.3. Molecular Subtyping Approach and MGMT Pyrosequencing
2.4. Statistical Analysis
3. Results
3.1. Clinical Data
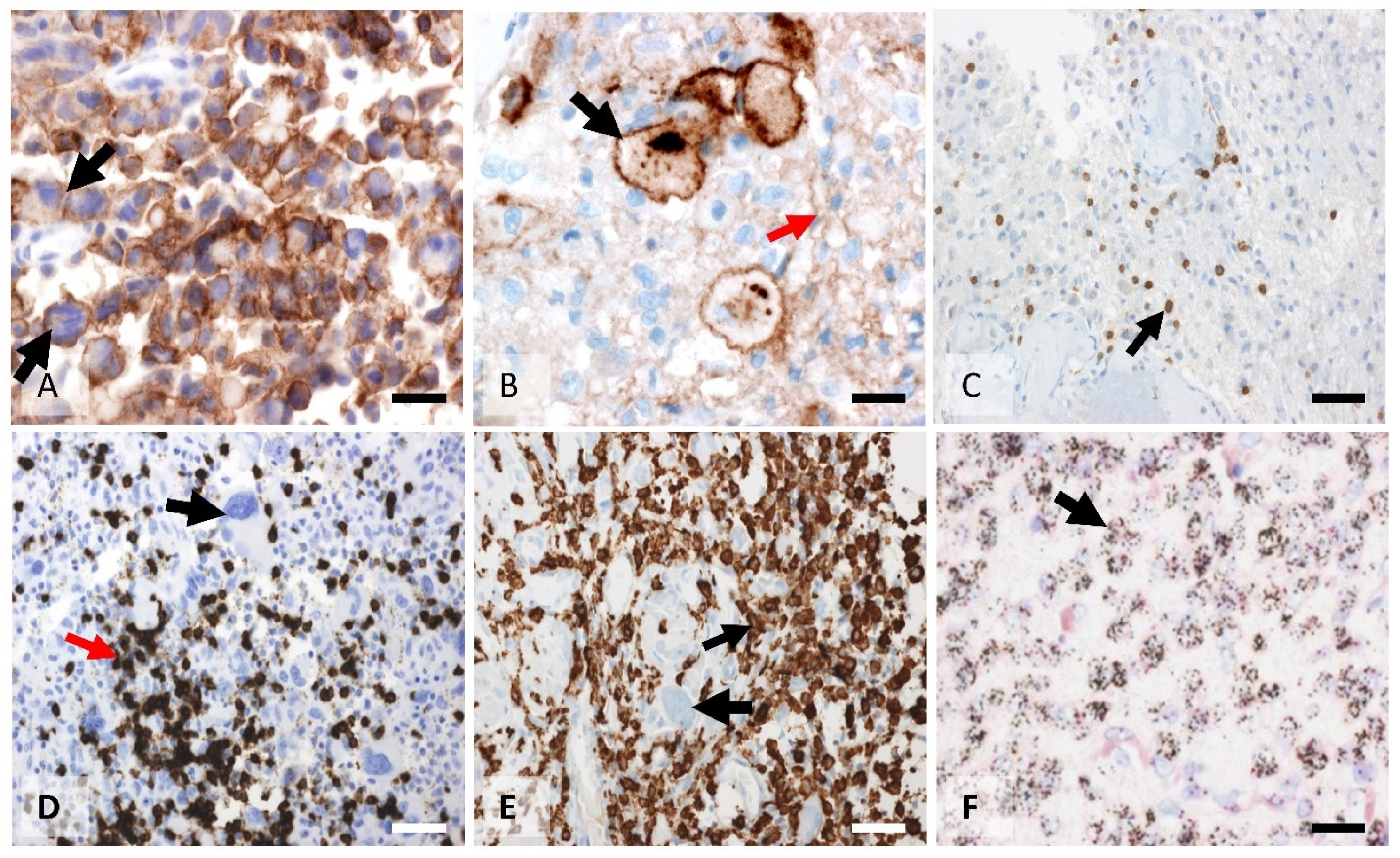
3.2. Morphological Characterization of the Macrophage Tissue Immune Barrier and Quantification of the CD163 Values
3.3. Non-Significant Statistical Differences between Biomarkers in the Vaccination and Non-Vaccination Groups Permits Studying All Cases as a Single Group
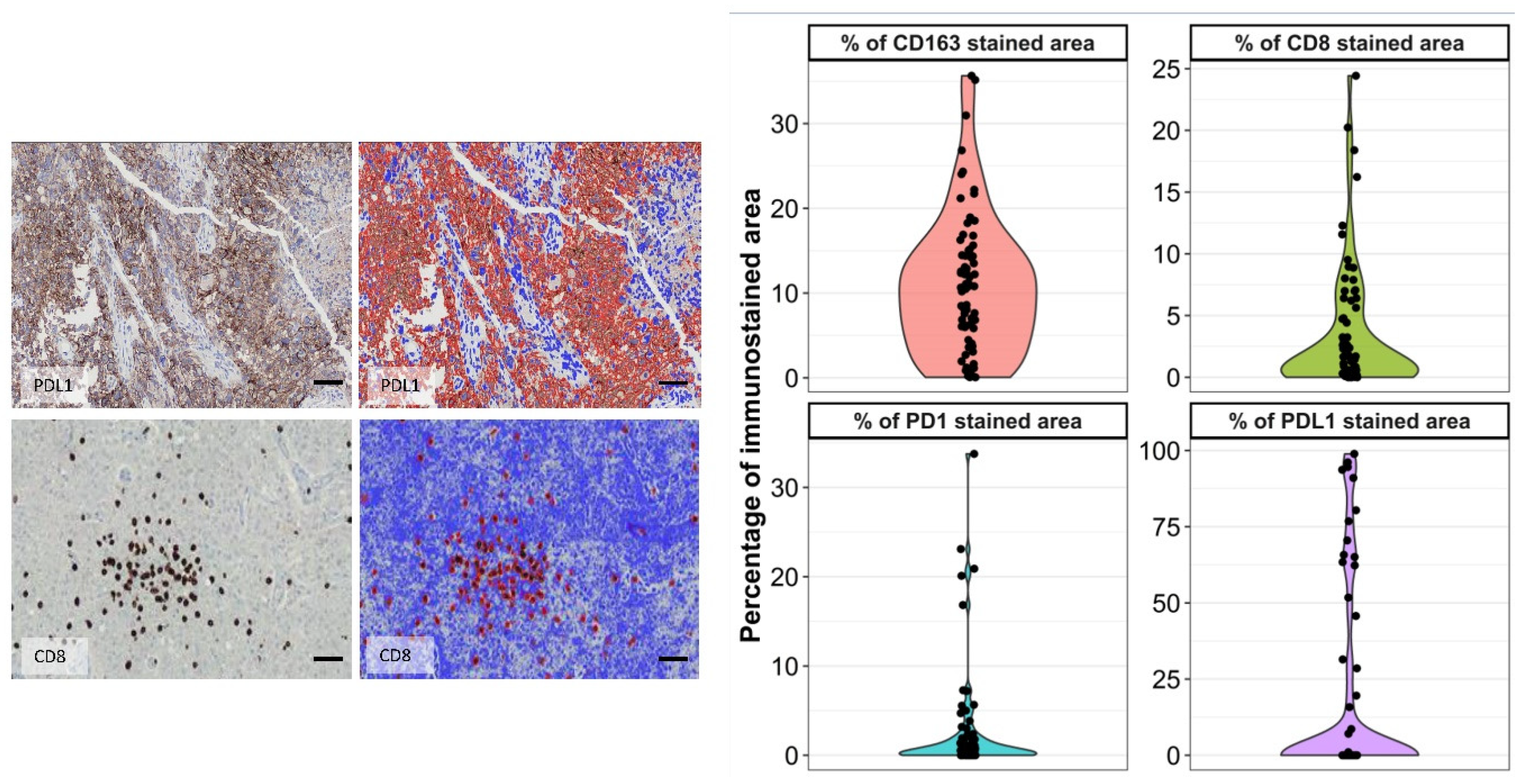
3.4. CD8, PD1, and PDL1 Characterization and Quantification
3.5. CD163 Values Showed a Direct Correlation with Either CD8 or PDL1/ PD1 Infiltration
3.6. Macrophage Infiltration Correlated with the Non-Classical Subtype of GBM
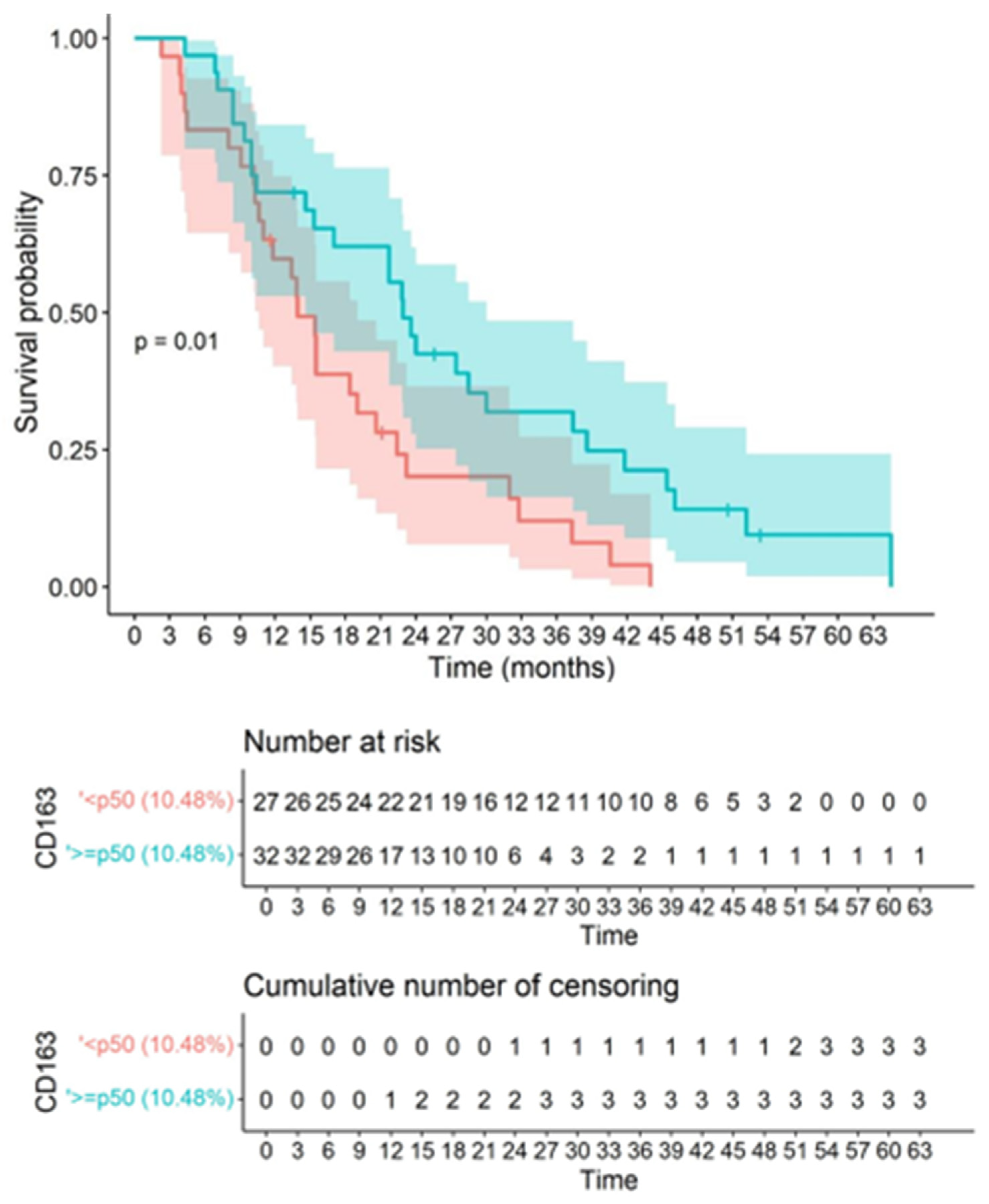
3.7. CD163 Macrophage and CD8 Infiltration Correlated with Survival
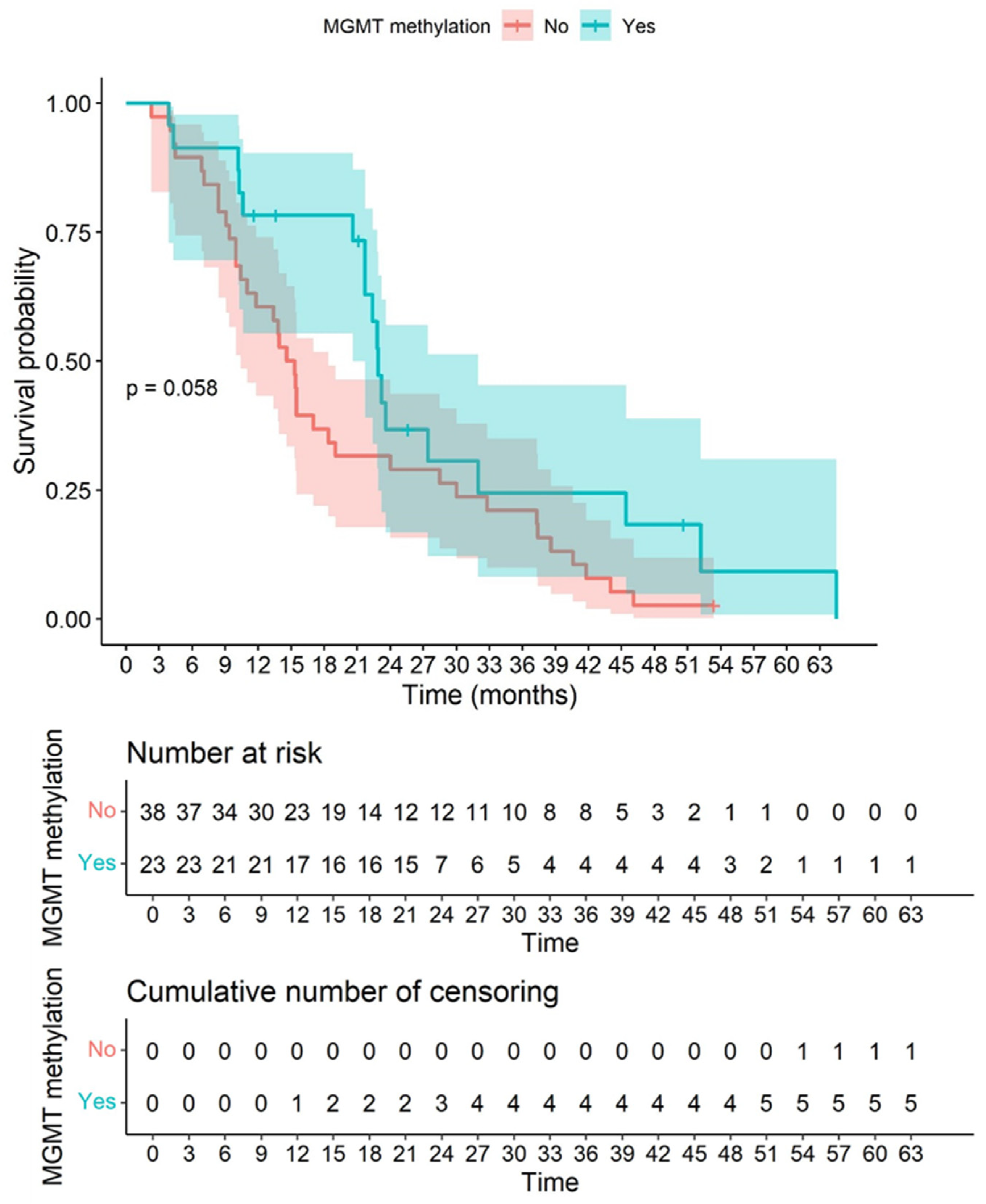
3.8. CD163 and CD8 Are Independent Biomarkers of Overall Survival and Not Related to Other Relevant Clinical Parameters Such as Age, MGMT Methylation Status, and Dendritic Cell Vaccination
4. Discussion
4.1. Automated Digital Image Analysis Is a Relevant Tool in the Objective Quantification of TILs and Reduces the Inter- and Intra-Observer Variability Seen in the Conventional Semiquantitative Approach
4.2. CD163-Positive Macrophage Infiltration in the Tumor Forms Dense Immune Clusters among the Tumor and Effector Cells
4.3. Quantified CD163-Positive Macrophage Infiltration Could Be Postulated as a Significant Independent Biomarker of Survival
4.4. The Beneficial Effect of Vaccination on Survival despite a Strongly Immunosuppressive Microenvironment
4.5. Higher CD8 Intratumor Infiltration Is Associated with a Hostile Microenvironment Created by the Tumor and Is a Significant Independent Prognostic Biomarker That Correlates with Shorter Survival
4.6. Microglia/Macrophage Infiltration Is Higher in the Non-Classical Subtype of GBM
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vanderwalde, A.; Spetzler, D.; Xiao, N.; Gatalica, Z.; Marshall, J. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med. 2018, 7, 746–756. [Google Scholar] [CrossRef] [Green Version]
- Rutledge, W.C.; Kong, J.; Gao, J.; Gutman, D.A.; Cooper, L.A.; Appin, C.; Park, Y.; Scarpace, L.; Mikkelsen, T.; Cohen, M.L.; et al. Tumor-infiltrating lymphocytes in glioblastoma are associated with specific genomic alterations and related to transcriptional class. Clin. Cancer Res. 2013, 19, 4951–4960. [Google Scholar] [CrossRef] [Green Version]
- Heimberger, A.B.; Abou-Ghazal, M.; Reina-Ortiz, C.; Yang, D.S.; Sun, W.; Qiao, W.; Hiraoka, N.; Fuller, G.N. Incidence and prognostic impact of FoxP3+ regulatory T cells in human gliomas. Clin. Cancer Res. 2008, 14, 5166–5172. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Jill, P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [Green Version]
- Crespo, I.; Vital, A.L.; Gonzalez-Tablas, M.; Patino, M.; Otero, A.; Lopes, M.C.; de Oliveira, C.; Domingues, P.; Orfao, A.; Tabernero, M.D. Molecular and Genomic Alterations in Glioblastoma Multiforme. Am. J. Pathol. 2015, 185, 1820–1833. [Google Scholar] [CrossRef] [Green Version]
- Preusser, M.; Lim, M.; Hafler, D.A.; Reardon, D.A.; Sampson, J.H. Prospects of immune checkpoint modulators in the treatment of glioblastoma. Nat. Rev. Neurol. 2015, 11, 504–514. [Google Scholar] [CrossRef] [Green Version]
- Schalper, K.A.; Rodriguez-Ruiz, M.; Diez-Valle, R.; López-Janeiro, A.; Porciuncula, A.; Idoate, M.A.; Inogés, S.; De Andrea, C.; De Cerio, A.L.-D.; Tejada, S.; et al. Neoadjuvant nivolumab modifies the tumor immune microenvironment in resectable glioblastoma. Nat. Med. 2019, 25, 470–476. [Google Scholar] [CrossRef]
- Garber, S.T.; Hashimoto, Y.; Weathers, S.P.; Xiu, J.; Gatalica, Z.; Verhaak, R.G.; Zhou, S.; Fuller, G.N.; Khasraw, M.; de Groot, J.; et al. Immune checkpoint blockade as a potential therapeutic target: Surveying CNS malignancies. Neuro Oncol. 2016, 18, 1357–1366. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, M.; Kamphorst, A.O.; Im, S.J.; Kissick, H.T.; Pillai, R.N.; Ramalingam, S.S.; Araki, K.; Ahmed, R. CD8 T Cell Exhaustion in Chronic Infection and Cancer: Opportunities for Interventions. Annu. Rev. Med. 2018, 69, 301–318. [Google Scholar] [CrossRef]
- Friebel, E.; Kapolou, K.; Unger, S.; Núñez, N.G.; Utz, S.; Rushing, E.J.; Regli, L.; Weller, M.; Greter, M.; Tugues, S.; et al. Cellular and Molecular Identity of Tumor-Associated Macrophages in Glioblastoma. Single-Cell Mapping of Human Brain Cancer Reveals Tumor-Specific Instruction of Tissue-Invading Leukocytes. Cell 2020, 181, 1626–1642.e20. [Google Scholar] [CrossRef]
- Skytthe, M.K.; Graversen, J.H.; Moestrup, S.K. Targeting of CD163+ Macrophages in Inflammatory and Malignant Diseases. Int. J. Mol. Sci. 2020, 31, 5497. [Google Scholar] [CrossRef]
- Chen, T.; Chen, J.; Zhu, Y.; Li, Y.; Wang, Y.; Chen, H.; Wang, J.; Li, X.; Liu, Y.; Li, B.; et al. CD163, a novel therapeutic target, regulates the proliferation and stemness of glioma cells via casein kinase 2. Oncogene 2019, 38, 1183–1199. [Google Scholar] [CrossRef]
- Komohara, Y.; Hirahara, J.; Horikawa, T.; Kawamura, K.; Kiyota, E.; Sakashita, N.; Araki, N.; Takeya, M. AM-3K, an anti-macrophage antibody, recognizes CD163, a molecule associated with an anti-inflammatory macrophage phenotype. J. Histochem. Cytochem. 2006, 5, 763–771. [Google Scholar] [CrossRef]
- Zeiner, P.S.; Preusse, C.; Golebiewska, A.; Zinke, J.; Iriondo, A.; Muller, A.; Kaoma, T.; Filipski, K.; Müller-Eschner, M.; Bernatz, S.; et al. Distribution and prognostic impact of microglia/macrophage subpopulations in gliomas. Brain Pathol. 2019, 29, 513–529. [Google Scholar] [CrossRef]
- Inogés, S.; Tejada, S.; de Cerio, A.L.; Gállego Pérez-Larraya, J.; Espinós, J.; Idoate, M.A.; Domínguez, P.D.; de Eulate, R.G.; Aristu, J.; Bendandi, M.; et al. A phase II trial of autologous dendritic cell vaccination and radiochemotherapy following fluorescence-guided surgery in newly diagnosed glioblastoma patients. J. Transl. Med. 2017, 15, 104. [Google Scholar] [CrossRef] [Green Version]
- Han, S.; Zhang, C.; Li, Q.; Dong, J.; Liu, Y.; Huang, Y.; Jiang, T.; Wu, A. Tumour-infiltrating CD4+ and CD8+ lymphocytes as predictors of clinical outcome in glioma. Br. J. Cancer 2014, 110, 2560–2568. [Google Scholar] [CrossRef]
- Nduom, E.K.; Wei, J.; Yaghi, N.K.; Huang, N.; Kong, L.Y.; Gabrusiewicz, K.; Ling, X.; Zhou, S.; Ivan, C.; Chen, J.Q.; et al. PD-L1 expression and prognostic impact in glioblastoma. Neuro Oncol. 2016, 18, 195–205. [Google Scholar] [CrossRef] [Green Version]
- Wulf, M.A.; Bode, B.; Zimmermann, D.; Rufibach, K.; Weder, W.; Moch, H.; Soltermann, A.; Tischler, V. Silver-enhanced in situ hybridization for determination of EGFR copy number alterations in non-small cell lung cancer. Am. J. Surg. Pathol. 2012, 36, 1801–1808. [Google Scholar] [CrossRef]
- Stålhammar, G.; Fuentes Martinez, N.; Lippert, M.; Tobin, N.P.; Mølholm, I.; Kis, L.; Rosin, G.; Rantalainen, M.; Pedersen, L.; Bergh, J.; et al. Digital image analysis outperforms manual biomarker assessment in breast cancer. Mod. Pathol. 2016, 2, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Higgins, C. Applications and challenges of digital pathology and whole slide imaging. Biotech. Histochem. 2015, 90, 341–347. [Google Scholar] [CrossRef]
- Hanna, M.G.; Pantanowitz, L.; Evans, A.J. Overview of contemporary guidelines in digital pathology: What is available in 2015 and what still needs to be addressed? J. Clin. Pathol. 2015, 68, 499–505. [Google Scholar] [CrossRef]
- Clarke, E.L.; Treanor, D. Colour in Digital Pathology: A Review. Histopathology 2017, 70, 153–163. [Google Scholar] [CrossRef]
- Orrego, E.; Castaneda, C.A.; Castillo, M.; Bernabe, L.A.; Casavilca, S.; Chakravarti, A.; Meng, W.; Garcia-Corrochano, P.; Villa-Robles, M.R.; Zevallos, R.; et al. Distribution of tumor-infiltrating immune cells in glioblastoma. CNS Oncol. 2018, 7, CNS21. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Lage, M.; Lynch, T.M.; Bi, Y.; Cocito, C.; Way, G.P.; Pal, S.; Haller, J.; Yan, R.E.; Ziober, A.; Nguyen, A.; et al. Immune landscapes associated with different glioblastoma molecular subtypes. Acta Neuropathol. Commun. 2019, 7, 203. [Google Scholar] [CrossRef] [Green Version]
- Hussain, S.F.; Yang, D.; Suki, D.; Aldape, K.; Grimm, E.; Heimberger, A.B. The role of human glioma-infiltrating microglia/macrophages in mediating antitumor immune responses. Neuro Oncol. 2006, 8, 261–279. [Google Scholar] [CrossRef] [Green Version]
- Franco, R.; Fernandez-Suarez, D. Alternatively activated microglia and macrophages in the central nervous system. Prog. Neurobiol. 2015, 131, 65–86. [Google Scholar] [CrossRef]
- Komohara, Y.; Jinushi, M.; Takeya, M. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci. 2014, 105, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, B.C.; Showers, C.R.; Anderson, D.E.; Anderson, L.; Canoll, P.; Bruce, J.N.; Anderson, R.C. Tumor-associated macrophages in glioma: Friend or foe? J. Oncol. 2013, 2013, 486912. [Google Scholar] [CrossRef]
- Sasaki, A. Microglia and brain macrophages: An update. Neuropathology 2017, 37, 452–464. [Google Scholar] [CrossRef]
- Chen, Z.; Feng, X.; Herting, C.J.; Garcia, V.A.; Nie, K.; Pong, W.W.; Rasmussen, R.; Dwivedi, B.; Seby, S.; Wolf, S.A.; et al. Cellular and Molecular Identity of Tumor-Associated Macrophages in Glioblastoma. Cancer Res. 2017, 77, 2266–2278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buonfiglioli, A.; Hambardzumyan, D. Macrophages and microglia: The cerberus of glioblastoma. Acta Neuropathol. Commun. 2021, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Ajami, B.; Bennett, J.L.; Krieger, C.; Tetzlaff, W.; Rossi, F.M. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 2007, 10, 1538–1543. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Brandenburg, S.; Turkowski, K.; Müller, S.; Vajkoczy, P. Resident microglia, and not peripheral macrophages, are the main source of brain tumor mononuclear cells. Int. J. Cancer 2015, 137, 278–288. [Google Scholar] [CrossRef]
- Felsenstein, M.; Blank, A.; Bungert, A.D.; Mueller, A.; Ghori, A.; Kremenetskaia, I.; Rung, O.; Broggini, T.; Turkowski, K.; Scherschinski, L.; et al. CCR2 of Tumor Microenvironmental Cells Is a Relevant Modulator of Glioma Biology. Cancers 2020, 12, 1882. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Y.; Qi, Y.; Huang, Y.; Hu, F.; Dong, F.; Shu, K.; Lei, T. Signal Pathways Involved in the Interaction Between Tumor-Associated Macrophages/TAMs and Glioblastoma Cells. Front Oncol. 2022, 12, 822085. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, L.; Zhang, I.Y.; Liang, J.; Wang, H.; Ouyang, M.; Wu, S.; da Fonseca, A.C.C.; Weng, L.; Yamamoto, Y.; et al. RAGE Expression in Tumor-Associated Macrophages Promotes Angiogenesis in Glioma. Cancer Res. 2014, 74, 7285–7297. [Google Scholar] [CrossRef] [Green Version]
- Hajji, N.; Garcia-Revilla, J.; Soto, M.S.; Perryman, R.; Symington, J.; Quarles, C.C.; Healey, D.R.; Guo, Y.; Orta-Vázquez, M.L.; Mateos-Cordero, S.; et al. Arginine deprivation alters microglial polarity and synergizes with radiation to eradicate non-arginine-auxotrophic glioblastoma tumors. J. Clin. Investig. 2022, 132, e142137. [Google Scholar] [CrossRef]
- Díaz, L.R.; Saavedra-López, E.; Romarate, L.; Mitxitorena, I.; Casanova, P.V.; Cribaro, G.P.; Gallego, J.M.; Pérez-Vallés, A.; Forteza-Vila, J.; Alfaro-Cervello, C.; et al. Imbalance of immunological synapse-kinapse states reflects tumor escape to immunity in glioblastoma. JCI Insight 2018, 20, 18. [Google Scholar] [CrossRef]
- Komohara, Y.; Ohnishi, K.; Kuratsu, J.; Takeya, M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J. Pathol. 2008, 216, 15–24. [Google Scholar] [CrossRef]
- Sørensen, M.D.; Dahlrot, R.H.; Boldt, H.B.; Hansen, S.; Kristensen, B.W. Tumour-associated microglia/macrophages predict poor prognosis in high-grade gliomas and correlate with an aggressive tumour subtype. Neuropathol. Appl. Neurobiol. 2018, 44, 85–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Zhang, C.; Maimela, N.R.; Yang, L.; Zhang, Z.; Ping, Y.; Huang, L.; Zhang, Y. Molecular and clinical characterization of CD163 expression via large-scale analysis in glioma. Oncoimmunology 2019, 17, 1601478. [Google Scholar] [CrossRef]
- Barberi, T.; Martin, A.; Suresh, R.; Barakat, D.J.; Harris-Bookman, S.; Drake, C.G.; Lim, M.; Friedman, A.D. Absence of host NF-κB p50 induces murine glioblastoma tumor regression, increases survival, and decreases T-cell induction of tumor-associated macrophage M2 polarization. Cancer Immunol. Immunother. 2018, 67, 1491–1503. [Google Scholar] [CrossRef] [PubMed]
- Pyonteck, S.M.; Akkari, L.; Schuhmacher, A.J.; Bowman, R.L.; Sevenich, L.; Quail, D.F.; Quail, D.F.; Olson, O.C.; Quick, M.L.; Huse, J.T.; et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 2013, 19, 1264–1272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woroniecka, K.; Fecci, P.E. T-cell exhaustion in glioblastoma. Oncotarget 2018, 9, 35287–35288. [Google Scholar] [CrossRef]
- Yang, I.; Tihan, T.; Han, S.J.; Wrensch, M.R.; Wiencke, J.; Sughrue, M.E.; Parsa, A.T. CD8+ T-cell infiltrate in newly diagnosed glioblastoma is associated with long-term survival. J. Clin. Neurosci. 2010, 17, 1381–1385. [Google Scholar] [CrossRef] [Green Version]
- Kmiecik, J.; Poli, A.; Brons, N.H.; Waha, A.; Eide, G.E.; Enger, P.Ø.; Zimmer, J.; Chekenya, M. Elevated CD3+ and CD8+ tumor-infiltrating immune cells correlate with prolonged survival in glioblastoma patients despite integrated immunosuppressive mechanisms in the tumor microenviron-ment and at the systemic level. J. Neuroimmunol. 2013, 264, 71–83. [Google Scholar] [CrossRef] [Green Version]
- Safdari, H.; Hochberg, F.H.; Richardson, E.P., Jr. Prognostic value of round cell (lymphocyte) infiltration in malignant gliomas. Surg. Neurol. 1985, 23, 221–226. [Google Scholar] [CrossRef]
- Woroniecka, K.; Chongsathidkiet, P.; Rhodin, K.; Kemeny, H.; Dechant, C.; Farber, S.H.; Elsamadicy, A.A.; Cui, X.; Koyama, S.; Jackson, C.; et al. T-Cell Exhaustion Signatures Vary with Tumor Type and Are Severe in Glioblastoma. Clin. Cancer Res. 2018, 24, 4175–4186. [Google Scholar] [CrossRef] [Green Version]
- Peranzoni, E.; Lemoine, J.; Vimeux, L.; Feuillet, V.; Barrin, S.; Kantari-Mimoun, C.; Bercovici, N.; Guérin, M.; Biton, J.; Ouakrim, H.; et al. Macrophages impede CD8 T cells from reaching tumor cells and limit the efficacy of anti-PD-1 treatment. Proc. Natl. Acad. Sci. USA 2018, 115, E4041–E4050. [Google Scholar] [CrossRef] [Green Version]
- Berghoff, A.S.; Kiesel, B.; Widhalm, G.; Rajky, O.; Ricken, G.; Wöhrer, A.; Dieckmann, K.; Filipits, M.; Brandstetter, A.; Weller, M.; et al. Programmed death ligand 1 expression and tumor-infiltrating lymphocytes in glioblastoma. Neuro Oncol. 2015, 17, 1064–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, C.; Chen, G.; Zhao, H.; Li, Y.; Chen, J.; Zhang, H.; Li, S.; Zhao, Y.; Chen, F.; Li, W.; et al. PD-L1 Expression in Glioblastoma, the Clinical and Prognostic Significance: A Systematic Literature Review and Meta-Analysis. Front Oncol. 2020, 10, 1015. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; de Carvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 10, 42–56. [Google Scholar] [CrossRef] [Green Version]
- Cooper, L.A.D.; Gutman, D.A.; Chisolm, C.; Appin, C.; Kong, J.; Rong, Y.; Kurc, T.; Van Meir, E.G.; Saltz, J.H.; Moreno, C.S.; et al. The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. Am. J. Pathol. 2012, 180, 2108–2119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doucette, T.; Rao, G.; Rao, A.; Shen, L.; Aldape, K.; Wei, J.; Dziurzynski, K.; Gilbert, M.; Heimberger, A.B. Immune heterogeneity of glioblastoma subtypes: Extrapolation from the cancer genome atlas. Cancer Immunol. Res. 2013, 1, 112–122. [Google Scholar] [CrossRef] [Green Version]
- White, K.; Connor, K.; Clerkin, J.; Murphy, B.M.; Salvucci, M.; O’Farrell, A.C.; Rehm, M.; O’Brien, D.; Prehn, J.H.M.; Niclou, S.P.; et al. New hints towards a precision medicine strategy for IDH wild-type glioblastoma. Ann. Oncol. 2020, 31, 1679–1692. [Google Scholar] [CrossRef]
- Pollack, B.P.; Sapkota, B.; Cartee, T.V. Epidermal growth factor receptor inhibition augments the expression of MHC class I and II genes. Clin. Cancer Res. 2011, 17, 4400–4413. [Google Scholar] [CrossRef] [Green Version]
- Rizzardi, A.E.; Johnson, A.T.; Vogel, R.I.; Pambuccian, S.E.; Henriksen, J.; Skubitz, A.P.; Metzger, G.J.; Schmechel, S.C. Quantitative comparison of immunohistochemical staining measured by digital image analysis versus pathologist visual scoring. Diagn. Pathol. 2012, 7, 42. [Google Scholar] [CrossRef] [Green Version]







| Parameters | Data |
|---|---|
| Males (n, %) | 42 (55) |
| Age (years) (median, p25–75) | 61 (52–68) |
| Dendritic cell vaccination (n, %) | 33 (42.1) |
| Overall survival (months) (median, p25–75) | 15.45 (9.7–27.9) |
| Karnofsky at diagnosis (median, p25–75) | 80 (80–90) |
| Percentage of tumor resection (mean range) | 98.9 (90.5–100) |
| Relapse (n, %) | 26 (34) |
| >1 resection | 25 (33) |
| Parameters. | Dendritic Cell Vaccination | p-Value | |
|---|---|---|---|
| No | Yes | ||
| %CD163 (mean) (sd) 1 | 11.59 (9.26) | 9.64 (6.30) | 0.144 |
| %CD8 (median) (IQR) 2 | 7.25 (1.95–11.06) | 4.72 (0.79–8.46) | 0.494 |
| %PDL1 (median) (IQR) 2 | 7.1 (0.62–16.73) | 1.36 (0.39–1.83) | 0.061 |
| %PD1 (median) (IQR) 2 | 2.12 (1.5–4.51) | 1.53 (0.3–19.12) | 0.460 |
| Variables | %CD163 | %CD8 | %PDL1 | %PD1 |
|---|---|---|---|---|
| %CD163 | 1 | 0.563 * | 0.544 * | 0.414 * |
| p (<0.001) | p (0.016) | p (0.005) | ||
| %CD8 | 1 | 0.511 * | 0.391 * | |
| p (0.026) | p (0.007) | |||
| %PDL1 | 1 | 0.079 | ||
| p (0.770) | ||||
| %PD1 | 1 |
| Parameter | Hazard Ratio | 95%CI | p-Value |
|---|---|---|---|
| Univariable model | |||
| %CD163 ≥ p50 (10.48%) | 2.04 | 1.15–3.61 | 0.014 * |
| %CD8 ≥ p50 (1.18%) | 1.87 | 1.08–3.24 | 0.025 * |
| %PDL1 ≥ p50 (1.79%) | 1.55 | 0.57–4.23 | 0.392 |
| %PD1 ≥ p50 (1.31%) | 1.12 | 0.57–2.22 | 0.738 |
| MGMT methylation | 0.57 | 0.32–1.03 | 0.062 |
| Dendritic cell vaccine | 0.49 | 0.28–0.85 | 0.012 * |
| EGFR (not classical) | 1.41 | 0.83–2.42 | 0.207 |
| Tumor resection | 0.88 | 0.77–1.03 | 0.111 |
| KPS | 1.00 | 0.98–1.03 | 0.900 |
| Age > p50 (60 years old) | 2.18 | 1.23–3.88 | 0.008 * |
| Multivariable model | |||
| %CD163 ≥ p50 (10.48%) | 2.50 | 1.29–4.85 | 0.007 * |
| %CD8 ≥ p50 (1.18%) | 2.05 | 1.02–4.13 | 0.045 * |
| MGMT methylation | 0.39 | 0.19–0.79 | 0.009 * |
| Dendritic cell vaccine | 0.50 | 0.26–0.94 | 0.032 * |
| Age > p50 (60 years old) | 2.74 | 1.48–5.07 | 0.001 * |
| Model validation | |||
| Concordance = 0.74 (SE = 0.035) | |||
| Likelihood ratio test | <0.001 ** | ||
| Wald test | <0.001 ** | ||
| Score (log-rank) test | <0.001 ** | ||
| Proportional hazards validation | |||
| %CD163 ≥ p50 (10.48%) | 0.85 *** | ||
| %CD8 ≥ p50 (1.18%) | 0.22 *** | ||
| MGMT methylation | 0.60 *** | ||
| Dendritic cell vaccine | 0.40 *** | ||
| Age > p50 (60 years old) | 0.37 *** | ||
| Global | 0.67 *** | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Idoate Gastearena, M.A.; López-Janeiro, Á.; Lecumberri Aznarez, A.; Arana-Iñiguez, I.; Guillén-Grima, F. A Quantitative Digital Analysis of Tissue Immune Components Reveals an Immunosuppressive and Anergic Immune Response with Relevant Prognostic Significance in Glioblastoma. Biomedicines 2022, 10, 1753. https://doi.org/10.3390/biomedicines10071753
Idoate Gastearena MA, López-Janeiro Á, Lecumberri Aznarez A, Arana-Iñiguez I, Guillén-Grima F. A Quantitative Digital Analysis of Tissue Immune Components Reveals an Immunosuppressive and Anergic Immune Response with Relevant Prognostic Significance in Glioblastoma. Biomedicines. 2022; 10(7):1753. https://doi.org/10.3390/biomedicines10071753
Chicago/Turabian StyleIdoate Gastearena, Miguel A., Álvaro López-Janeiro, Arturo Lecumberri Aznarez, Iñigo Arana-Iñiguez, and Francisco Guillén-Grima. 2022. "A Quantitative Digital Analysis of Tissue Immune Components Reveals an Immunosuppressive and Anergic Immune Response with Relevant Prognostic Significance in Glioblastoma" Biomedicines 10, no. 7: 1753. https://doi.org/10.3390/biomedicines10071753
APA StyleIdoate Gastearena, M. A., López-Janeiro, Á., Lecumberri Aznarez, A., Arana-Iñiguez, I., & Guillén-Grima, F. (2022). A Quantitative Digital Analysis of Tissue Immune Components Reveals an Immunosuppressive and Anergic Immune Response with Relevant Prognostic Significance in Glioblastoma. Biomedicines, 10(7), 1753. https://doi.org/10.3390/biomedicines10071753








