Patient-Derived Lung Tumoroids—An Emerging Technology in Drug Development and Precision Medicine
Abstract
:1. Introduction
2. Current Experimental Models for Drug Development in NSCLC and Their Limits
2.1. Lung Cancer Cell Lines for In Vitro Studies
2.2. Murine Models for In Vivo Studies
- (a)
- Patient-derived tumor xenografts (PDXs)
- (b) Syngeneic murine models
- (c) Genetically engineered mouse models (GEMMs)
3. Limits of Current Preclinical Models in Lung Cancer Research
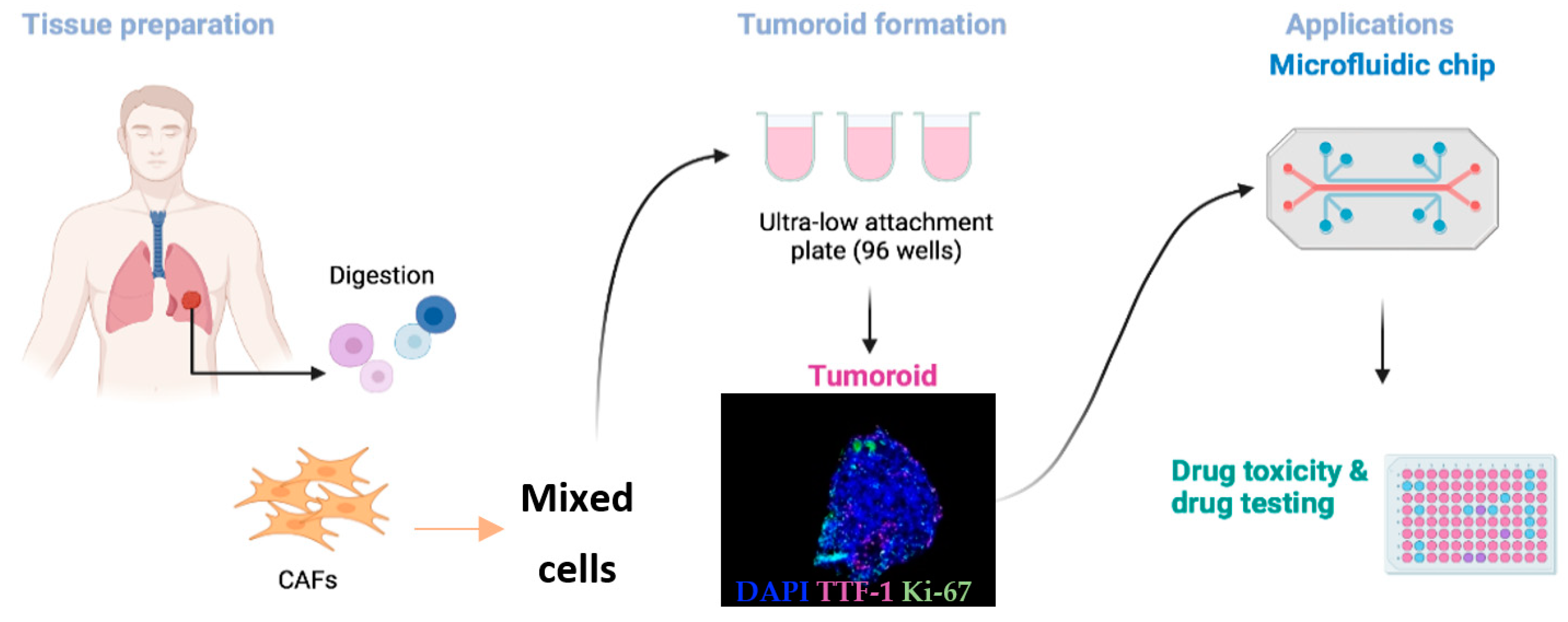
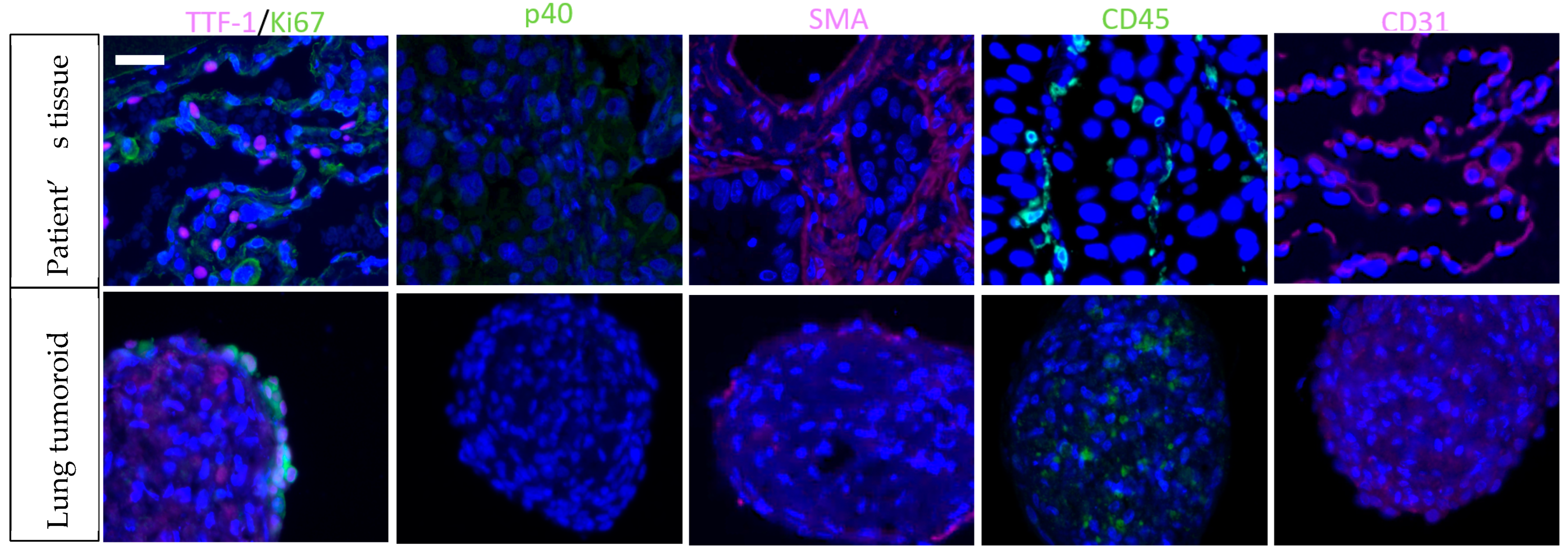
4. Tumoroids: A Next-Generation Preclinical Model
5. Towards the Optimization of Lung Tumoroids
6. Application to Drug Discovery, Screening, and Study of Mode of Action
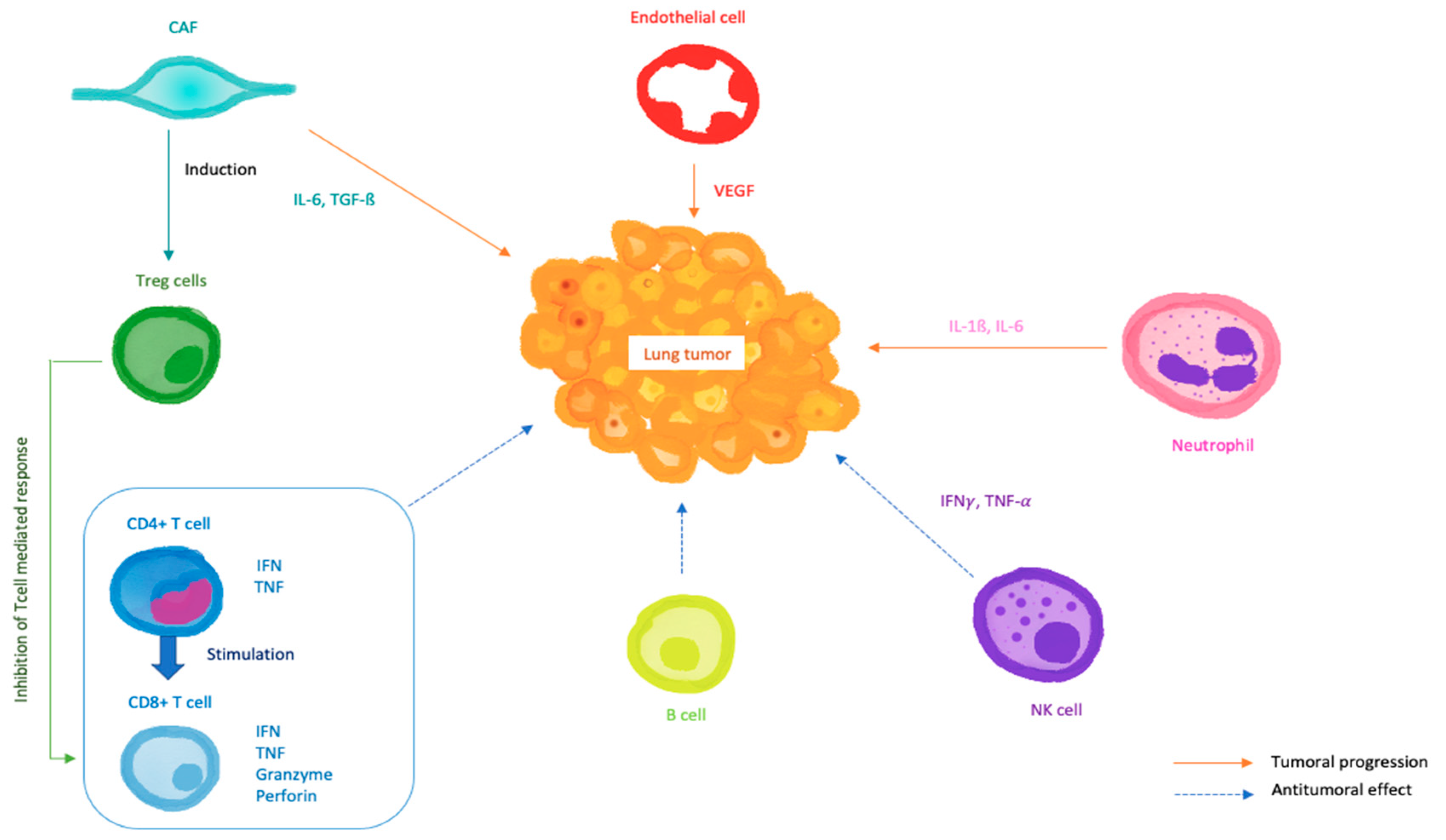
7. Modelling the Tumor Microenvironment (TME)
8. Looking for Biomarkers in Lung Cancer Therapeutic Management and Lung Cancer Progression
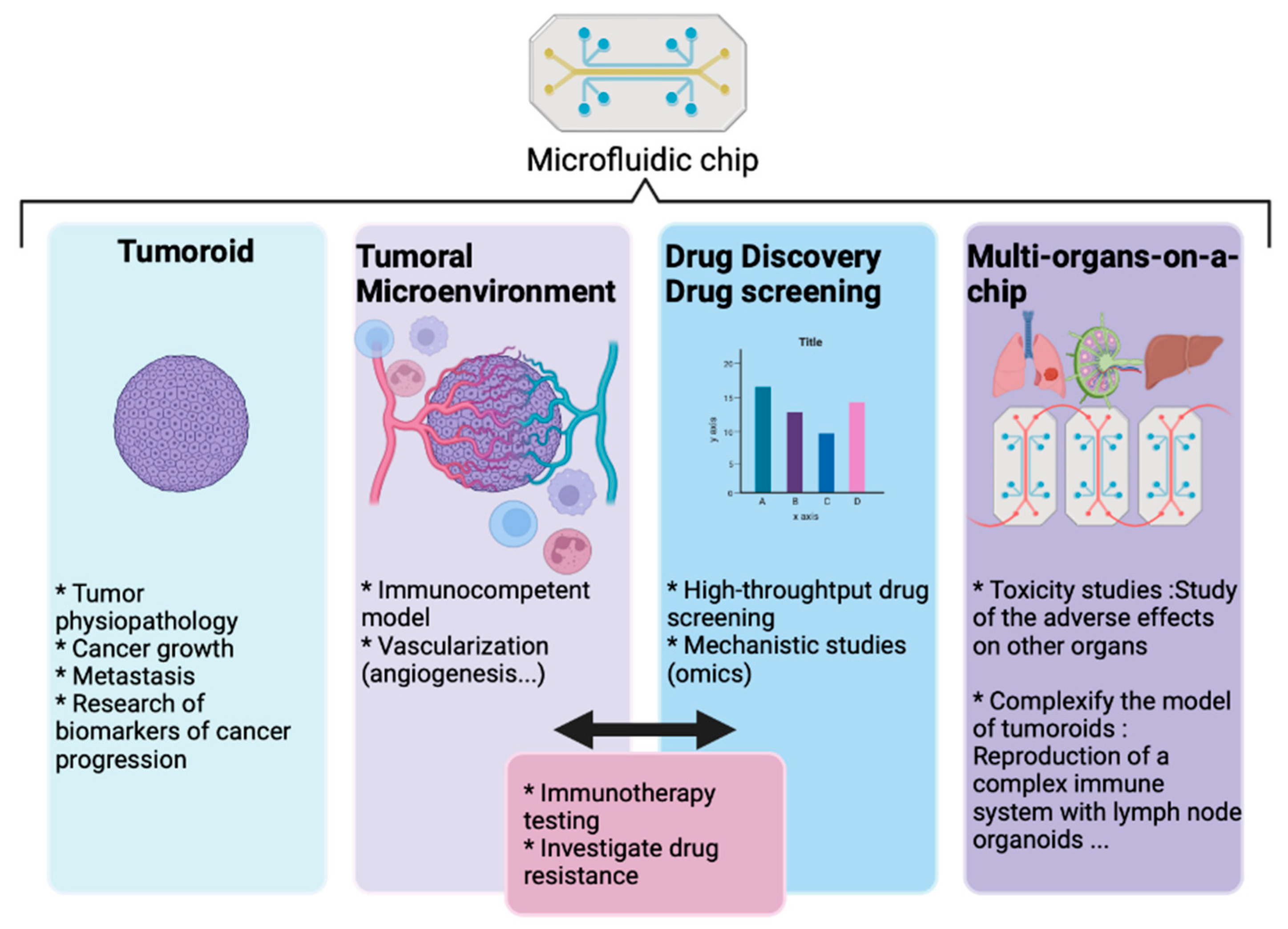
9. Convergence of Technologies into Microfluidic Systems
10. Discussion and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non–Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin. Proc. 2019, 94, 1623–1640. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The Biology and Management of Non-Small Cell Lung Cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, H.; Sen, T.; Rudin, C.M. Targeted Therapies and Biomarkers in Small Cell Lung Cancer. Front. Oncol. 2020, 10, 741. [Google Scholar] [CrossRef]
- National Cancer Institute. Percent of Cases & 5-Year Relative Survival by Stage at Diagnosis: Lung and Bronchus Cancer; US National Cancer Institute: Rockville, MD, USA, 2011.
- Benzaquen, J.; Boutros, J.; Marquette, C.; Delingette, H.; Hofman, P. Lung Cancer Screening, towards a Multidimensional Approach: Why and How? Cancers 2019, 11, 212. [Google Scholar] [CrossRef] [Green Version]
- de Koning, H.J.; van der Aalst, C.M.; de Jong, P.A.; Scholten, E.T.; Nackaerts, K.; Heuvelmans, M.A.; Lammers, J.-W.J.; Weenink, C.; Yousaf-Khan, U.; Horeweg, N.; et al. Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N. Engl. J. Med. 2020, 382, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2018: Cancer Statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Ray, M.R.; Jablons, D.; He, B. Lung Cancer Therapeutics That Target Signaling Pathways: An Update. Expert Rev. Respir. Med. 2010, 4, 631–645. [Google Scholar] [CrossRef] [Green Version]
- Chan, B.A.; Hughes, B.G.M. Targeted Therapy for Non-Small Cell Lung Cancer: Current Standards and the Promise of the Future. Transl. Lung Cancer Res. 2015, 4, 19. [Google Scholar]
- Wu, Y.-L.; Herbst, R.S.; Mann, H.; Rukazenkov, Y.; Marotti, M.; Tsuboi, M. ADAURA: Phase III, Double-Blind, Randomized Study of Osimertinib Versus Placebo in EGFR Mutation-Positive Early-Stage NSCLC after Complete Surgical Resection. Clin. Lung Cancer 2018, 19, e533–e536. [Google Scholar] [CrossRef]
- Broderick, S.R. Adjuvant and Neoadjuvant Immunotherapy in Non-Small Cell Lung Cancer. Thorac. Surg. Clin. 2020, 30, 215–220. [Google Scholar] [CrossRef]
- Zappa, C.; Mousa, S.A. Non-Small Cell Lung Cancer: Current Treatment and Future Advances. Transl. Lung Cancer Res. 2016, 5, 288–300. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.-F.; Chen, Y.; Song, S.-Y.; Wang, T.-J.; Ji, W.-J.; Li, S.-W.; Liu, N.; Yan, C.-X. Immune-Related Adverse Events Associated with Anti-PD-1/PD-L1 Treatment for Malignancies: A Meta-Analysis. Front. Pharmacol. 2017, 8, 730. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Wang, L.; Jiang, M.; Zhao, D.; Wang, Y.; Zhang, F.; Li, J.; Zhang, X. A Review About Pembrolizumab in First-Line Treatment of Advanced NSCLC: Focus on KEYNOTE Studies. Cancer Manag. Res. 2020, 12, 6493–6509. [Google Scholar] [CrossRef] [PubMed]
- Suresh, K.; Naidoo, J.; Lin, C.T.; Danoff, S. Immune Checkpoint Immunotherapy for Non-Small Cell Lung Cancer. Chest 2018, 154, 1416–1423. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Wahab, N.; Shah, M.; Suarez-Almazor, M.E. Adverse Events Associated with Immune Checkpoint Blockade in Patients with Cancer: A Systematic Review of Case Reports. PLoS ONE 2016, 11, e0160221. [Google Scholar] [CrossRef]
- Sajjad, H.; Imtiaz, S.; Noor, T.; Siddiqui, Y.H.; Sajjad, A.; Zia, M. Cancer Models in Preclinical Research: A Chronicle Review of Advancement in Effective Cancer Research. Anim. Models Exp. Med. 2021, 4, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Gazdar, A.F.; Girard, L.; Lockwood, W.W.; Lam, W.L.; Minna, J.D. Lung Cancer Cell Lines as Tools for Biomedical Discovery and Research. J. Natl. Cancer Inst. 2010, 102, 1310–1321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-David, U.; Siranosian, B.; Ha, G.; Tang, H.; Oren, Y.; Hinohara, K.; Strathdee, C.A.; Dempster, J.; Lyons, N.J.; Burns, R.; et al. Genetic and Transcriptional Evolution Alters Cancer Cell Line Drug Response. Nature 2018, 560, 325–330. [Google Scholar] [CrossRef]
- Liu, Y.; Mi, Y.; Mueller, T.; Kreibich, S.; Williams, E.G.; Van Drogen, A.; Borel, C.; Frank, M.; Germain, P.-L.; Bludau, I.; et al. Multi-Omic Measurements of Heterogeneity in HeLa Cells across Laboratories. Nat. Biotechnol. 2019, 37, 314–322. [Google Scholar] [CrossRef]
- Gazdar, A.F.; Gao, B.; Minna, J.D. Lung Cancer Cell Lines: Useless Artifacts or Invaluable Tools for Medical Science? Lung Cancer 2010, 68, 309–318. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Korn, J.M.; Ferretti, S.; Monahan, J.E.; Wang, Y.; Singh, M.; Zhang, C.; Schnell, C.; Yang, G.; Zhang, Y.; et al. High-Throughput Screening Using Patient-Derived Tumor Xenografts to Predict Clinical Trial Drug Response. Nat. Med. 2015, 21, 1318–1325. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Pham, N.-A.; Tong, J.; Sakashita, S.; Allo, G.; Kim, L.; Yanagawa, N.; Raghavan, V.; Wei, Y.; To, C.; et al. Molecular Heterogeneity of Non-Small Cell Lung Carcinoma Patient-Derived Xenografts Closely Reflect Their Primary Tumors: Molecular Landscape of NSCLC Patient-Derived Xenografts. Int. J. Cancer 2017, 140, 662–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Mun, H.; Sung, C.O.; Cho, E.J.; Jeon, H.-J.; Chun, S.-M.; Jung, D.J.; Shin, T.H.; Jeong, G.S.; Kim, D.K.; et al. Patient-Derived Lung Cancer Organoids as In Vitro Cancer Models for Therapeutic Screening. Nat. Commun. 2019, 10, 3991. [Google Scholar] [CrossRef]
- Hynds, R.E.; Frese, K.K.; Pearce, D.R.; Grönroos, E.; Dive, C.; Swanton, C. Progress towards Non-Small-Cell Lung Cancer Models That Represent Clinical Evolutionary Trajectories. Open Biol. 2021, 11, 200247. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Pitt, J.M.; Daillère, R.; Smyth, M.J.; Kroemer, G. Mouse Models in Oncoimmunology. Nat. Rev. Cancer 2016, 16, 759–773. [Google Scholar] [CrossRef]
- Nolan, K.; Verzosa, G.; Cleaver, T.; Tippimanchai, D.; DePledge, L.N.; Wang, X.-J.; Young, C.; Le, A.; Doebele, R.; Li, H.; et al. Development of Syngeneic Murine Cell Lines for Use in Immunocompetent Orthotopic Lung Cancer Models. Cancer Cell Int. 2020, 20, 417. [Google Scholar] [CrossRef]
- Foggetti, G.; Li, C.; Cai, H.; Hellyer, J.A.; Lin, W.-Y.; Ayeni, D.; Hastings, K.; Choi, J.; Wurtz, A.; Andrejka, L.; et al. Genetic Determinants of EGFR-Driven Lung Cancer Growth and Therapeutic Response In Vivo. Cancer Discov. 2021, 11, 1736–1753. [Google Scholar] [CrossRef]
- Starrett, J.H.; Guernet, A.A.; Cuomo, M.E.; Poels, K.E.; van Rosenburgh, I.K.A.; Nagelberg, A.; Farnsworth, D.; Price, K.S.; Khan, H.; Ashtekar, K.D.; et al. Drug Sensitivity and Allele Specificity of First-Line Osimertinib Resistance EGFR Mutations. Cancer Res. 2020, 80, 2017–2030. [Google Scholar] [CrossRef] [Green Version]
- Wilding, J.L.; Bodmer, W.F. Cancer Cell Lines for Drug Discovery and Development. Cancer Res. 2014, 74, 2377–2384. [Google Scholar] [CrossRef] [Green Version]
- Kunnumakkara, A.B.; Bordoloi, D.; Sailo, B.L.; Roy, N.K.; Thakur, K.K.; Banik, K.; Shakibaei, M.; Gupta, S.C.; Aggarwal, B.B. Cancer Drug Development: The Missing Links. Exp. Biol. Med. 2019, 244, 663–689. [Google Scholar] [CrossRef]
- Fitzgerald, A.A.; Li, E.; Weiner, L.M. 3D Culture Systems for Exploring Cancer Immunology. Cancers 2020, 13, 56. [Google Scholar] [CrossRef] [PubMed]
- Kodack, D.P.; Farago, A.F.; Dastur, A.; Held, M.A.; Dardaei, L.; Friboulet, L.; von Flotow, F.; Damon, L.J.; Lee, D.; Parks, M.; et al. Primary Patient-Derived Cancer Cells and Their Potential for Personalized Cancer Patient Care. Cell Rep. 2017, 21, 3298–3309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wensink, G.E.; Elias, S.G.; Mullenders, J.; Koopman, M.; Boj, S.F.; Kranenburg, O.W.; Roodhart, J.M.L. Patient-Derived Organoids as a Predictive Biomarker for Treatment Response in Cancer Patients. NPJ Precis. Oncol. 2021, 5, 30. [Google Scholar] [CrossRef]
- Li, Z.; Qian, Y.; Li, W.; Liu, L.; Yu, L.; Liu, X.; Wu, G.; Wang, Y.; Luo, W.; Fang, F.; et al. Human Lung Adenocarcinoma-Derived Organoid Models for Drug Screening. IScience 2020, 23, 101411. [Google Scholar] [CrossRef]
- Dijkstra, K.K.; Monkhorst, K.; Schipper, L.J.; Hartemink, K.J.; Smit, E.F.; Kaing, S.; de Groot, R.; Wolkers, M.C.; Clevers, H.; Cuppen, E.; et al. Challenges in Establishing Pure Lung Cancer Organoids Limit Their Utility for Personalized Medicine. Cell Rep. 2020, 31, 107588. [Google Scholar] [CrossRef]
- Shi, R.; Radulovich, N.; Ng, C.; Liu, N.; Notsuda, H.; Cabanero, M.; Martins-Filho, S.N.; Raghavan, V.; Li, Q.; Mer, A.S.; et al. Organoid Cultures as Preclinical Models of Non–Small Cell Lung Cancer. Clin. Cancer Res. 2020, 26, 1162–1174. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Wang, H.; Ding, Q.; Xing, Y.; Xu, Z.; Lu, C.; Luo, D.; Xu, L.; Xia, W.; Zhou, C.; et al. Establishment of Patient-Derived Tumor Spheroids for Non-Small Cell Lung Cancer. PLoS ONE 2018, 13, e0194016. [Google Scholar] [CrossRef]
- Sachs, N.; Papaspyropoulos, A.; Zomer-van Ommen, D.D.; Heo, I.; Böttinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakobachvili, N.; Amatngalim, G.D.; et al. Long-term Expanding Human Airway Organoids for Disease Modeling. EMBO J. 2019, 38, e100300. [Google Scholar] [CrossRef]
- Chen, J.; Chu, X.; Zhang, J.; Nie, Q.; Tang, W.; Su, J.; Yan, H.; Zheng, H.; Chen, Z.; Chen, X.; et al. Genomic Characteristics and Drug Screening among Organoids Derived from Non-Small cell Lung Cancer Patients. Thorac. Cancer 2020, 11, 2279–2290. [Google Scholar] [CrossRef]
- Delom, F.; Begiristain, I.; Grenier, T.; Begueret, H.; Soulet, F.; Siegfried, G.; Khatib, A.-M.; Robert, J.; Fessart, D. Patients Lung Derived Tumoroids (PLDTs) to Model Therapeutic Response. Biochim. Biophys. Acta BBA-Mol. Cell Res. 2020, 1867, 118808. [Google Scholar] [CrossRef]
- Li, Y.F.; Gao, Y.; Liang, B.W.; Cao, X.Q.; Sun, Z.J.; Yu, J.H.; Liu, Z.D.; Han, Y. Patient-Derived Organoids of Non-Small Cells Lung Cancer and Their Application for Drug Screening. Neoplasma 2020, 67, 430–437. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Sui, X.; Song, F.; Li, Y.; Li, K.; Chen, Z.; Yang, F.; Chen, X.; Zhang, Y.; Wang, X.; et al. Lung Cancer Organoids Analyzed on Microwell Arrays Predict Drug Responses of Patients within a Week. Nat. Commun. 2021, 12, 2581. [Google Scholar] [CrossRef] [PubMed]
- Yokota, E.; Iwai, M.; Yukawa, T.; Yoshida, M.; Naomoto, Y.; Haisa, M.; Monobe, Y.; Takigawa, N.; Guo, M.; Maeda, Y.; et al. Clinical Application of a Lung Cancer Organoid (Tumoroid) Culture System. NPJ Precis. Oncol. 2021, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Seitlinger, J.; Nounsi, A.; Idoux-Gillet, Y.; Santos Pujol, E.; Lê, H.; Grandgirard, E.; Olland, A.; Lindner, V.; Zaupa, C.; Balloul, J.-M.; et al. Vascularization of Patient-Derived Tumoroid from Non-Small-Cell Lung Cancer and Its Microenvironment. Biomedicines 2022, 10, 1103. [Google Scholar] [CrossRef]
- Porter, R.J.; Murray, G.I.; McLean, M.H. Current Concepts in Tumour-Derived Organoids. Br. J. Cancer 2020, 123, 1209–1218. [Google Scholar] [CrossRef]
- Gunti, S.; Hoke, A.T.K.; Vu, K.P.; London, N.R. Organoid and Spheroid Tumor Models: Techniques and Applications. Cancers 2021, 13, 874. [Google Scholar] [CrossRef]
- Rodrigues, J.; Heinrich, M.A.; Teixeira, L.M.; Prakash, J. 3D In Vitro Model (R)Evolution: Unveiling Tumor–Stroma Interactions. Trends Cancer 2021, 7, 249–264. [Google Scholar] [CrossRef]
- Yatabe, Y.; Dacic, S.; Borczuk, A.C.; Warth, A.; Russell, P.A.; Lantuejoul, S.; Beasley, M.B.; Thunnissen, E.; Pelosi, G.; Rekhtman, N.; et al. Best Practices Recommendations for Diagnostic Immunohistochemistry in Lung Cancer. J. Thorac. Oncol. 2019, 14, 377–407. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Wang, X.; Chen, Y.; Fang, Z. In-Vitro Photothermal Therapy Using Plant Extract Polyphenols Functionalized Graphene Sheets for Treatment of Lung Cancer. J. Photochem. Photobiol. B 2020, 204, 111587. [Google Scholar] [CrossRef]
- Rosenkranz, A.; Perini, G.; Aguilar-Hurtado, J.Y.; Zambrano, D.F.; Wang, B.; Niccolini, B.; Henriques, P.C.; Rosa, E.; De Maio, F.; Delogu, G.; et al. Laser-Mediated Antibacterial Effects of Few- and Multi-Layer Ti3C2Tx MXenes. Appl. Surf. Sci. 2021, 567, 150795. [Google Scholar] [CrossRef]
- Tatullo, M.; Marrelli, B.; Benincasa, C.; Aiello, E.; Makeeva, I.; Zavan, B.; Ballini, A.; De Vito, D.; Spagnuolo, G. Organoids in Translational Oncology. J. Clin. Med. 2020, 9, 2774. [Google Scholar] [CrossRef] [PubMed]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, Transcriptome and Proteome: The Rise of Omics Data and Their Integration in Biomedical Sciences. Brief. Bioinform. 2018, 19, 286–302. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Wang, Y.; Duan, W.; Xia, S.; Wei, S.; Liu, W.; Wang, Q. Proteomic Reveals Reasons for Acquired Drug Resistance in Lung Cancer Derived Brain Metastasis Based on a Newly Established Multi-Organ Microfluidic Chip Model. Front. Bioeng. Biotechnol. 2020, 8, 612091. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yu, L.; Li, Z.; Li, W.; Huang, W. Patient-Derived Organoid (PDO) Platforms to Facilitate Clinical Decision Making. J. Transl. Med. 2021, 19, 40. [Google Scholar] [CrossRef]
- Peng, D.; Gleyzer, R.; Tai, W.-H.; Kumar, P.; Bian, Q.; Isaacs, B.; da Rocha, E.L.; Cai, S.; DiNapoli, K.; Huang, F.W.; et al. Evaluating the Transcriptional Fidelity of Cancer Models. Genome Med. 2021, 13, 73. [Google Scholar] [CrossRef]
- Ma, X.; Yang, S.; Jiang, H.; Wang, Y.; Xiang, Z. Transcriptomic Analysis of Tumor Tissues and Organoids Reveals the Crucial Genes Regulating the Proliferation of Lung Adenocarcinoma. J. Transl. Med. 2021, 19, 368. [Google Scholar] [CrossRef]
- Wang, X.; Xiao, H.; Wu, D.; Zhang, D.; Zhang, Z. MiR-335-5p Regulates Cell Cycle and Metastasis in Lung Adenocarcinoma by Targeting CCNB2. Onco. Targets Ther. 2020, 13, 6255–6263. [Google Scholar] [CrossRef]
- Weeber, F.; Ooft, S.N.; Dijkstra, K.K.; Voest, E.E. Tumor Organoids as a Pre-Clinical Cancer Model for Drug Discovery. Cell Chem. Biol. 2017, 24, 1092–1100. [Google Scholar] [CrossRef]
- Finnberg, N.K.; Gokare, P.; Lev, A.; Grivennikov, S.I.; MacFarlane, A.W.; Campbell, K.S.; Winters, R.M.; Kaputa, K.; Farma, J.M.; Abbas, A.E.-S.; et al. Application of 3D Tumoroid Systems to Define Immune and Cytotoxic Therapeutic Responses Based on Tumoroid and Tissue Slice Culture Molecular Signatures. Oncotarget 2017, 8, 66747–66757. [Google Scholar] [CrossRef] [Green Version]
- Altorki, N.K.; Markowitz, G.J.; Gao, D.; Port, J.L.; Saxena, A.; Stiles, B.; McGraw, T.; Mittal, V. The Lung Microenvironment: An Important Regulator of Tumour Growth and Metastasis. Nat. Rev. Cancer 2019, 19, 9–31. [Google Scholar] [CrossRef] [Green Version]
- Jenkins, R.W.; Barbie, D.A.; Flaherty, K.T. Mechanisms of Resistance to Immune Checkpoint Inhibitors. Br. J. Cancer 2018, 118, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Mazieres, J.; Drilon, A.; Lusque, A.; Mhanna, L.; Cortot, A.B.; Mezquita, L.; Thai, A.A.; Mascaux, C.; Couraud, S.; Veillon, R.; et al. Immune Checkpoint Inhibitors for Patients with Advanced Lung Cancer and Oncogenic Driver Alterations: Results from the IMMUNOTARGET Registry. Ann. Oncol. 2019, 30, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
- Boucherit, N.; Gorvel, L.; Olive, D. 3D Tumor Models and Their Use for the Testing of Immunotherapies. Front. Immunol. 2020, 11, 603640. [Google Scholar] [CrossRef] [PubMed]
- Frigola, J.; Navarro, A.; Carbonell, C.; Callejo, A.; Iranzo, P.; Cedrés, S.; Martinez-Marti, A.; Pardo, N.; Saoudi-Gonzalez, N.; Martinez, D.; et al. Molecular Profiling of Long-Term Responders to Immune Checkpoint Inhibitors in Advanced Non-small Cell Lung Cancer. Mol. Oncol. 2021, 15, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Pai-Scherf, L.; Blumenthal, G.M.; Li, H.; Subramaniam, S.; Mishra-Kalyani, P.S.; He, K.; Zhao, H.; Yu, J.; Paciga, M.; Goldberg, K.B.; et al. FDA Approval Summary: Pembrolizumab for Treatment of Metastatic Non-Small Cell Lung Cancer: First-Line Therapy and Beyond. Oncologist 2017, 22, 1392–1399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gandhi, L.; Rodríguez-Abreu, D.; Gadgeel, S.; Esteban, E.; Felip, E.; De Angelis, F.; Domine, M.; Clingan, P.; Hochmair, M.J.; Powell, S.F.; et al. Pembrolizumab plus Chemotherapy in Metastatic Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 2078–2092. [Google Scholar] [CrossRef]
- Kazandjian, D.; Suzman, D.L.; Blumenthal, G.; Mushti, S.; He, K.; Libeg, M.; Keegan, P.; Pazdur, R. FDA Approval Summary: Nivolumab for the Treatment of Metastatic Non-Small Cell Lung Cancer with Progression on or after Platinum-Based Chemotherapy. Oncologist 2016, 21, 634–642. [Google Scholar] [CrossRef] [Green Version]
- Vellanki, P.J.; Mulkey, F.; Jaigirdar, A.A.; Rodriguez, L.; Wang, Y.; Xu, Y.; Zhao, H.; Liu, J.; Howe, G.; Wang, J.; et al. FDA Approval Summary: Nivolumab with Ipilimumab and Chemotherapy for Metastatic Non-Small Cell Lung Cancer, A Collaborative Project Orbis Review. Clin. Cancer Res. 2021, 27, 3522–3527. [Google Scholar] [CrossRef]
- US Food and Drug Administration FDA. Approves Durvalumab after Chemoradiation for Unresectable Stage III NSCLC; FDA: Silver Spring, MD, USA, 2019.
- Herbst, R.S.; Giaccone, G.; de Marinis, F.; Reinmuth, N.; Vergnenegre, A.; Barrios, C.H.; Morise, M.; Felip, E.; Andric, Z.; Geater, S.; et al. Atezolizumab for First-Line Treatment of PD-L1–Selected Patients with NSCLC. N. Engl. J. Med. 2020, 383, 1328–1339. [Google Scholar] [CrossRef]
- Sezer, A.; Kilickap, S.; Gümüş, M.; Bondarenko, I.; Özgüroğlu, M.; Gogishvili, M.; Turk, H.M.; Cicin, I.; Bentsion, D.; Gladkov, O.; et al. Cemiplimab Monotherapy for First-Line Treatment of Advanced Non-Small-Cell Lung Cancer with PD-L1 of at Least 50%: A Multicentre, Open-Label, Global, Phase 3, Randomised, Controlled Trial. Lancet 2021, 397, 592–604. [Google Scholar] [CrossRef]
- Bremnes, R.M.; Busund, L.-T.; Kilvær, T.L.; Andersen, S.; Richardsen, E.; Paulsen, E.E.; Hald, S.; Khanehkenari, M.R.; Cooper, W.A.; Kao, S.C.; et al. The Role of Tumor-Infiltrating Lymphocytes in Development, Progression, and Prognosis of Non–Small Cell Lung Cancer. J. Thorac. Oncol. 2016, 11, 789–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dijkstra, K.K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; van de Haar, J.; Fanchi, L.F.; Slagter, M.; van der Velden, D.L.; Kaing, S.; Kelderman, S.; et al. Generation of Tumor-Reactive T Cells by Co-Culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 2018, 174, 1586–1598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, N.; Hoshi, H.; Higa, A.; Hiyama, G.; Tamura, H.; Ogawa, M.; Takagi, K.; Goda, K.; Okabe, N.; Muto, S.; et al. An In Vitro System for Evaluating Molecular Targeted Drugs Using Lung Patient-Derived Tumor Organoids. Cells 2019, 8, 481. [Google Scholar] [CrossRef] [Green Version]
- Powley, I.R.; Patel, M.; Miles, G.; Pringle, H.; Howells, L.; Thomas, A.; Kettleborough, C.; Bryans, J.; Hammonds, T.; MacFarlane, M.; et al. Patient-Derived Explants (PDEs) as a Powerful Preclinical Platform for Anti-Cancer Drug and Biomarker Discovery. Br. J. Cancer 2020, 122, 735–744. [Google Scholar] [CrossRef] [Green Version]
- Di Liello, R.; Ciaramella, V.; Barra, G.; Venditti, M.; Della Corte, C.M.; Papaccio, F.; Sparano, F.; Viscardi, G.; Iacovino, M.L.; Minucci, S.; et al. Ex Vivo Lung Cancer Spheroids Resemble Treatment Response of a Patient with NSCLC to Chemotherapy and Immunotherapy: Case Report and Translational Study. ESMO Open 2019, 4, e000536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Votanopoulos, K.I.; Forsythe, S.; Sivakumar, H.; Mazzocchi, A.; Aleman, J.; Miller, L.; Levine, E.; Triozzi, P.; Skardal, A. Model of Patient-Specific Immune-Enhanced Organoids for Immunotherapy Screening: Feasibility Study. Ann. Surg. Oncol. 2020, 27, 1956–1967. [Google Scholar] [CrossRef] [PubMed]
- Majem, M.; Juan, O.; Insa, A.; Reguart, N.; Trigo, J.M.; Carcereny, E.; García-Campelo, R.; García, Y.; Guirado, M.; Provencio, M. SEOM Clinical Guidelines for the Treatment of Non-Small Cell Lung Cancer. Clin. Transl. Oncol. 2019, 21, 3–17. [Google Scholar] [CrossRef] [Green Version]
- Holmes, M.; Mahar, A.; Lum, T.; Boyer, M.; Kao, S.; Cooper, W. P1.09-26 Prevalence of PD-L1 Expression Rates in Different NSCLC Specimens. J. Thorac. Oncol. 2019, 14, S506. [Google Scholar] [CrossRef]
- Sacher, A.G.; Gandhi, L. Biomarkers for the Clinical Use of PD-1/PD-L1 Inhibitors in Non–Small-Cell Lung Cancer: A Review. JAMA Oncol. 2016, 2, 1217. [Google Scholar] [CrossRef]
- Jain, P.; Jain, C.; Velcheti, V. Role of Immune-Checkpoint Inhibitors in Lung Cancer. Ther. Adv. Respir. Dis. 2018, 12, 175346581775007. [Google Scholar] [CrossRef] [Green Version]
- Jung, D.J.; Shin, T.H.; Kim, M.; Sung, C.O.; Jang, S.J.; Jeong, G.S. A One-Stop Microfluidic-Based Lung Cancer Organoid Culture Platform for Testing Drug Sensitivity. Lab Chip 2019, 19, 2854–2865. [Google Scholar] [CrossRef] [PubMed]
- Maulana, T.I.; Kromidas, E.; Wallstabe, L.; Cipriano, M.; Alb, M.; Zaupa, C.; Hudecek, M.; Fogal, B.; Loskill, P. Immunocompetent Cancer-on-Chip Models to Assess Immuno-Oncology Therapy. Adv. Drug Deliv. Rev. 2021, 173, 281–305. [Google Scholar] [CrossRef] [PubMed]
- Abou Khouzam, R.; Brodaczewska, K.; Filipiak, A.; Zeinelabdin, N.A.; Buart, S.; Szczylik, C.; Kieda, C.; Chouaib, S. Tumor Hypoxia Regulates Immune Escape/Invasion: Influence on Angiogenesis and Potential Impact of Hypoxic Biomarkers on Cancer Therapies. Front. Immunol. 2021, 11, 613114. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Ching, H.; Yoon, J.-K.; Jeon, N.L.; Kim, Y. Microvascularized Tumor Organoids-on-Chips: Advancing Preclinical Drug Screening with Pathophysiological Relevance. Nano Converg. 2021, 8, 12. [Google Scholar] [CrossRef]
- Yildiz-Ozturk, E.; Saglam-Metiner, P.; Yesil-Celiktas, O. Lung Carcinoma Spheroids Embedded in a Microfluidic Platform. Cytotechnology 2021, 73, 457–471. [Google Scholar] [CrossRef]
- Schuster, B.; Junkin, M.; Kashaf, S.S.; Romero-Calvo, I.; Kirby, K.; Matthews, J.; Weber, C.R.; Rzhetsky, A.; White, K.P.; Tay, S. Automated Microfluidic Platform for Dynamic and Combinatorial Drug Screening of Tumor Organoids. Nat. Commun. 2020, 11, 5271. [Google Scholar] [CrossRef]
- “Organ-on-a-Chip”-Based Physiologically Relevant Pharmacokinetic Models—Science Direct. Available online: https://www.sciencedirect.com/science/article/pii/B9780128144251000243 (accessed on 24 April 2022).
- Hassell, B.A.; Goyal, G.; Lee, E.; Sontheimer-Phelps, A.; Levy, O.; Chen, C.S.; Ingber, D.E. Human Organ Chip Models Recapitulate Orthotopic Lung Cancer Growth, Therapeutic Responses, and Tumor Dormancy In Vitro. Cell Rep. 2017, 21, 508–516. [Google Scholar] [CrossRef] [Green Version]
- Ruppen, J.; Wildhaber, F.D.; Strub, C.; Hall, S.R.R.; Schmid, R.A.; Geiser, T.; Guenat, O.T. Towards Personalized Medicine: Chemosensitivity Assays of Patient Lung Cancer Cell Spheroids in a Perfused Microfluidic Platform. Lab Chip 2015, 15, 3076–3085. [Google Scholar] [CrossRef] [Green Version]
- Valente, K.P.; Khetani, S.; Kolahchi, A.R.; Sanati-Nezhad, A.; Suleman, A.; Akbari, M. Microfluidic Technologies for Anticancer Drug Studies. Drug Discov. Today 2017, 22, 1654–1670. [Google Scholar] [CrossRef]
- Feng, J.; Wu, T.; Cheng, Q.; Ma, H.; Ren, X.; Wang, X.; Lee, J.Y.; Wei, Q.; Ju, H. A Microfluidic Cathodic Photoelectrochemical Biosensor Chip for the Targeted Detection of Cytokeratin 19 Fragments 21-1. Lab Chip 2021, 21, 378–384. [Google Scholar] [CrossRef]
- Nixon, N.A.; Khan, O.F.; Imam, H.; Tang, P.A.; Monzon, J.; Li, H.; Sun, G.; Ezeife, D.; Parimi, S.; Dowden, S.; et al. Drug Development for Breast, Colorectal, and Non-Small Cell Lung Cancers from 1979 to 2014: Cancer Drug Development from 1979–2014. Cancer 2017, 123, 4672–4679. [Google Scholar] [CrossRef] [PubMed]
- Mansinho, A.; Boni, V.; Miguel, M.; Calvo, E. New Designs in Early Clinical Drug Development. Ann. Oncol. 2019, 30, 1460–1465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werner, R.S.; Kirschner, M.B.; Opitz, I. Primary Lung Cancer Organoids for Personalized Medicine—Are They Ready for Clinical Use? Cancers 2021, 13, 4832. [Google Scholar] [CrossRef] [PubMed]
- Sura, R.; Van Vleet, T.; Berridge, B.R. Microphysiological Systems: A Pathologist’s Perspective. Vet. Pathol. 2020, 57, 358–368. [Google Scholar] [CrossRef]

| Technologies | Advantages | Limits | References | |
|---|---|---|---|---|
| In vitro | Cancer cell lines | Pure population of tumor cells Replicative ability Large diversity of genomic backgrounds |
| [17,18,19,20] |
| In vivo | PDXs | Closer to patients’ primary tissues |
| [17,22,23,24,25] |
| Syngeneic models | Functional immune system |
| [27] | |
| GEMMs | Functional immune system Inducible model |
| [17,28,29] | |
| Primary Tumor Histology, (Mutations) * | Technology Name | Culture Time | Applications | Ref. |
|---|---|---|---|---|
| NS | Patient-derived tumor spheroid (PDS) | 120 days | Mechanistic studies Resistant models Drug screening | [38] |
| ADK, SCC, LCC | Lung cancer organoids | >1 year | Drug screening | [39] |
| ADK, SCC, LCC | Patient-derived lung cancer organoids | >6 months | Patient-specific drugs screening Living biobank as support to xenograft model | [24] |
| ADK, SCC | NSCLC organoids | 3 months | Drug screening | [37] |
| ADK, SCC NSCLC (EGFR, KRAS) | Patient-derived organoids models (PDOs) | NS | Genomic analyses Production of treatment response | [40] |
| NSCLC (EGFR, KRAS) | Patient lung-derived tumoroids (PLDTs) | NS | Drug screening | [41] |
| ADK, SCC, LCC, NSCLC | Lung cancer organoids | NS | Personalized medicine | [36] |
| NS | Patient-derived organoids (PDOs) | 2–3 months | Drug screening Comparative analysis | [42] |
| ADK | Lung ADK (LADC)-derived organoid model | >50–200 days | Transcriptome analysis Biomarkers discovery Drug screening Living biobank | [35] |
| ADK and SCC | Lung cancer organoids | 6 days | Drug screening | [43] |
| ADK and SCC primary or metastatic NSCLC | Patient-derived tumoroids (PDTs) | >13 months | Generation of cell lines | [44] |
| ADK | Patient-derived tumoroids (PDTs) | 4 days | Mimic the tumor vascular network PDTs ready to use in microfluidic device for drug screening | [45] |
| Drug | Target(S) | Indications | FDA Approval | Ref. |
|---|---|---|---|---|
| Pembrolizumab KEYTRUDA® | PD-1 |
| 2015 2015 2017 | [66] [67] |
| Nivolumab OPDIVO® | PD-1 |
| 2015 2020 | [68] [69] |
| Durvalumab IMFINZI® | PD-L1 | Unresectable stage III NSCLC patients that have not progressed after chemoradiation therapy | 2018 | [70] |
| Atezolizumab TECENTRIQv | PD-L1 | First-line treatment in metastatic NSCLC with PD-L1 > 50% | 2020 | [71] |
| Cemiplimab-rwlcLIBTAYO® | PD-1 | First-line treatment in locally or metastatic advanced stage NSCLC (no eligible to surgical resection nor definitive chemoradiation) with PD-L1 > 50% | 2021 | [72] |
| Microfluidic Model | Applications | Ref. |
|---|---|---|
| Microfluidic device for lung cancer organoids | Drug screening
| [83] |
| Lung carcinoma spheroid based microfluidic platform | Drug assessment of panaxatriol in fluidic conditions with a perfusion function on cancer cells and healthy cells | [87] |
| Human organ chip model | Recapitulation of human cancer with its specific microenvironment Assessment of tyrosine kinase inhibitors’ (TKI) responses to physical cues mimicking breathing motions
| [90] |
| Lung cancer cell spheroids in a perfused microfluidic platform | Cell viability assessment of chemotherapeutical drug | [91] |
| Detection of cytokeratin 19 fragments | Biomarkers study of diagnosis and prognosis | [93] |
| Chip for study of lung cancer brain metastasis | Study metastasis | [54] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lê, H.; Seitlinger, J.; Lindner, V.; Olland, A.; Falcoz, P.-E.; Benkirane-Jessel, N.; Quéméneur, E. Patient-Derived Lung Tumoroids—An Emerging Technology in Drug Development and Precision Medicine. Biomedicines 2022, 10, 1677. https://doi.org/10.3390/biomedicines10071677
Lê H, Seitlinger J, Lindner V, Olland A, Falcoz P-E, Benkirane-Jessel N, Quéméneur E. Patient-Derived Lung Tumoroids—An Emerging Technology in Drug Development and Precision Medicine. Biomedicines. 2022; 10(7):1677. https://doi.org/10.3390/biomedicines10071677
Chicago/Turabian StyleLê, Hélène, Joseph Seitlinger, Véronique Lindner, Anne Olland, Pierre-Emmanuel Falcoz, Nadia Benkirane-Jessel, and Eric Quéméneur. 2022. "Patient-Derived Lung Tumoroids—An Emerging Technology in Drug Development and Precision Medicine" Biomedicines 10, no. 7: 1677. https://doi.org/10.3390/biomedicines10071677
APA StyleLê, H., Seitlinger, J., Lindner, V., Olland, A., Falcoz, P.-E., Benkirane-Jessel, N., & Quéméneur, E. (2022). Patient-Derived Lung Tumoroids—An Emerging Technology in Drug Development and Precision Medicine. Biomedicines, 10(7), 1677. https://doi.org/10.3390/biomedicines10071677







