Multi-Channel Vision Transformer for Epileptic Seizure Prediction
Abstract
:1. Introduction
2. Related Work
3. Materials and Methods
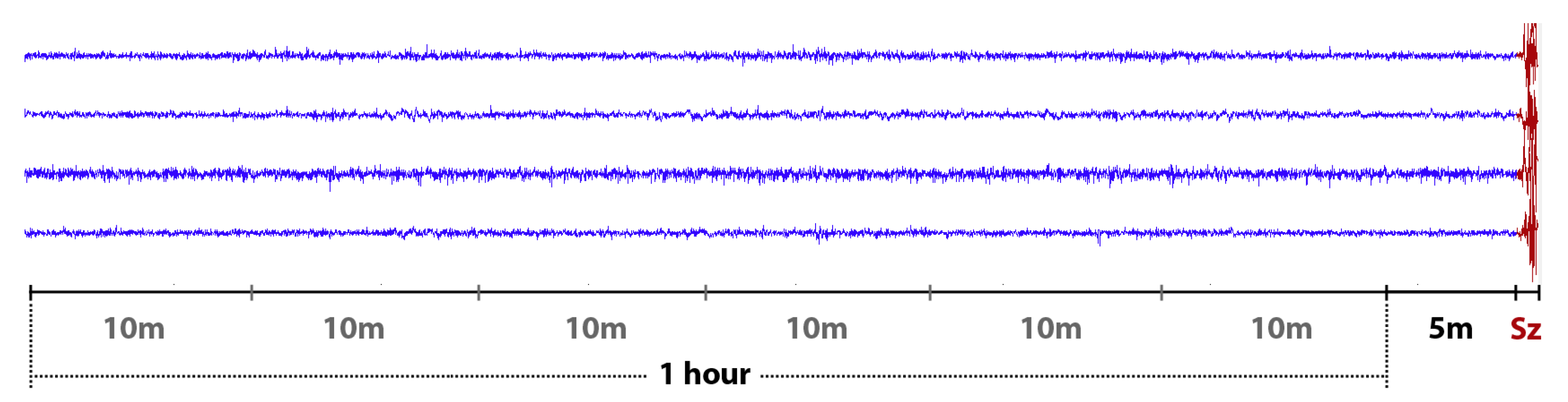
3.1. Datasets
3.2. Methodology
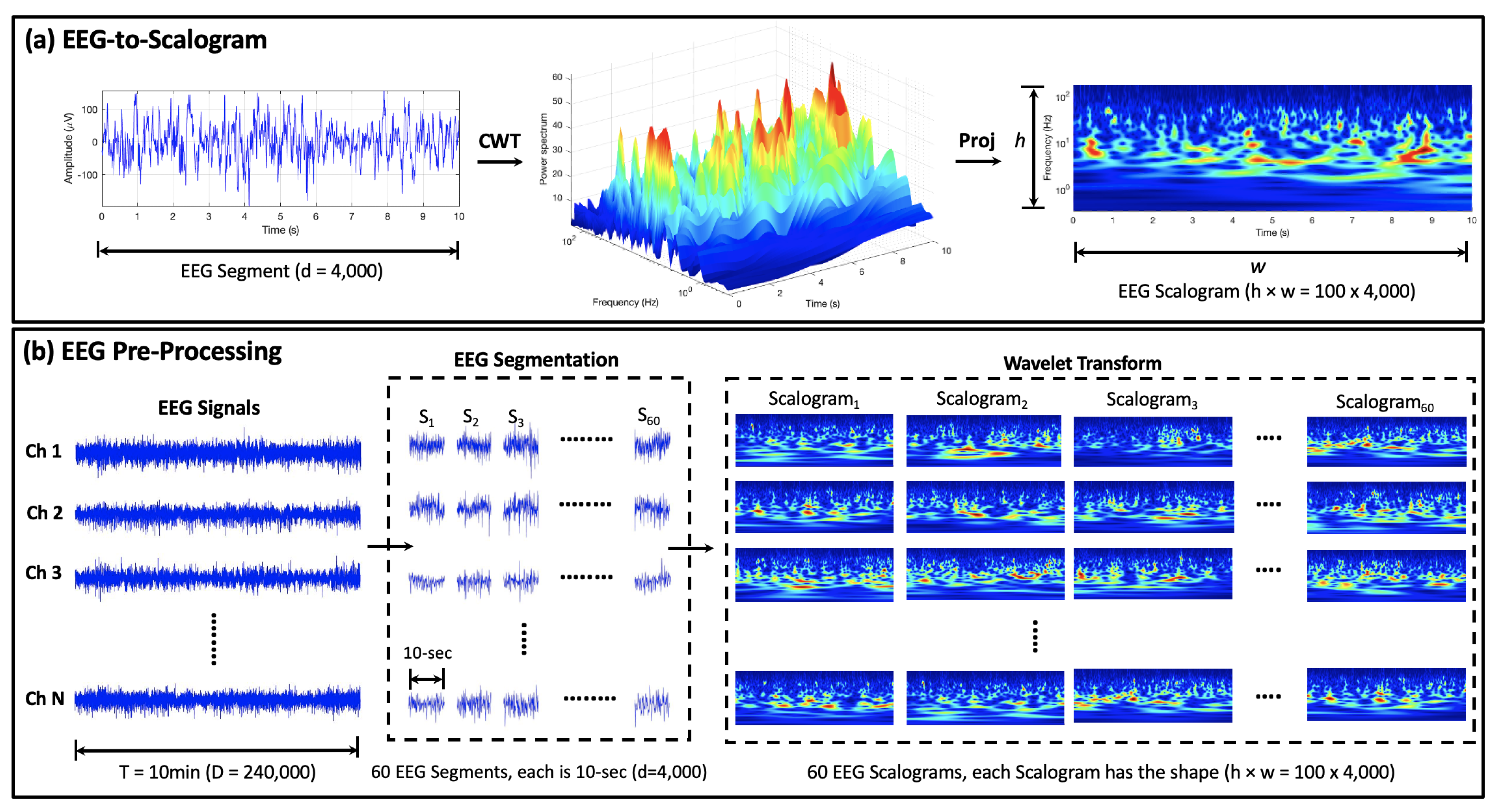
3.2.1. EEG Pre-Processing
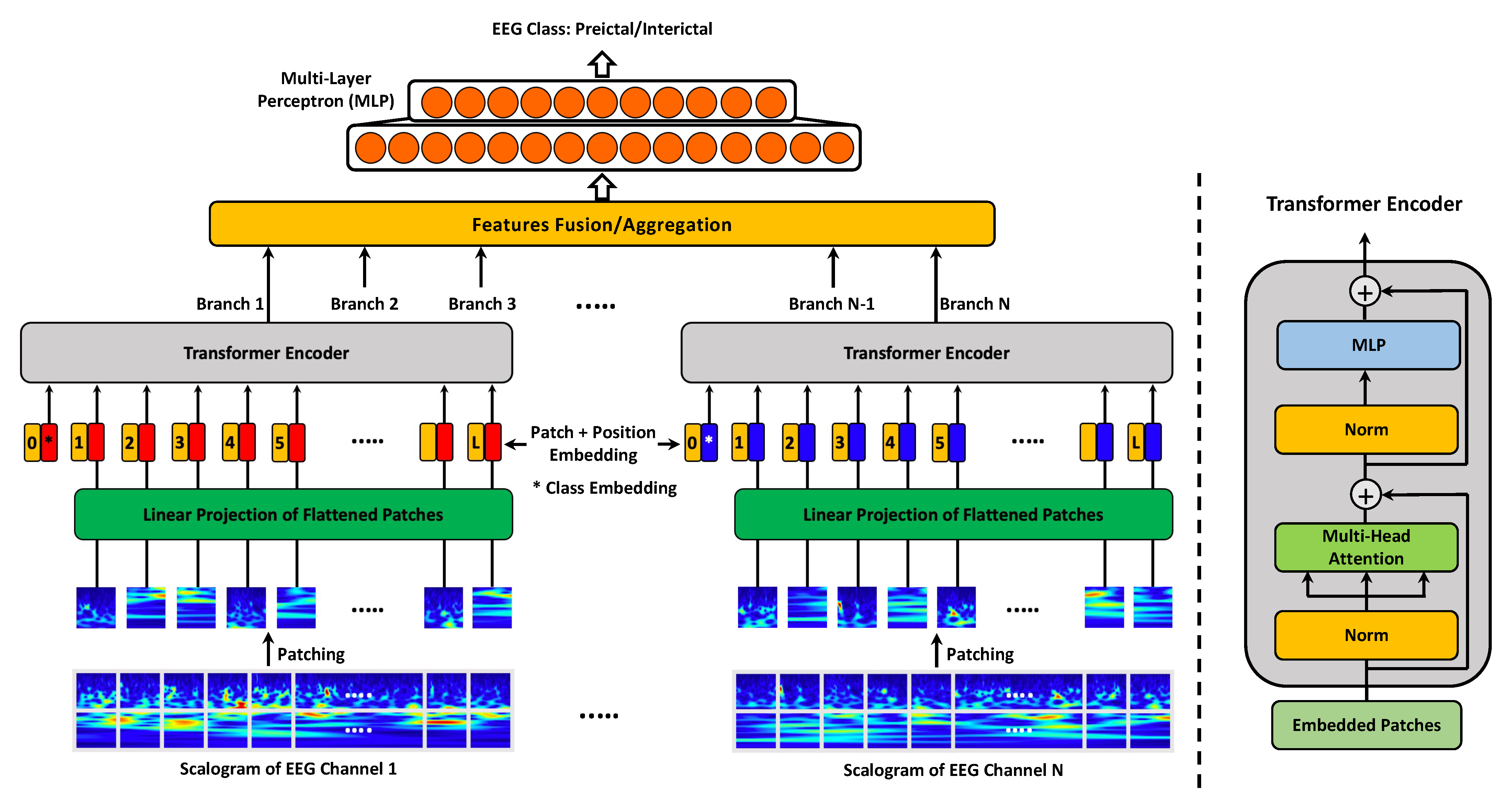
3.2.2. MViT for EEG Representation Learning
3.2.3. Performance Evaluation
4. Results and Discussion
4.1. MViT Prediction Performance on Surface Pediatric EEG
4.2. MViT Prediction Performance on Invasive Human and Canine EEG
4.3. MViT Prediction Performance on Invasive Human EEG
| Authors/ Team | Year | EEG Features | Classifier | SENS (%) | AUC Score Public/Private |
|---|---|---|---|---|---|
| Cook et al. [15] ☆ | 2013 | Signal energy | Decision tree, kNN | 33.67 | - |
| Karoly et al. [62] ☆ | 2017 | Signal energy, circadian profile | Logistic regression | 52.67 | - |
| Kiral-Kornek et al. [16] ☆ | 2018 | EEG Spectrogram, circadian profile | CNN | 77.36 | - |
| Not-so-random | 2018 | Hurst exponent, spectral power, | Extreme gradient | - | 0.853/0.807 |
| -anymore [33] | distribution attributes, fractal dimensions, | boosting, | |||
| AR error, and cross-frequency coherence | kNN, SVM | ||||
| Arete | 2018 | Correlation, entropy, zero-crossings, | Extremely | - | 0.783/0.799 |
| Associates [33] | distribution statistics, and spectral power | randomized trees | |||
| GarethJones [33] | 2018 | Distribution statistics, spectral power, | SVM | - | 0.815/0.797 |
| signal RMS, correlation, and spectral edge | tree ensemble | ||||
| QingnanTang [33] | 2018 | Spectral power, spectral entropy | Gradient boosting, | - | 0.854/0.791 |
| correlation, and spectral edge power | SVM | ||||
| Nullset [33] | 2018 | Hjorth parameters, spectral power, | Random Forest, | - | 0.844/0.746 |
| spectral edge, spectral entropy, | adaptive boosting, | ||||
| Shannon entropy, and fractal dimensions | and gradient boosting | ||||
| Reuben et al. [63] | 2019 | Preictal probabilities from | MLP | - | 0.815/- |
| the top 8 teams in [33] | |||||
| Varnosfaderani et al. [64] | 2021 | Temporal features, statistical moments, | LSTM | 86.80 | 0.920/- |
| and spectral power | |||||
| Hussein et al. [29] | 2021 | EEG Scalogram | SDCN | 89.52 | 0.883/- |
| Zhao et al. [61] | 2022 | Raw EEG | CNN | 85.19–86.27 | 0.914–0.933/- |
| Proposed Method | 2022 | EEG Scalogram | MViT | 91.15 | 0.924/- |
5. Clinical Significance and Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AES | American Epilepsy Society |
| AUC | Area under the ROC Curve |
| CHB | Children’s Hospital Boston |
| CNN | Convolutional Neural Networks |
| CWT | Continuous Wavelet Transform |
| DCAE | Deep Convolutional AutoEncoder |
| DFT | Directed Transfer Function |
| EEG | Electroencephalogram |
| FPR | False Positive Rate |
| FFT | Fast Fourier Transform |
| GCN | Graph Convolutional Network |
| iEEG | intracranial EEG |
| LassoGLM | Lasso regularization of Generalized Linear Models |
| LN | Layer normalization |
| LSTM | Long Short-Term Memory |
| MLP | Multi-Layer Perceptron |
| MViT | Multi-Channel Vision Transformer |
| MSA | Multi-head Self Attention |
| NLP | Natural Language Processing |
| PCA | Principal Component Analysis |
| Proj | Projection |
| RNN | Recurrent Neural Networks |
| ROC | Receiver Operating Characteristic |
| SAS | Seizure Advisory System |
| SENS | Sensitivity |
| STFT | Short-Time Fourier Transform |
| SPEC | Specificity |
| SVM | Support Vector Machine |
| ViT | Vision Transformer |
References
- Rogers, G. Epilepsy: The facts. Prim. Health Care Res. Dev. 2010, 11, 413. [Google Scholar] [CrossRef]
- Acharya, U.R.; Sree, S.V.; Swapna, G.; Martis, R.J.; Suri, J.S. Automated EEG analysis of epilepsy: A review. Knowl.-Based Syst. 2013, 45, 147–165. [Google Scholar] [CrossRef]
- French, J.A. Refractory epilepsy: Clinical overview. Epilepsia 2007, 48, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Téllez-Zenteno, J.F.; Ronquillo, L.H.; Moien-Afshari, F.; Wiebe, S. Surgical outcomes in lesional and non-lesional epilepsy: A systematic review and meta-analysis. Epilepsy Res. 2010, 89, 310–318. [Google Scholar] [CrossRef]
- D’Alessandro, M.; Esteller, R.; Vachtsevanos, G.; Hinson, A.; Echauz, J.; Litt, B. Epileptic seizure prediction using hybrid feature selection over multiple intracranial EEG electrode contacts: A report of four patients. IEEE Trans. Biomed. Eng. 2003, 50, 603–615. [Google Scholar] [CrossRef]
- Gadhoumi, K.; Lina, J.M.; Mormann, F.; Gotman, J. Seizure prediction for therapeutic devices: A review. J. Neurosci. Methods 2016, 260, 270–282. [Google Scholar] [CrossRef]
- Assi, E.B.; Nguyen, D.K.; Rihana, S.; Sawan, M. Towards accurate prediction of epileptic seizures: A review. Biomed. Signal Process. Control 2017, 34, 144–157. [Google Scholar] [CrossRef]
- Iasemidis, L.; Principe, J.; Sackellares, J. Measurement and quantification of spatiotemporal dynamics of human epileptic seizures. Nonlinear Biomed. Signal Process. 2000, 2, 294–318. [Google Scholar]
- Stafstrom, C.E.; Carmant, L. Seizures and epilepsy: An overview for neuroscientists. Cold Spring Harb. Perspect. Med. 2015, 5, a022426. [Google Scholar] [CrossRef]
- Aarabi, A.; Fazel-Rezai, R.; Aghakhani, Y. EEG seizure prediction: Measures and challenges. In Proceedings of the 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Minneapolis, MN, USA, 2–6 September 2009; pp. 1864–1867. [Google Scholar]
- Bandarabadi, M.; Teixeira, C.A.; Rasekhi, J.; Dourado, A. Epileptic seizure prediction using relative spectral power features. Clin. Neurophysiol. 2015, 126, 237–248. [Google Scholar] [CrossRef]
- Vahabi, Z.; Amirfattahi, R.; Shayegh, F.; Ghassemi, F. Online epileptic seizure prediction using wavelet-based bi-phase correlation of electrical signals tomography. Int. J. Neural Syst. 2015, 25, 1550028. [Google Scholar] [CrossRef] [PubMed]
- Sackellares, J.C. Seizure prediction. Epilepsy Curr. 2008, 8, 55–59. [Google Scholar] [CrossRef] [Green Version]
- Kawaguchi, K.; Kaelbling, L.P.; Bengio, Y. Generalization in deep learning. arXiv 2017, arXiv:1710.05468. [Google Scholar]
- Cook, M.J.; O’Brien, T.J.; Berkovic, S.F.; Murphy, M.; Morokoff, A.; Fabinyi, G.; D’Souza, W.; Yerra, R.; Archer, J.; Litewka, L.; et al. Prediction of seizure likelihood with a long-term, implanted seizure advisory system in patients with drug-resistant epilepsy: A first-in-man study. Lancet Neurol. 2013, 12, 563–571. [Google Scholar] [CrossRef]
- Kiral-Kornek, I.; Roy, S.; Nurse, E.; Mashford, B.; Karoly, P.; Carroll, T.; Payne, D.; Saha, S.; Baldassano, S.; O’Brien, T.; et al. Epileptic seizure prediction using big data and deep learning: Toward a mobile system. EBioMedicine 2018, 27, 103–111. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui, M.K.; Morales-Menendez, R.; Huang, X.; Hussain, N. A review of epileptic seizure detection using machine learning classifiers. Brain Inform. 2020, 7, 5. [Google Scholar] [CrossRef]
- Park, Y.; Luo, L.; Parhi, K.K.; Netoff, T. Seizure prediction with spectral power of EEG using cost-sensitive support vector machines. Epilepsia 2011, 52, 1761–1770. [Google Scholar] [CrossRef]
- Shiao, H.T.; Cherkassky, V.; Lee, J.; Veber, B.; Patterson, E.E.; Brinkmann, B.H.; Worrell, G.A. SVM-based system for prediction of epileptic seizures from iEEG signal. IEEE Trans. Biomed. Eng. 2016, 64, 1011–1022. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, M.A.; Khan, N.A.; Majeed, W. Computer assisted analysis system of electroencephalogram for diagnosing epilepsy. In Proceedings of the 2014 22nd International Conference on Pattern Recognition (ICPR), Stockholm, Sweden, 24–28 August 2014; pp. 3386–3391. [Google Scholar]
- Zabihi, M.; Kiranyaz, S.; Ince, T.; Gabbouj, M. Patient-specific epileptic seizure detection in long-term EEG recording in paediatric patients with intractable seizures. In Proceedings of the IET Intelligent Signal Processing Conference 2013 (ISP 2013), London, UK, 2–3 December 2013. [Google Scholar]
- Williamson, J.R.; Bliss, D.W.; Browne, D.W.; Narayanan, J.T. Seizure prediction using EEG spatiotemporal correlation structure. Epilepsy Behav. 2012, 25, 230–238. [Google Scholar] [CrossRef] [Green Version]
- Khan, H.; Marcuse, L.; Fields, M.; Swann, K.; Yener, B. Focal Onset Seizure Prediction Using Convolutional Networks. IEEE Trans. Biomed. Eng. 2018, 65, 2109–2118. [Google Scholar] [CrossRef] [Green Version]
- Truong, N.D.; Nguyen, A.D.; Kuhlmann, L.; Bonyadi, M.R.; Yang, J.; Ippolito, S.; Kavehei, O. Convolutional neural networks for seizure prediction using intracranial and scalp electroencephalogram. Neural Netw. 2018, 105, 104–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Wang, D.; Du, C.; Li, K.; Zhang, J.; Liu, Z.; Tao, Y.; Wang, M.; Cao, Z.; Yan, X. Seizure prediction using directed transfer function and convolution neural network on intracranial EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2020, 28, 2711–2720. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, A.R.; Erturk, S. Seizure Prediction in Scalp EEG Using 3D Convolutional Neural Networks With an Image-Based Approach. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 2284–2293. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.L.; Xiao, B.; Hsaio, W.H.; Tseng, V.S. Epileptic Seizure Prediction With Multi-View Convolutional Neural Networks. IEEE Access 2019, 7, 170352–170361. [Google Scholar] [CrossRef]
- Qi, Y.; Ding, L.; Wang, Y.; Pan, G. Learning Robust Features in Nonstationary Brain Signals by Domain Adaptation Networks for Seizure Prediction. Trans. Neural Syst. Rehabil. Eng. 2021. [Google Scholar] [CrossRef]
- Hussein, R.; Lee, S.; Ward, R.; McKeown, M.J. Semi-dilated convolutional neural networks for epileptic seizure prediction. Neural Netw. 2021, 139, 212–222. [Google Scholar] [CrossRef]
- Lian, Q.; Qi, Y.; Pan, G.; Wang, Y. Learning graph in graph convolutional neural networks for robust seizure prediction. J. Neural Eng. 2020, 17, 035004. [Google Scholar] [CrossRef]
- Shoeb, A.H. Application of Machine Learning to Epileptic Seizure Onset Detection and Treatment. Ph.D. Thesis, Massachusetts Institute of Technology, Cambridge, MA, USA, 2009. [Google Scholar]
- Brinkmann, B.H.; Wagenaar, J.; Abbot, D.; Adkins, P.; Bosshard, S.C.; Chen, M.; Tieng, Q.M.; He, J.; Muñoz-Almaraz, F.; Botella-Rocamora, P.; et al. Crowdsourcing reproducible seizure forecasting in human and canine epilepsy. Brain 2016, 139, 1713–1722. [Google Scholar] [CrossRef]
- Kuhlmann, L.; Karoly, P.; Freestone, D.R.; Brinkmann, B.H.; Temko, A.; Barachant, A.; Li, F.; Titericz, G., Jr.; Lang, B.W.; Lavery, D.; et al. Epilepsyecosystem.org: Crowd-sourcing reproducible seizure prediction with long-term human intracranial EEG. Brain 2018, 141, 2619–2630. [Google Scholar] [CrossRef] [Green Version]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Azami, H.; Mohammadi, K.; Hassanpour, H. An improved signal segmentation method using genetic algorithm. Int. J. Comput. Appl. 2011, 29, 5–9. [Google Scholar] [CrossRef]
- Hassanpour, H.; Shahiri, M. Adaptive segmentation using wavelet transform. In Proceedings of the Electrical Engineering, ICEE’07, Lahore, Pakistan, 11–12 April 2007; pp. 1–5. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 1–11. [Google Scholar]
- Zheng, S.; Lu, J.; Zhao, H.; Zhu, X.; Luo, Z.; Wang, Y.; Fu, Y.; Feng, J.; Xiang, T.; Torr, P.H.; et al. Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 6881–6890. [Google Scholar]
- Zhang, Z.; Parhi, K.K. Low-complexity seizure prediction from iEEG/sEEG using spectral power and ratios of spectral power. IEEE Trans. Biomed. Circuits Syst. 2016, 10, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Cho, D.; Min, B.; Kim, J.; Lee, B. EEG-based prediction of epileptic seizures using phase synchronization elicited from noise-assisted multivariate empirical mode decomposition. IEEE Trans. Neural Syst. Rehabil. Eng. 2016, 25, 1309–1318. [Google Scholar] [CrossRef]
- Usman, S.M.; Usman, M.; Fong, S. Epileptic seizures prediction using machine learning methods. Comput. Math. Methods Med. 2017, 2017, 9074759. [Google Scholar] [CrossRef] [Green Version]
- Tsiouris, K.M.; Pezoulas, V.C.; Zervakis, M.; Konitsiotis, S.; Koutsouris, D.D.; Fotiadis, D.I. A Long Short-Term Memory deep learning network for the prediction of epileptic seizures using EEG signals. Comput. Biol. Med. 2018, 99, 24–37. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, Y.; Yang, P.; Chen, W.; Lo, B. Epilepsy seizure prediction on EEG using common spatial pattern and convolutional neural network. IEEE J. Biomed. Health Inform. 2019, 24, 465–474. [Google Scholar] [CrossRef]
- Daoud, H.; Bayoumi, M.A. Efficient epileptic seizure prediction based on deep learning. IEEE Trans. Biomed. Circuits Syst. 2019, 13, 804–813. [Google Scholar] [CrossRef]
- Usman, S.M.; Khalid, S.; Aslam, M.H. Epileptic Seizures Prediction Using Deep Learning Techniques. IEEE Access 2020, 8, 2169–3536. [Google Scholar]
- Barkın Büyükçakır, B.; Elmaz, F.; Mutlu, A.Y. Hilbert Vibration Decomposition-based epileptic seizure prediction with neural network. Comput. Biol. Med. 2020, 119, 103665. [Google Scholar] [CrossRef]
- Xu, Y.; Yang, J.; Zhao, S.; Wu, H.; Sawan, M. An End-to-End Deep Learning Approach for Epileptic Seizure Prediction. In Proceedings of the 2020 2nd IEEE International Conference on Artificial Intelligence Circuits and Systems (AICAS), Genova, Italy, 31 August–4 September 2020; pp. 266–270. [Google Scholar]
- Dissanayake, T.; Fernando, T.; Denman, S.; Sridharan, S.; Fookes, C. Deep Learning for Patient-Independent Epileptic Seizure Prediction Using Scalp EEG Signals. IEEE Sens. J. 2021, 21, 9377–9388. [Google Scholar] [CrossRef]
- Jana, R.; Mukherjee, I. Deep learning based efficient epileptic seizure prediction with EEG channel optimization. Biomed. Signal Process. Control 2021, 68, 102767. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Guo, Y.Z.; Liao, X.F.; Hu, B.; Yu, T. Spatio-Temporal-Spectral Hierarchical Graph Convolutional Network With Semisupervised Active Learning for Patient-Specific Seizure Prediction. IEEE Trans. Cybern. 2021, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Usman, S.M.; Khalid, S.; Bashir, Z. Epileptic seizure prediction using scalp electroencephalogram signals. Biocybern. Biomed. Eng. 2021, 41, 211–220. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, J.; Sun, Q.; Lu, J.; Ma, X. An Effective Dual Self-Attention Residual Network for Seizure Prediction. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 1604–1613. [Google Scholar] [CrossRef]
- Dissanayake, T.; Fernando, T.; Denman, S.; Sridharan, S.; Fookes, C. Geometric Deep Learning for Subject Independent Epileptic Seizure Prediction Using Scalp EEG Signals. IEEE J. Biomed. Health Inform. 2022, 26, 527–538. [Google Scholar] [CrossRef]
- Gao, Y.; Chen, X.; Liu, A.; Liang, D.; Wu, L.; Qian, R.; Xie, H.; Zhang, Y. Pediatric Seizure Prediction in Scalp EEG Using a Multi-Scale Neural Network With Dilated Convolutions. IEEE J. Transl. Eng. Health Med. 2022, 10, 4900209. [Google Scholar] [CrossRef]
- Zhang, X.; Li, H. Patient-Specific Seizure prediction from Scalp EEG Using Vision Transformer. In Proceedings of the 2022 IEEE 6th Information Technology and Mechatronics Engineering Conference (ITOEC), Chongqing China, 4–6 March 2022; pp. 1663–1667. [Google Scholar]
- Eberlein, M.; Hildebrand, R.; Tetzlaff, R.; Hoffmann, N.; Kuhlmann, L.; Brinkmann, B.; Müller, J. Convolutional neural networks for epileptic seizure prediction. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 3–6 December 2018; pp. 2577–2582. [Google Scholar]
- Ma, X.; Qiu, S.; Zhang, Y.; Lian, X.; He, H. Predicting epileptic seizures from intracranial EEG using LSTM-based multi-task learning. In Proceedings of the Chinese Conference on Pattern Recognition and Computer Vision (PRCV), Guangzhou, China, 23–26 November 2018; pp. 157–167. [Google Scholar]
- Korshunova, I.; Kindermans, P.J.; Degrave, J.; Verhoeven, T.; Brinkmann, B.H.; Dambre, J. Towards improved design and evaluation of epileptic seizure predictors. IEEE Trans. Biomed. Eng. 2018, 65, 502–510. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Parhi, K.K. Seizure Prediction using Convolutional Neural Networks and Sequence Transformer Networks. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Virtual, 1–5 November 2021; pp. 6483–6486. [Google Scholar]
- Usman, S.M.; Khalid, S.; Bashir, S. A deep learning based ensemble learning method for epileptic seizure prediction. Comput. Biol. Med. 2021, 136, 104710. [Google Scholar] [CrossRef]
- Zhao, S.; Yang, J.; Sawan, M. Energy-Efficient Neural Network for Epileptic Seizure Prediction. IEEE Trans. Biomed. Eng. 2022, 69, 401–411. [Google Scholar] [CrossRef]
- Karoly, P.J.; Ung, H.; Grayden, D.B.; Kuhlmann, L.; Leyde, K.; Cook, M.J.; Freestone, D.R. The circadian profile of epilepsy improves seizure forecasting. Brain 2017, 140, 2169–2182. [Google Scholar] [CrossRef] [PubMed]
- Reuben, C.; Karoly, P.; Freestone, D.R.; Temko, A.; Barachant, A.; Li, F.; Titericz, G., Jr.; Lang, B.W.; Lavery, D.; Roman, K.; et al. Ensembling crowdsourced seizure prediction algorithms using long-term human intracranial EEG. Epilepsia 2019, 61, e7–e12. [Google Scholar] [CrossRef] [PubMed]
- Varnosfaderani, S.M.; Rahman, R.; Sarhan, N.J.; Kuhlmann, L.; Asano, E.; Luat, A.; Alhawari, M. A Two-Layer LSTM Deep Learning Model for Epileptic Seizure Prediction. In Proceedings of the 2021 IEEE 3rd International Conference on Artificial Intelligence Circuits and Systems (AICAS), Washington, DC, USA, 6–9 June 2021. [Google Scholar]
- Khan, S.; Naseer, M.; Hayat, M.; Zamir, S.W.; Khan, F.S.; Shah, M. Transformers in vision: A survey. ACM Comput. Surv. (CSUR) 2021. [Google Scholar] [CrossRef]
- Chefer, H.; Gur, S.; Wolf, L. Transformer interpretability beyond attention visualization. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 782–791. [Google Scholar]
- Naseer, M.M.; Ranasinghe, K.; Khan, S.H.; Hayat, M.; Shahbaz Khan, F.; Yang, M.H. Intriguing properties of vision transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 23296–23308. [Google Scholar]
| Authors | Year | EEG Features | Classifier | SENS (%) | SPEC (%) | ACC (%) | FPR (/h) |
|---|---|---|---|---|---|---|---|
| Zhang and Parhi [39] | 2016 | Spectral power | SVM | 98.7 | - | - | 0.04 |
| Cho et al. [40] | 2016 | Phase locking value | SVM | 82.4 | 82.8 | - | - |
| Usman et al. [41] | 2017 | Statistical and spectral moments | SVM | 92.2 | - | - | - |
| Khan et al. [23] | 2018 | Wavelet coefficients | CNN | 86.6 | - | - | 0.147 |
| Truong et al. [24] | 2018 | EEG Spectrogram | CNN | 81.2 | - | - | 0.16 |
| Tsiouris et al. [42] | 2018 | Spectral power, statistical moments | LSTM | 99.3–99.8 | 99.3–99.9 | - | 0.02–0.11 |
| Ozcan et al. [26] | 2018 | Spectral power, statistical moments | 3D CNN | 85.7 | - | - | 0.096 |
| Zhang et al. [43] | 2019 | Common spatial patterns | CNN | 92.0 | - | 90.0 | 0.12 |
| Daoud et al. [44] | 2019 | Multi-channel time series | LSTM | 99.7 | 99.6 | 99.7 | 0.004 |
| Usman et al. [45] | 2020 | EEG Spectrogram + CNN features | SVM | 92.7 | 90.8 | - | - |
| Büyükçakır et al. [46] | 2020 | Statiscal moments, spectral power | MLP | 89.8 | - | - | 0.081 |
| Xu et al. [47] | 2020 | Raw EEG | CNN | 98.8 | - | - | 0.074 |
| Dissanayake et al. [48] | 2021 | Mel-frequency cepstral coefficients | Siamese NN | 92.5 | 89.9 | 91.5 | - |
| Hussein et al. [29] | 2021 | Scalogram | SDCN | 98.9 | - | - | - |
| Jana et al. [49] | 2021 | Raw EEG | CNN | 92.0 | 86.4 | - | 0.136 |
| Li et al. [50] | 2021 | Spectral-temporal features | GCN | 95.5 | - | - | 0.109 |
| Usman et al. [51] | 2021 | EEG Spectrogram | LSTM | 93.0 | 92.5 | - | - |
| Yang et al. [52] | 2021 | EEG Spectrogram | Residual network | 89.3 | 93.0 | 92.1 | - |
| Dissanayake et al. [53] | 2022 | Mel frequency cepstral coefficients | GNN | 94.5 | 94.2 | 95.4 | - |
| Gao et al. [54] | 2022 | Raw EEG | Dilated CNN | 93.3 | - | - | 0.007 |
| Zhang et al. [55] | 2022 | EEG Spectrogram | ViT | 59.2–97.0 | 65.8–94.6 | - | - |
| Proposed Method | 2022 | EEG Scalogram | MViT | 99.8 | 99.7 | 99.8 | 0.004 |
| Authors/ Team | Year | EEG Features | Classifier | SENS (%) | AUC Score Public/Private |
|---|---|---|---|---|---|
| Medrr [32] | 2016 | N/A | N/A | - | 0.903/0.840 |
| QMSDP [32] | 2016 | Correlation, Hurst exponent, | LassoGLM, | - | 0.859/0.820 |
| fractal dimensions, | Bagged SVM, | ||||
| Spectral entropy | Random Forest | ||||
| Birchwood [32] | 2016 | Covariance, spectral power | SVM | - | 0.839/0.801 |
| ESAI CEU-UCH [32] | 2016 | Spectral power, | Neural Network, | - | 0.825/0.793 |
| correlation, PCA | kNN | ||||
| Michael Hills [32] | 2016 | Spectral power, correlation, | SVM | - | 0.862/0.793 |
| spectral entropy, fractal dimensions | |||||
| Truong et al. [24] | 2018 | EEG Spectrogram | CNN | 75.0 | - |
| Eberlein et al. [56] | 2018 | Multi-channel time series | CNN | - | 0.843/- |
| Ma et al. [57] | 2018 | Spectral power, correlation | LSTM | - | 0.894/- |
| Korshunova et al. [58] | 2018 | Spectral power | CNN | - | 0.780/0.760 |
| Liu et al. [27] | 2019 | PCA, spectral power | Multi-view CNN | - | 0.837/0.842 |
| Qi et al. [28] | 2019 | Spectral power, variance, correlation | Multi-scale CNN | - | 0.829/0.774 |
| Chen et al. [59] | 2021 | EEG Spectrogram | CNN | 82.00 | 0.746/- |
| Hussein et al. [29] | 2021 | EEG Scalogram | SDCN | 88.45 | 0.928/0.856 |
| Usman et al. [60] | 2021 | statistical and spectral moments | Ensemble of SVM, | 94.20 | - |
| CNN, and LSTM | |||||
| Zhao et al. [61] | 2022 | Raw EEG | CNN | 91.77–93.48 | 0.953–0.977/- |
| Proposed Method | 2022 | EEG Scalogram | MViT | 90.28 | 0.940/0.885 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussein, R.; Lee, S.; Ward, R. Multi-Channel Vision Transformer for Epileptic Seizure Prediction. Biomedicines 2022, 10, 1551. https://doi.org/10.3390/biomedicines10071551
Hussein R, Lee S, Ward R. Multi-Channel Vision Transformer for Epileptic Seizure Prediction. Biomedicines. 2022; 10(7):1551. https://doi.org/10.3390/biomedicines10071551
Chicago/Turabian StyleHussein, Ramy, Soojin Lee, and Rabab Ward. 2022. "Multi-Channel Vision Transformer for Epileptic Seizure Prediction" Biomedicines 10, no. 7: 1551. https://doi.org/10.3390/biomedicines10071551
APA StyleHussein, R., Lee, S., & Ward, R. (2022). Multi-Channel Vision Transformer for Epileptic Seizure Prediction. Biomedicines, 10(7), 1551. https://doi.org/10.3390/biomedicines10071551






