The Antibacterial Effect of Platelets on Escherichia coli Strains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Platelet Preparation
2.2. Bacterial Preparation
2.3. Analysis of Platelet Activation by Flow Cytometry
2.4. Platelet Supernatant Effect on Bacterial Growth
2.5. Pangenome Analysis
2.6. O-antigen Strain Serotyping
2.7. Statistical Analysis
3. Results
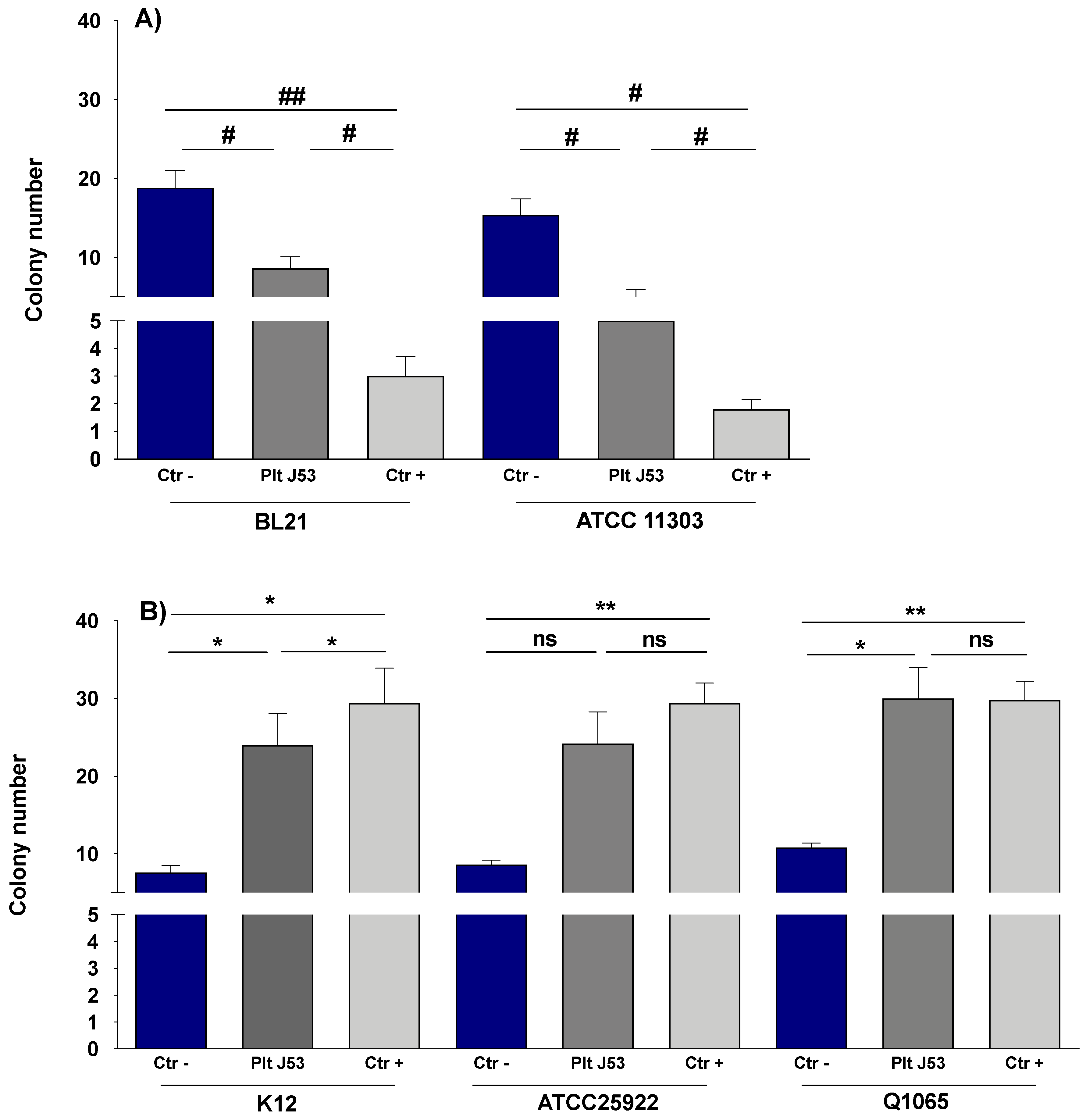
3.1. The Effect of Platelets on the Growth of E. coli Strains
3.2. The Effect of Platelet Supernatant on the Growth of E. coli Strains
3.3. The Effect of E. coli Strains on Platelet Activation
4. Pangenome Analysis
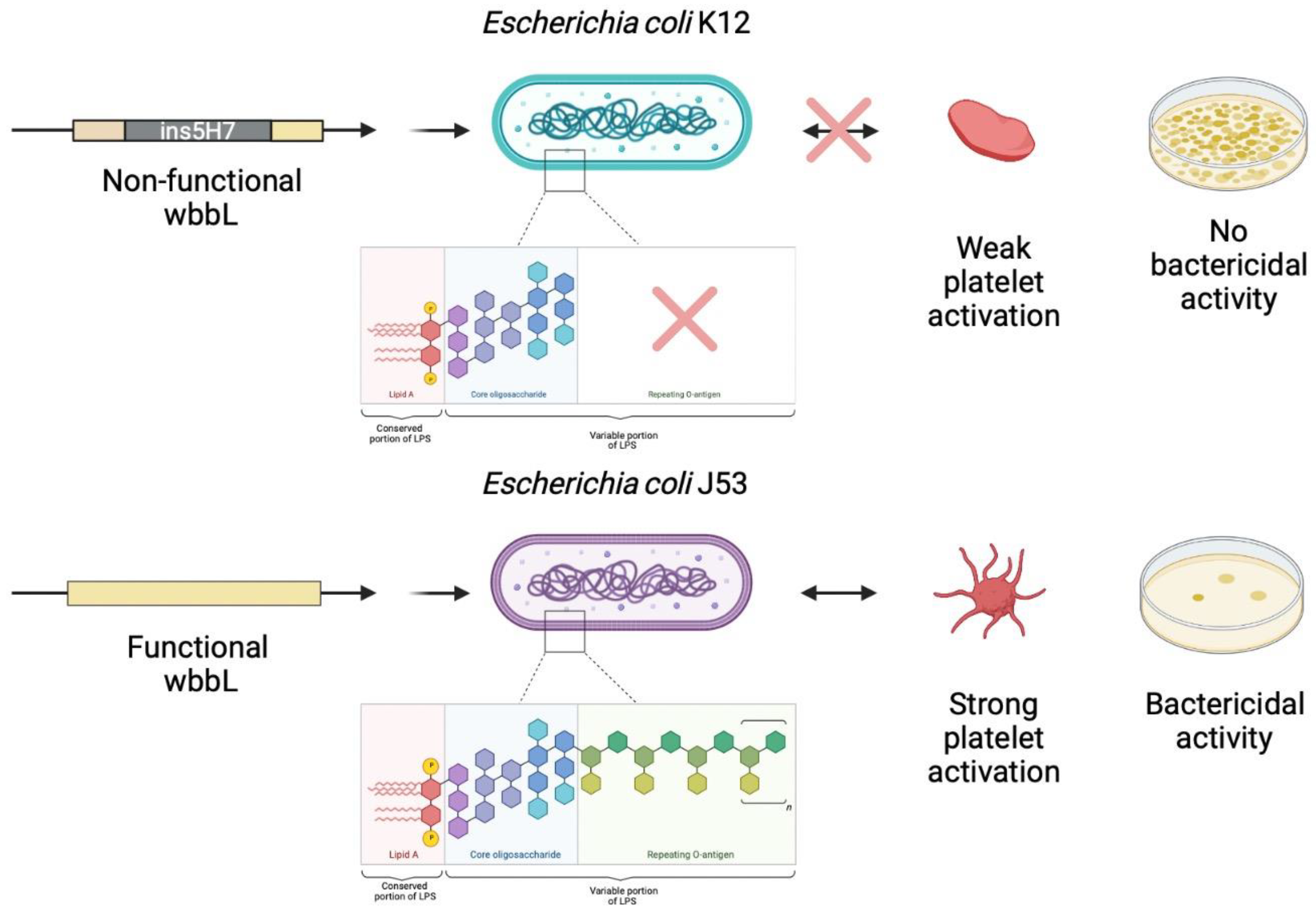
5. O-antigen Strain Serotyping
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nurden, A. Platelets, Inflammation and Tissue Regeneration. Thromb. Haemost. 2011, 105, S13–S33. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.R.; Storey, R.F. The Role of Platelets in Inflammation. Thromb. Haemost. 2015, 114, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Kerrigan, S.W.; Douglas, I.; Wray, A.; Heath, J.; Byrne, M.F.; Fitzgerald, D.; Cox, D. A Role for Glycoprotein Ib in Streptococcus Sanguis-Induced Platelet Aggregation. Blood 2002, 100, 509–516. [Google Scholar] [CrossRef] [PubMed]
- O’Seaghdha, M.; van Schooten, C.J.; Kerrigan, S.W.; Emsley, J.; Silverman, G.J.; Cox, D.; Lenting, P.J.; Foster, T.J. Staphylococcus Aureus Protein A Binding to von Willebrand Factor A1 Domain Is Mediated by Conserved IgG Binding Regions. FEBS J. 2006, 273, 4831–4841. [Google Scholar] [CrossRef]
- Cox, D.; Kerrigan, S.W.; Watson, S.P. Platelets and the Innate Immune System: Mechanisms of Bacterial-Induced Platelet Activation. J. Thromb. Haemost. JTH 2011, 9, 1097–1107. [Google Scholar] [CrossRef]
- Hamzeh-Cognasse, H.; Damien, P.; Chabert, A.; Pozzetto, B.; Cognasse, F.; Garraud, O. Platelets and Infections—Complex Interactions with Bacteria. Front. Immunol. 2015, 6, 82. [Google Scholar] [CrossRef] [Green Version]
- Kerrigan, S.W. Platelet Interactions with Bacteria. In The Non-Thrombotic Role of Platelets in Health and Disease; Kerrigan, S.W., Moran, N., Eds.; InTech: Dublin, Ireland, 2015. [Google Scholar]
- Rose, P.E.; Armour, J.A.; Williams, C.E.; Hill, F.G. Verotoxin and Neuraminidase Induced Platelet Aggregating Activity in Plasma: Their Possible Role in the Pathogenesis of the Haemolytic Uraemic Syndrome. J. Clin. Pathol. 1985, 38, 438–441. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.A.; Polanowska-Grabowska, R.K.; Fujii, J.; Obrig, T.; Gear, A.R.L. Shiga Toxin Binds to Activated Platelets. J. Thromb. Haemost. 2004, 2, 499–506. [Google Scholar] [CrossRef]
- Proulx, F.; Seidman, E.G.; Karpman, D. Pathogenesis of Shiga Toxin-Associated Hemolytic Uremic Syndrome. Pediatr. Res. 2001, 50, 163–171. [Google Scholar] [CrossRef] [Green Version]
- Flaumenhaft, R. Platelet Alpha-Granules: Basic Biology and Clinical Correlates. Blood Rev. 2009, 23, 177–189. [Google Scholar] [CrossRef] [Green Version]
- Portier, I.; Campbell, R.A. Role of Platelets in Detection and Regulation of Infection. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Hannachi, N.; Fournier, P.-E.; Martel, H.; Habib, G.; Camoin-Jau, L. Statins Potentiate the Antibacterial Effect of Platelets on Staphylococcus Aureus. Platelets 2021, 32, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, B.F.; Campbell, R.A.; Schwertz, H.; Cody, M.J.; Franks, Z.; Tolley, N.D.; Kahr, W.H.A.; Lindemann, S.; Seizer, P.; Yost, C.C.; et al. Novel Anti-Bacterial Activities of β-Defensin 1 in Human Platelets: Suppression of Pathogen Growth and Signaling of Neutrophil Extracellular Trap Formation. PLoS Pathog. 2011, 7, e1002355. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, M.R. Platelets in Defense against Bacterial Pathogens. Cell. Mol. Life Sci. CMLS 2010, 67, 525–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akashi, S.; Saitoh, S.; Wakabayashi, Y.; Kikuchi, T.; Takamura, N.; Nagai, Y.; Kusumoto, Y.; Fukase, K.; Kusumoto, S.; Adachi, Y.; et al. Lipopolysaccharide Interaction with Cell Surface Toll-like Receptor 4-MD-2: Higher Affinity than That with MD-2 or CD14. J. Exp. Med. 2003, 198, 1035–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, B.S.; Lee, J.-O. Recognition of Lipopolysaccharide Pattern by TLR4 Complexes. Exp. Mol. Med. 2013, 45, e66. [Google Scholar] [CrossRef] [Green Version]
- Vallance, T.M.; Zeuner, M.-T.; Williams, H.F.; Widera, D.; Vaiyapuri, S. Toll-Like Receptor 4 Signalling and Its Impact on Platelet Function, Thrombosis, and Haemostasis. Mediat. Inflamm. 2017, 2017, 9605894. [Google Scholar] [CrossRef] [Green Version]
- Moriarty, R.D.; Cox, A.; McCall, M.; Smith, S.G.J.; Cox, D. Escherichia Coli Induces Platelet Aggregation in an FcγRIIa-Dependent Manner. J. Thromb. Haemost. 2016, 14, 797–806. [Google Scholar] [CrossRef] [Green Version]
- Watson, C.N.; Kerrigan, S.W.; Cox, D.; Henderson, I.R.; Watson, S.P.; Arman, M. Human Platelet Activation by Escherichia Coli: Roles for FcγRIIA and Integrin AIIbβ3. Platelets 2016, 27, 535–540. [Google Scholar] [CrossRef] [Green Version]
- Matus, V.; Valenzuela, J.G.; Hidalgo, P.; Pozo, L.M.; Panes, O.; Wozniak, A.; Mezzano, D.; Pereira, J.; Sáez, C.G. Human Platelet Interaction with E. Coli O111 Promotes Tissue-Factor-Dependent Procoagulant Activity, Involving Toll like Receptor 4. PLoS ONE 2017, 12, e0185431. [Google Scholar] [CrossRef]
- Fejes, A.V.; Best, M.G.; van der Heijden, W.A.; Vancura, A.; Verschueren, H.; de Mast, Q.; Wurdinger, T.; Mannhalter, C. Impact of Escherichia Coli K12 and O18:K1 on Human Platelets: Differential Effects on Platelet Activation, RNAs and Proteins. Sci. Rep. 2018, 8, 16145. [Google Scholar] [CrossRef] [PubMed]
- Ståhl, A.; Svensson, M.; Mörgelin, M.; Svanborg, C.; Tarr, P.I.; Mooney, J.C.; Watkins, S.L.; Johnson, R.; Karpman, D. Lipopolysaccharide from Enterohemorrhagic Escherichia Coli Binds to Platelets through TLR4 and CD62 and Is Detected on Circulating Platelets in Patients with Hemolytic Uremic Syndrome. Blood 2006, 108, 167–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cattaneo, M.; Cerletti, C.; Harrison, P.; Hayward, C.P.M.; Kenny, D.; Nugent, D.; Nurden, P.; Rao, A.K.; Schmaier, A.H.; Watson, S.P.; et al. Recommendations for the Standardization of Light Transmission Aggregometry: A Consensus of the Working Party from the Platelet Physiology Subcommittee of SSC/ISTH. J. Thromb. Haemost. JTH 2013, 11, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Hadjadj, L.; Riziki, T.; Zhu, Y.; Li, J.; Diene, S.M.; Rolain, J.-M. Study of Mcr-1 Gene-Mediated Colistin Resistance in Enterobacteriaceae Isolated from Humans and Animals in Different Countries. Genes 2017, 8, 394. [Google Scholar] [CrossRef] [Green Version]
- Venter, P.; Lues, J.F.R. Extraction Methods for Lipopolysaccharides from Escherichia Coli ATCC 25922 for Quantitative Analysis by Capillary Electrophoresis. Int. J. Food Microbiol. 2003, 84, 245–250. [Google Scholar] [CrossRef]
- Pacífico, C.; Hilbert, M.; Sofka, D.; Dinhopl, N.; Pap, I.-J.; Aspöck, C.; Carriço, J.A.; Hilbert, F. Natural Occurrence of Escherichia Coli-Infecting Bacteriophages in Clinical Samples. Front. Microbiol. 2019, 10, 2484. [Google Scholar] [CrossRef]
- Serres, M.H.; Gopal, S.; Nahum, L.A.; Liang, P.; Gaasterland, T.; Riley, M. A Functional Update of the Escherichia Coli K-12 Genome. Genome Biol. 2001, 2, RESEARCH0035. [Google Scholar] [CrossRef]
- Yi, H.; Cho, Y.-J.; Yong, D.; Chun, J. Genome Sequence of Escherichia Coli J53, a Reference Strain for Genetic Studies. J. Bacteriol. 2012, 194, 3742–3743. [Google Scholar] [CrossRef] [Green Version]
- Jeong, H.; Kim, H.J.; Lee, S.J. Complete Genome Sequence of Escherichia Coli Strain BL21. Genome Announc. 2015, 3, e00134-15. [Google Scholar] [CrossRef] [Green Version]
- Hannachi, N.; Grac, L.; Baudoin, J.-P.; Fournier, P.-E.; Habib, G.; Camoin-Jau, L. Effect of Antiplatelet Agents on Platelet Antistaphylococcal Capacity: An in Vitro Study. Int. J. Antimicrob. Agents 2020, 55, 105890. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Bessonov, K.; Laing, C.; Robertson, J.; Yong, I.; Ziebell, K.; Gannon, V.P.J.; Nichani, A.; Arya, G.; Nash, J.H.E.; Christianson, S. ECTyper: In Silico Escherichia Coli Serotype and Species Prediction from Raw and Assembled Whole-Genome Sequence Data. Microb. Genom. 2021, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Tetzschner, A.M.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and Easy in Silico Serotyping of Escherichia Coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef] [Green Version]
- Palankar, R.; Kohler, T.P.; Krauel, K.; Wesche, J.; Hammerschmidt, S.; Greinacher, A. Platelets Kill Bacteria by Bridging Innate and Adaptive Immunity via Platelet Factor 4 and FcγRIIA. J. Thromb. Haemost. JTH 2018, 16, 1187–1197. [Google Scholar] [CrossRef] [Green Version]
- Tohidnezhad, M.; Varoga, D.; Wruck, C.J.; Podschun, R.; Sachweh, B.H.; Bornemann, J.; Bovi, M.; Sönmez, T.T.; Slowik, A.; Houben, A.; et al. Platelets Display Potent Antimicrobial Activity and Release Human Beta-Defensin 2. Platelets 2012, 23, 217–223. [Google Scholar] [CrossRef]
- Cieślik-Bielecka, A.; Bold, T.; Ziółkowski, G.; Pierchała, M.; Królikowska, A.; Reichert, P. Antibacterial Activity of Leukocyte- and Platelet-Rich Plasma: An in Vitro Study. BioMed Res. Int. 2018, 2018, 9471723. [Google Scholar] [CrossRef]
- Jiang, Z.; Georgel, P.; Du, X.; Shamel, L.; Sovath, S.; Mudd, S.; Huber, M.; Kalis, C.; Keck, S.; Galanos, C.; et al. CD14 Is Required for MyD88-Independent LPS Signaling. Nat. Immunol. 2005, 6, 565–570. [Google Scholar] [CrossRef]
- Huber, M.; Kalis, C.; Keck, S.; Jiang, Z.; Georgel, P.; Du, X.; Shamel, L.; Sovath, S.; Mudd, S.; Beutler, B.; et al. R-Form LPS, the Master Key to the Activation OfTLR4/MD-2-Positive Cells. Eur. J. Immunol. 2006, 36, 701–711. [Google Scholar] [CrossRef]
- Andonegui, G.; Kerfoot, S.M.; McNagny, K.; Ebbert, K.V.J.; Patel, K.D.; Kubes, P. Platelets Express Functional Toll-like Receptor-4. Blood 2005, 106, 2417–2423. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D.; Kubicek-Sutherland, J.Z. Mechanisms and Consequences of Bacterial Resistance to Antimicrobial Peptides. Drug Resist. Updates 2016, 26, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Diamond, G. The Role of Cationic Antimicrobial Peptides in Innate Host Defences. Trends Microbiol. 2000, 8, 402–410. [Google Scholar] [CrossRef]
- Jeong, H.; Barbe, V.; Lee, C.H.; Vallenet, D.; Yu, D.S.; Choi, S.-H.; Couloux, A.; Lee, S.-W.; Yoon, S.H.; Cattolico, L.; et al. Genome Sequences of Escherichia Coli B Strains REL606 and BL21(DE3). J. Mol. Biol. 2009, 394, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Bilge, S.S.; Vary, J.C.; Dowell, S.F.; Tarr, P.I. Role of the Escherichia Coli O157:H7 O Side Chain in Adherence and Analysis of an Rfb Locus. Infect. Immun. 1996, 64, 4795–4801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumura, Y.; Peirano, G.; Pitout, J.D.D. Complete Genome Sequence of Escherichia Coli J53, an Azide-Resistant Laboratory Strain Used for Conjugation Experiments. Genome Announc. 2018, 6, e00433-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niklaus, M.; Klingler, P.; Weber, K.; Koessler, A.; Boeck, M.; Kobsar, A.; Koessler, J. The Involvement of Toll-like Receptors 2 and 4 in Human Platelet Signalling Pathways. Cell. Signal. 2020, 76, 109817. [Google Scholar] [CrossRef]
- Baron, S.; Hadjadj, L.; Rolain, J.-M.; Olaitan, A.O. Molecular Mechanisms of Polymyxin Resistance: Knowns and Unknowns. Int. J. Antimicrob. Agents 2016, 48, 583–591. [Google Scholar] [CrossRef]


| Escherichia coli Strain | Origin | O-Antigen Type | Colistin Resistance Mechanism | MIC | References |
|---|---|---|---|---|---|
| IHU clinical isolates | |||||
| LH 1 | Human | O174 | mcr-1 gene | 7.8 mg/L | [25] |
| LH 30 | Human | O8 | mcr-1 gene | 3.9 mg/L | [25] |
| Q1066 | Human | O25 | Unknow mechanism | 7.8 mg/L | Unpublished |
| Q1065 | Human (Pharyngeal swab) | O9 | Unknow mechanism | 3.9 mg/L | Unpublished |
| Q6269 | Human (urine) | O175 | Unknow mechanism | 3.9 mg/L | Unpublished |
| Laboratory strains | |||||
| ATCC 25922 | Reference strain | O6 | - | 0.97 mg/L | [26] |
| ATCC 11303 | Reference strain | O7 | - | 0.48 mg/L | [27] |
| K12 | Human | - | - | 1.95 mg/L | [28] |
| J53 | Laboratory mutant of K12 | O16 | - | 0.97 mg/L | [29] |
| BL 21 DE3 | Laboratory mutant of K12 | O7 | - | 0.97 mg/L | [30] |
| Mean ± SD of MFI % of Platelets Stimulated with E. coli Strains and TRAP | |||
|---|---|---|---|
| E. coli Strains | Mean SD of Plt-E. coli | p-Value Plt-E. coli Compared to Plt | p-Value Summary of Plt-TRAP Compared to Plt |
| ATCC11303 | 124.9 ± 14.3 | 0.007 | ** |
| J53 | 190.7 ± 40.5 | 0.017 | ** |
| BL21DE3 | 134.5 ± 22.5 | 0.026 | ** |
| K12 | 106.3 ± 4 | 0.024 | ** |
| ATCC25922 | 109.1 ± 5.4 | 0.019 | ** |
| LH1 | 104.2 ± 17.9 | - | ** |
| LH30 | 96.5 ± 7.4 | - | ** |
| Q1065 | 99.4 ± 9.4 | - | * |
| Q1066 | 102.3 ± 7.8 | - | * |
| Q6269 | 100.4 ± 9 | - | * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ezzeroug Ezzraimi, A.; Hannachi, N.; Mariotti, A.; Rolland, C.; Levasseur, A.; Baron, S.A.; Rolain, J.-M.; Camoin-Jau, L. The Antibacterial Effect of Platelets on Escherichia coli Strains. Biomedicines 2022, 10, 1533. https://doi.org/10.3390/biomedicines10071533
Ezzeroug Ezzraimi A, Hannachi N, Mariotti A, Rolland C, Levasseur A, Baron SA, Rolain J-M, Camoin-Jau L. The Antibacterial Effect of Platelets on Escherichia coli Strains. Biomedicines. 2022; 10(7):1533. https://doi.org/10.3390/biomedicines10071533
Chicago/Turabian StyleEzzeroug Ezzraimi, Amina, Nadji Hannachi, Antoine Mariotti, Clara Rolland, Anthony Levasseur, Sophie Alexandra Baron, Jean-Marc Rolain, and Laurence Camoin-Jau. 2022. "The Antibacterial Effect of Platelets on Escherichia coli Strains" Biomedicines 10, no. 7: 1533. https://doi.org/10.3390/biomedicines10071533
APA StyleEzzeroug Ezzraimi, A., Hannachi, N., Mariotti, A., Rolland, C., Levasseur, A., Baron, S. A., Rolain, J.-M., & Camoin-Jau, L. (2022). The Antibacterial Effect of Platelets on Escherichia coli Strains. Biomedicines, 10(7), 1533. https://doi.org/10.3390/biomedicines10071533







