Murine Falcor/LL35 lncRNA Contributes to Glucose and Lipid Metabolism In Vitro and In Vivo
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Transfection
2.2. Antisense Oligonucleotides (ASOs) Description
2.3. Animal Care and Treatments
2.4. Insulin Tolerance Test
2.5. Histological Analysis
2.6. RNA Isolation, cDNA Synthesis and RT-qPCR
2.7. Western Blotting
2.8. Cell Viability Assay
2.9. Wound Healing and Migration Assay
2.10. Cell Cycle Analysis with Flow Cytometry
2.11. RNA-Seq Data Processing and Analysis
2.12. LC-MS-Based Proteomics
2.13. Seahorse Extracellular Flux Analysis
2.14. Mitochondria Staining with TMRE and Mito Green FM
2.15. Lipid Staining with Oil Red O for AML12
2.16. Cell Lines and Liver Tissue Extraction Protocol and Liquid Chromatography/Mass Spectrometry Analysis for Lipids and Metabolites Analysis
2.17. Post-Acquisition Processing and Statistical Analysis of Lipidome and Metabolome Data
2.18. Statistical Analysis
3. Results
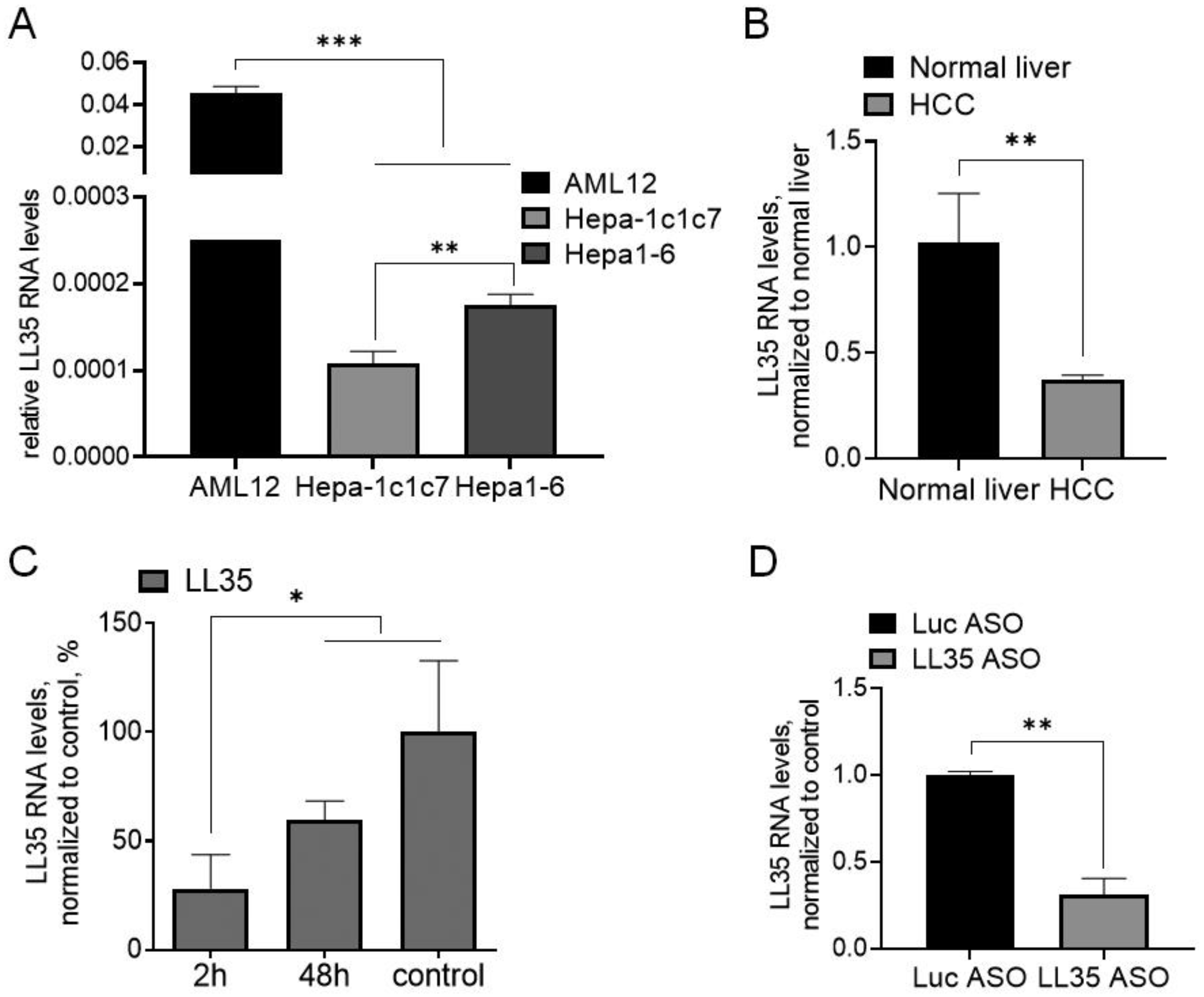
3.1. LL35 Expression in Murine Hepatocytes and Liver
3.2. LL35 Downregulation In Vitro and In Vivo
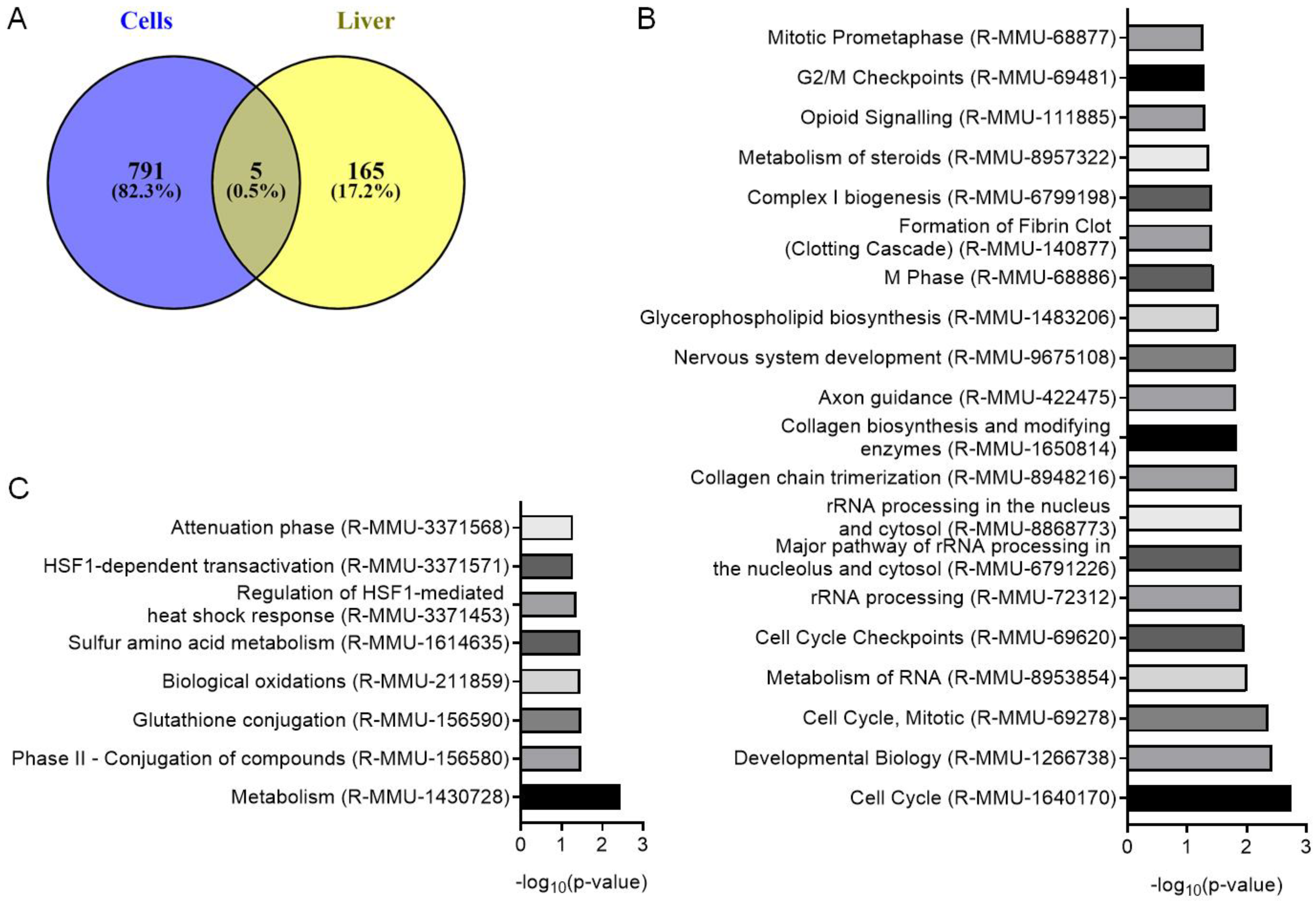
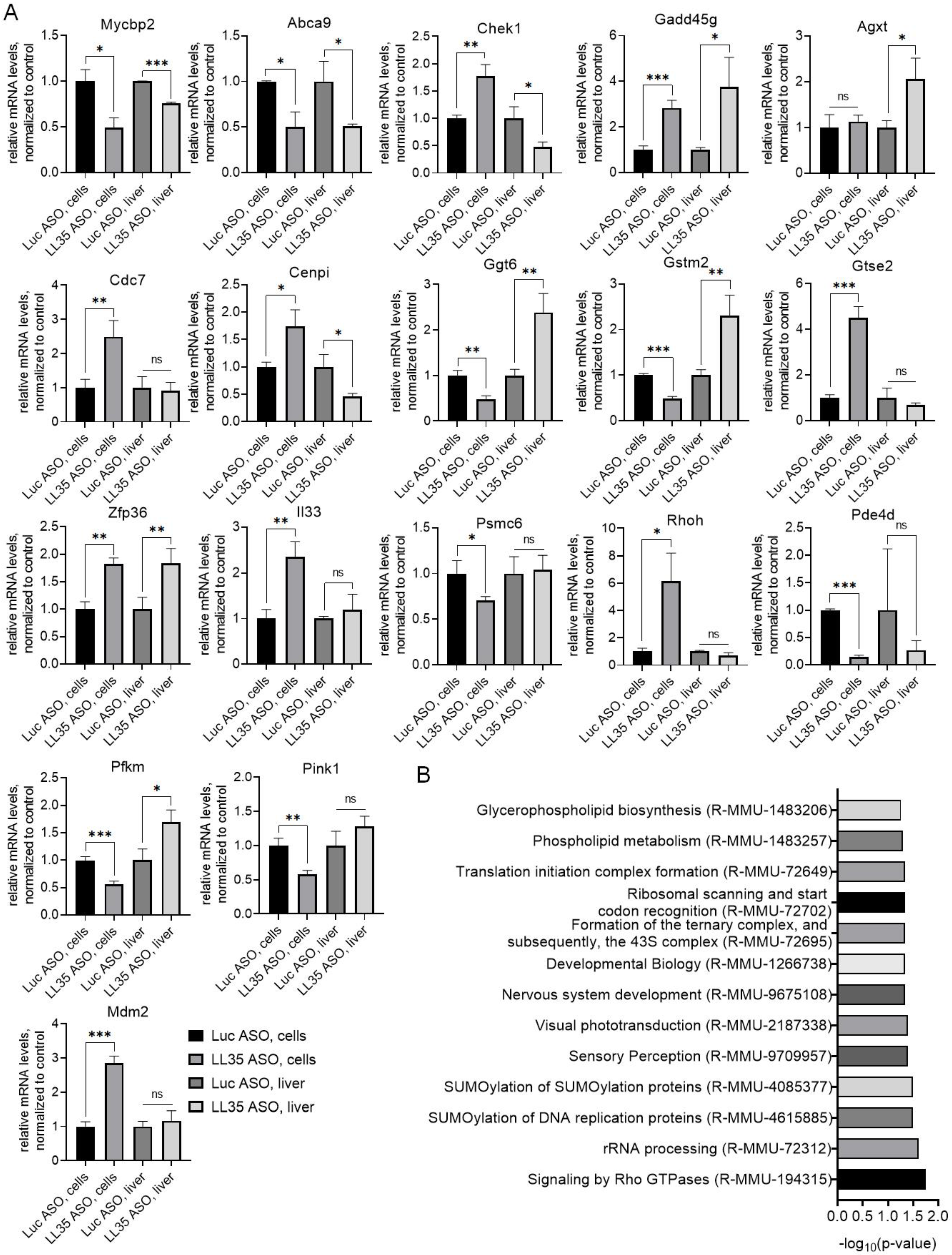
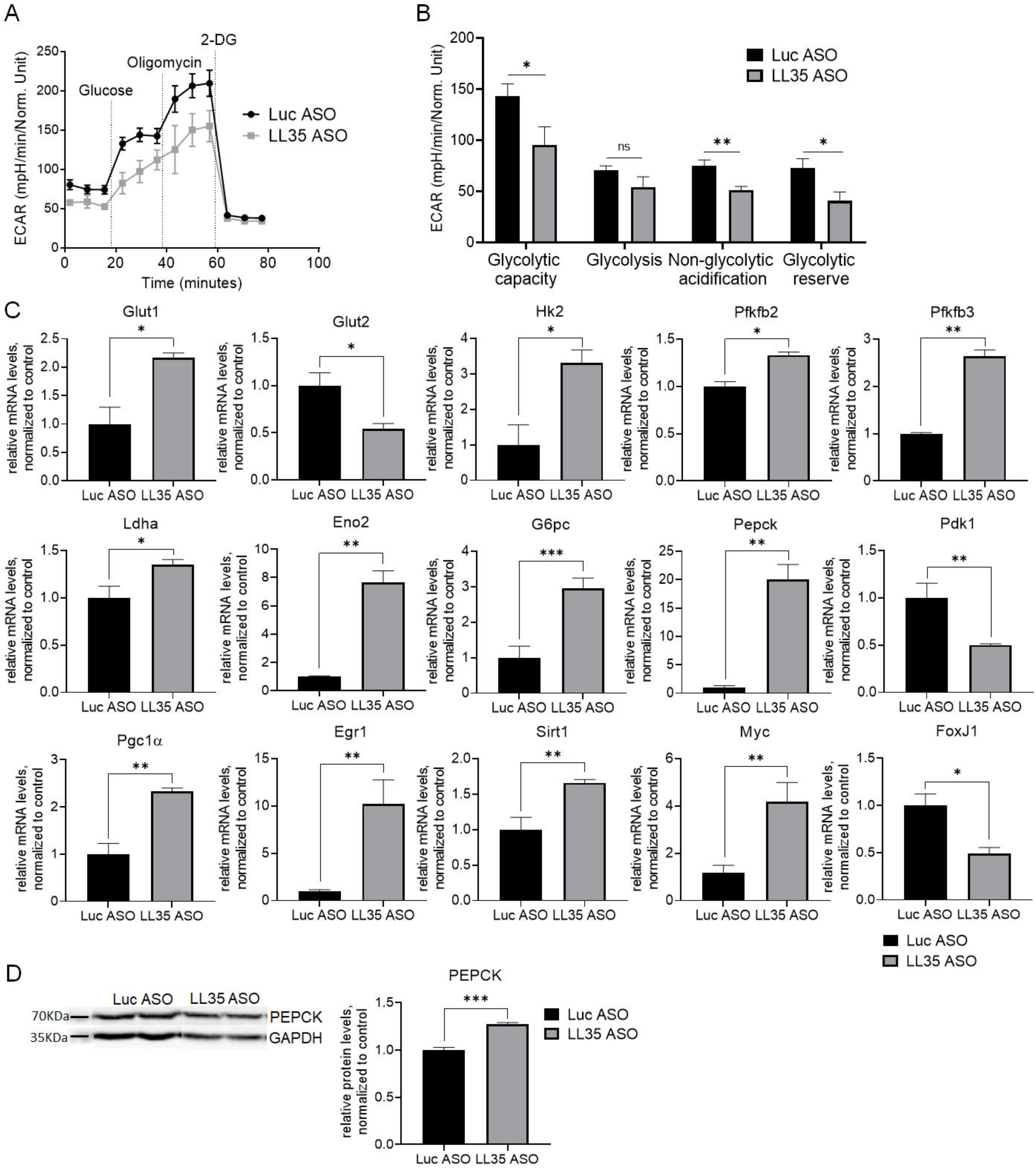
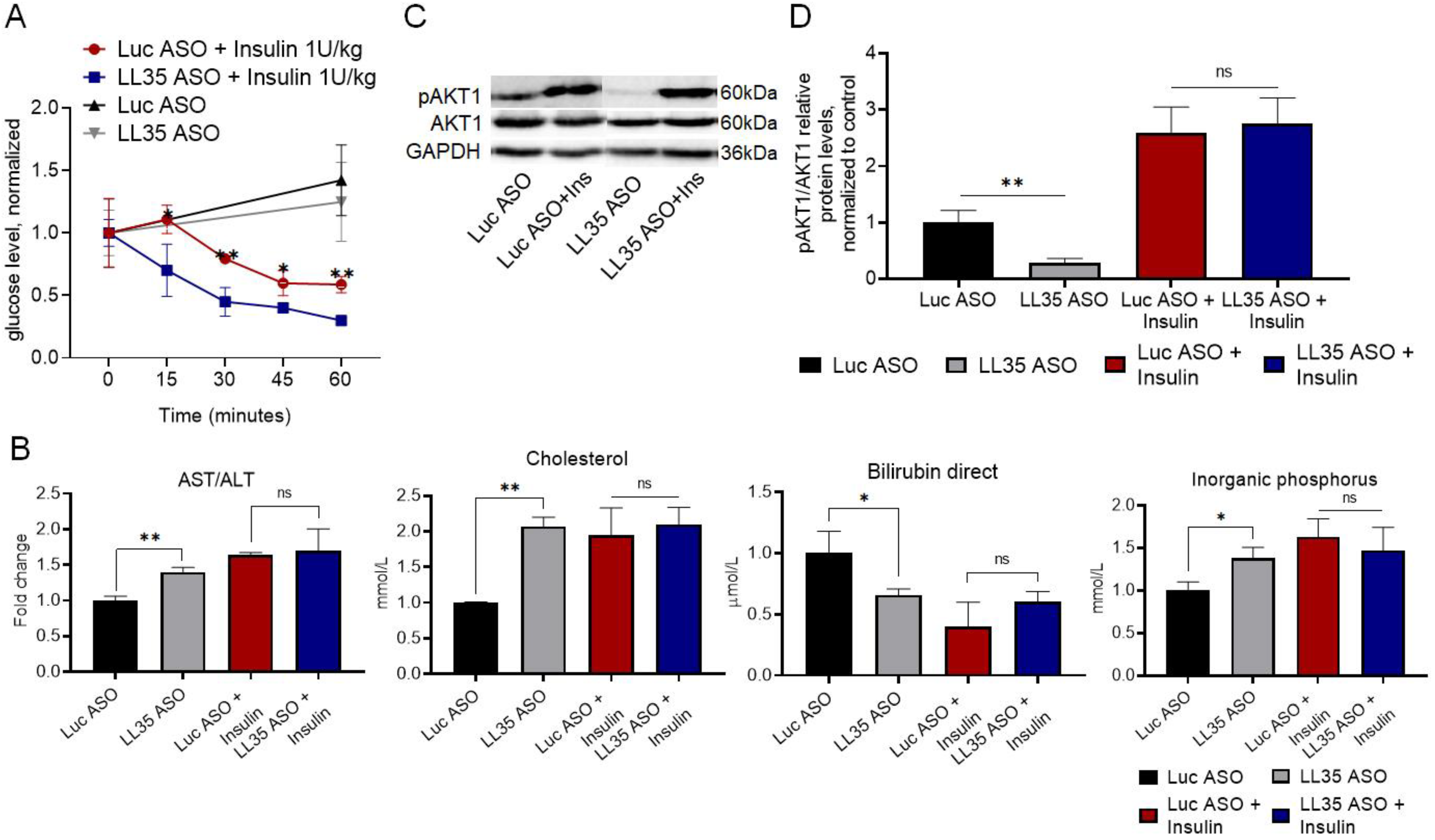
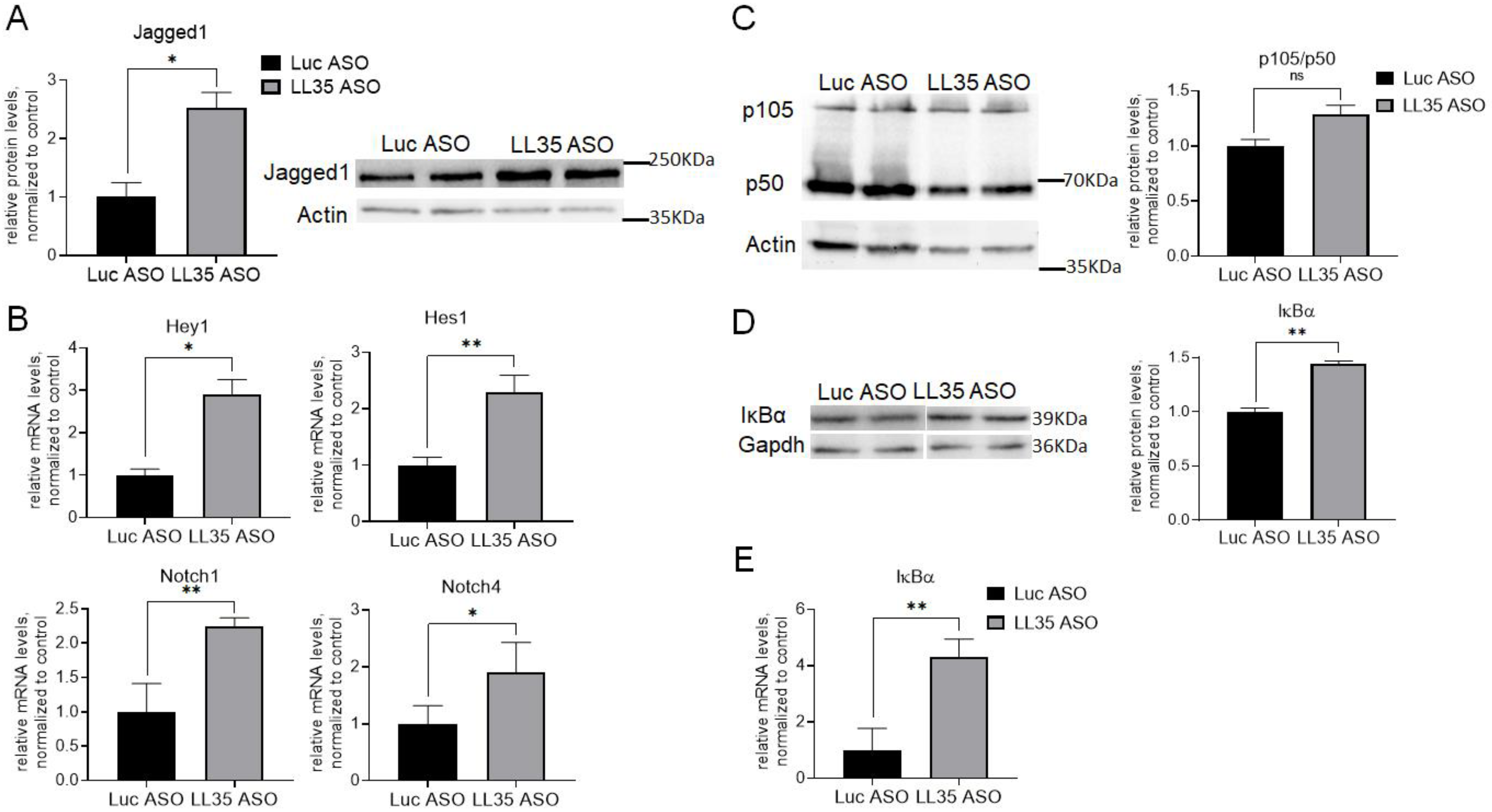
3.3. Analysis of Gene and Protein Expression after LL35 Knockdown In Vitro and In Vivo
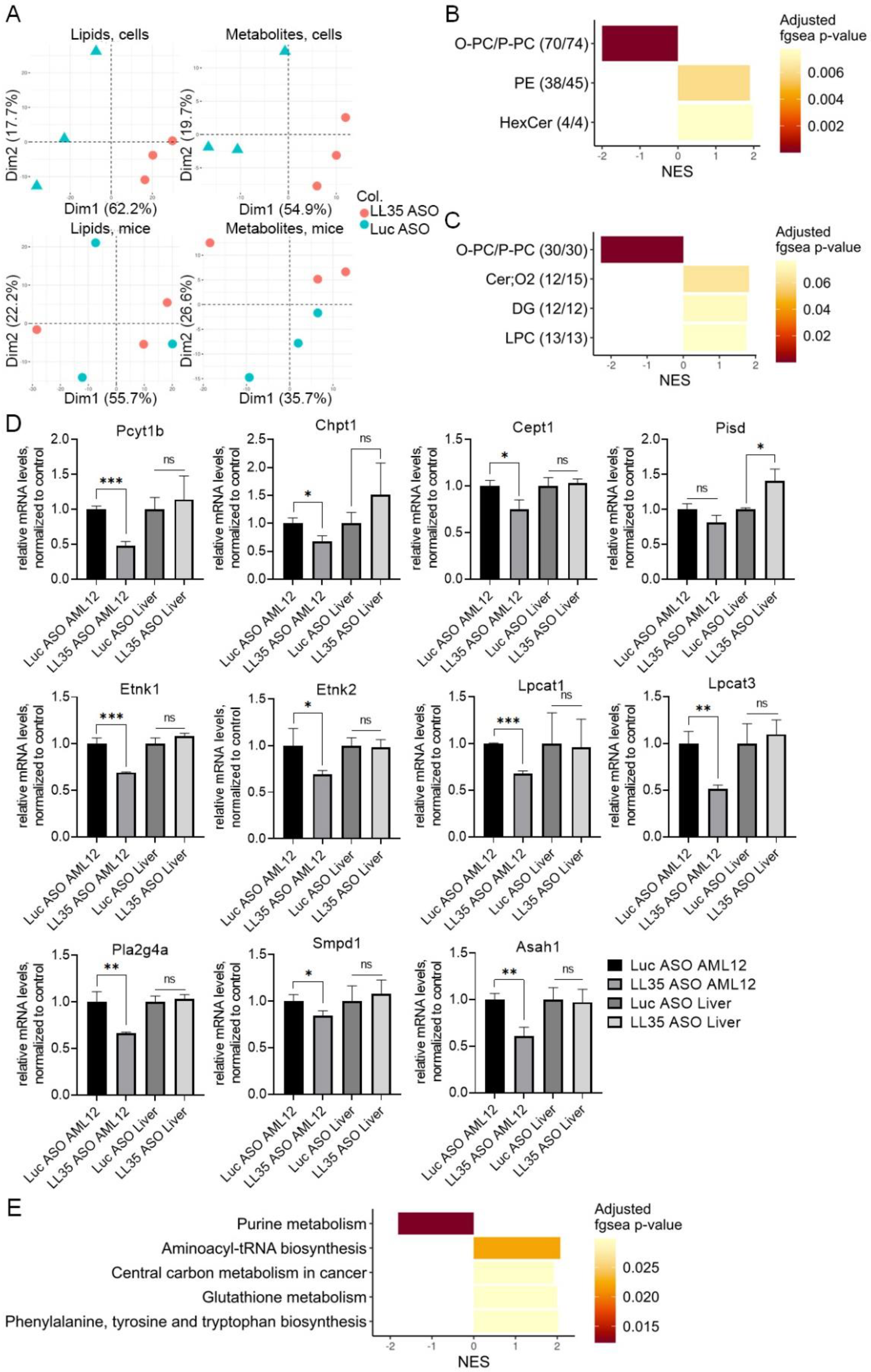
3.4. LL35 Downregulation Causes Changes in Lipidome and Metabolome Both in AML12 Cells and in Murine Liver
3.5. Inhibition of LL35 lncRNA Interferes with Glucose Metabolism In Vitro and In Vivo
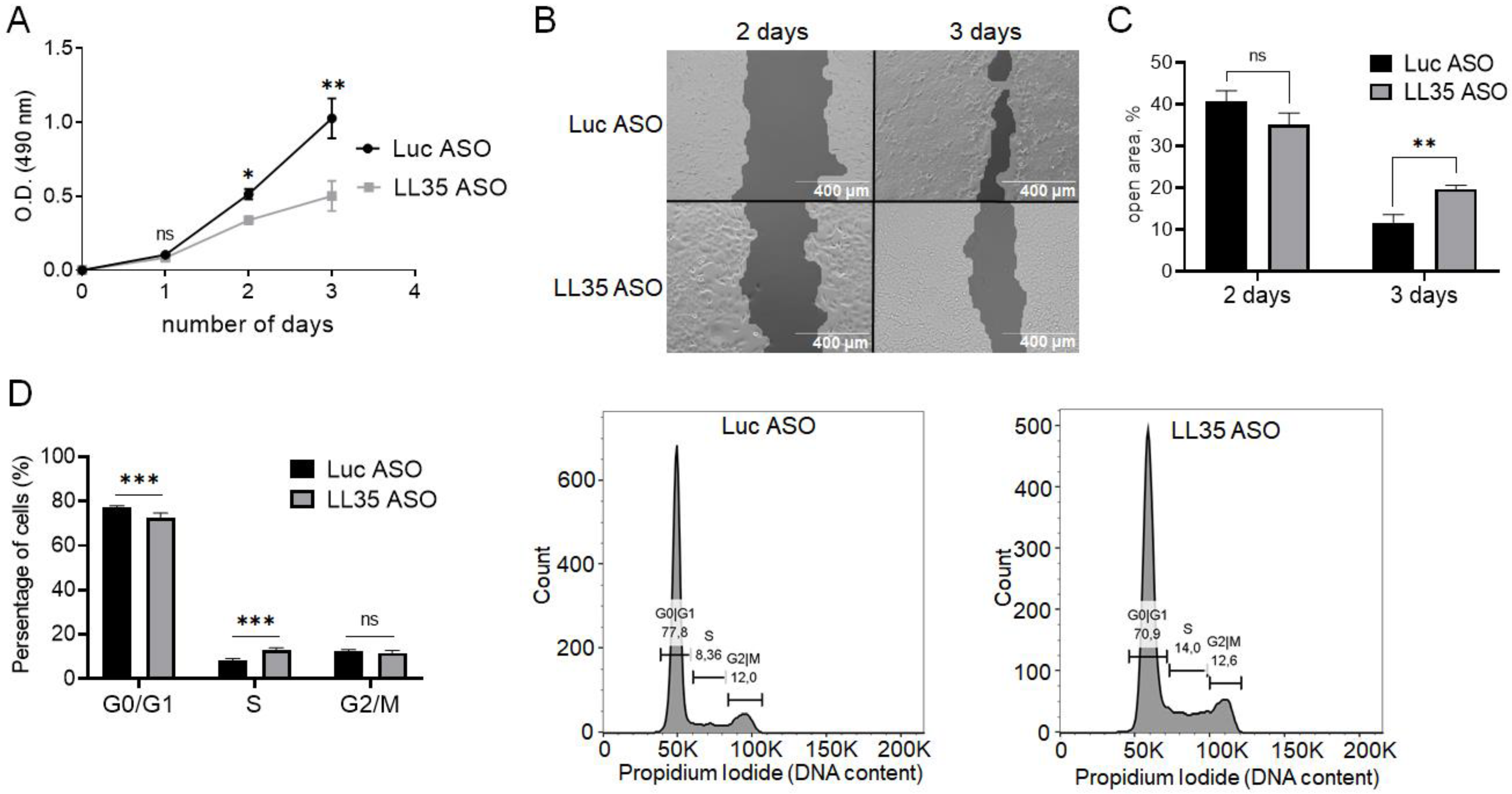
3.6. Depletion of LL35 lncRNA Decreases Cell Survival and Proliferation In Vitro
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.M.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of Transcription in Human Cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.B.; Uchida, S. Functional Characterization of Long Non-Coding RNAs. Curr. Opin. Cardiol. 2020, 35, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.A.; Ahmad, S.M.; Mumtaz, P.T.; Malik, A.A.; Dar, M.A.; Urwat, U.; Shah, R.A.; Ganai, N.A. Long Non-Coding RNAs: Mechanism of Action and Functional Utility. Non-Coding RNA Res. 2016, 1, 43–50. [Google Scholar] [CrossRef]
- Huang, B.; Zhang, R. Regulatory Non-Coding RNAs: Revolutionizing the RNA World. Mol. Biol. Rep. 2014, 41, 3915–3923. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-H. Crosstalk of LncRNA and Cellular Metabolism and Their Regulatory Mechanism in Cancer. Int. J. Mol. Sci. 2020, 21, 2947. [Google Scholar] [CrossRef]
- Okazaki, Y.; Furuno, M.; Kasukawa, T.; Adachi, J.; Bono, H.; Kondo, S.; Nikaido, I.; Osato, N.; Saito, R.; Suzuki, H.; et al. Analysis of the Mouse Transcriptome Based on Functional Annotation of 60,770 Full-Length CDNAs. Nature 2002, 420, 563–573. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin Signature Reveals over a Thousand Highly Conserved Large Non-Coding RNAs in Mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Tupy, J.L.; Bailey, A.M.; Dailey, G.; Evans-Holm, M.; Siebel, C.W.; Misra, S.; Celniker, S.E.; Rubin, G.M. Identification of Putative Noncoding Polyadenylated Transcripts in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2005, 102, 5495–5500. [Google Scholar] [CrossRef]
- Nam, J.-W.; Bartel, D.P. Long Noncoding RNAs in C. elegans. Genome Res. 2012, 22, 2529–2540. [Google Scholar] [CrossRef]
- Pauli, A.; Valen, E.; Lin, M.F.; Garber, M.; Vastenhouw, N.L.; Levin, J.Z.; Fan, L.; Sandelin, A.; Rinn, J.L.; Regev, A.; et al. Systematic Identification of Long Noncoding RNAs Expressed during Zebrafish Embryogenesis. Genome Res. 2012, 22, 577–591. [Google Scholar] [CrossRef]
- Hezroni, H.; Koppstein, D.; Schwartz, M.G.; Avrutin, A.; Bartel, D.P.; Ulitsky, I. Principles of Long Noncoding RNA Evolution Derived from Direct Comparison of Transcriptomes in 17 Species. Cell Rep. 2015, 11, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Feyder, M.; Goff, L.A. Investigating Long Noncoding RNAs Using Animal Models. J. Clin. Investig. 2016, 126, 2783–2791. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhai, S.; Xu, Z.; Xie, J.; Zhang, J.; Wang, X.; Peng, C.; Li, H.; Chen, H.; Shen, B.; Deng, X. Epigenetic Silencing of LncRNA LINC00261 Promotes C-Myc-Mediated Aerobic Glycolysis by Regulating MiR-222-3p/HIPK2/ERK Axis and Sequestering IGF2BP1. Oncogene 2021, 40, 277–291. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Sang, L.; Du, S. Long Noncoding RNA LINC00261 Regulates Endometrial Carcinoma Progression by Modulating MiRNA/FOXO1 Expression. Cell Biochem. Funct. 2018, 36, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Li, Y.; Zang, X.; Jiang, Z.; Ning, H.; Li, J. Effect of TLR2 on the Proliferation of Inflammation-Related Colorectal Cancer and Sporadic Colorectal Cancer. Cancer Cell Int. 2020, 20, 95. [Google Scholar] [CrossRef]
- Lin, K.; Jiang, H.; Zhuang, S.-S.; Qin, Y.-S.; Qiu, G.-D.; She, Y.-Q.; Zheng, J.-T.; Chen, C.; Fang, L.; Zhang, S.-Y. Long Noncoding RNA LINC00261 Induces Chemosensitization to 5-Fluorouracil by Mediating Methylation-Dependent Repression of DPYD in Human Esophageal Cancer. FASEB J. 2019, 33, 1972–1988. [Google Scholar] [CrossRef]
- Zhang, M.; Gao, F.; Yu, X.; Zhang, Q.; Sun, Z.; He, Y.; Guo, W. LINC00261: A Burgeoning Long Noncoding RNA Related to Cancer. Cancer Cell Int. 2021, 21, 274. [Google Scholar] [CrossRef]
- Shi, J.; Ma, H.; Wang, H.; Zhu, W.; Jiang, S.; Dou, R.; Yan, B. Overexpression of LINC00261 Inhibits Non-Small Cell Lung Cancer Cells Progression by Interacting with MiR-522-3p and Suppressing Wnt Signaling. J. Cell. Biochem. 2019, 120, 18378–18387. [Google Scholar] [CrossRef]
- Gao, J.; Qin, W.; Kang, P.; Xu, Y.; Leng, K.; Li, Z.; Huang, L.; Cui, Y.; Zhong, X. Up-Regulated LINC00261 Predicts a Poor Prognosis and Promotes a Metastasis by EMT Process in Cholangiocarcinoma. Pathol. Res. Pract. 2020, 216, 152733. [Google Scholar] [CrossRef]
- Guo, G.; Dai, S.; Chen, Q. Long Noncoding RNA LINC00261 Reduces Proliferation and Migration of Breast Cancer Cells via the NME1-EMT Pathway. Cancer Manag. Res. 2020, 12, 3081–3089. [Google Scholar] [CrossRef]
- Wang, Z.K.; Yang, L.; Wu, L.L.; Mao, H.; Zhou, Y.H.; Zhang, P.F.; Dai, G.H. Long Non-Coding RNA LINC00261 Sensitizes Human Colon Cancer Cells to Cisplatin Therapy. Braz. J. Med. Biol. Res. 2017, 51, e6793. [Google Scholar] [CrossRef] [PubMed]
- Sergeeva, O.V.; Korinfskaya, S.A.; Kurochkin, I.I.; Zatsepin, T.S. Long Noncoding RNA LL35/Falcor Regulates Expression of Transcription Factor Foxa2 in Hepatocytes in Normal and Fibrotic Mouse Liver. Acta Nat. 2019, 11, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Swarr, D.T.; Herriges, M.; Li, S.; Morley, M.; Fernandes, S.; Sridharan, A.; Zhou, S.; Garcia, B.A.; Stewart, K.; Morrisey, E.E. The Long Noncoding RNA Falcor Regulates Foxa2 Expression to Maintain Lung Epithelial Homeostasis and Promote Regeneration. Genes Dev. 2019, 33, 656–668. [Google Scholar] [CrossRef]
- Ulashchik, E.A.; Martynenko-Makaev, Y.V.; Akhlamionok, T.P.; Melnik, D.M.; Shmanai, V.V.; Zatsepin, T.S. Synthesis of GalNAc-Oligonucleotide Conjugates Using GalNAc Phosphoramidite and Triple-GalNAc CPG Solid Support. In Design and Delivery of SiRNA Therapeutics; Ditzel, H.J., Tuttolomondo, M., Kauppinen, S., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2021; pp. 101–118. ISBN 978-1-07-161298-9. [Google Scholar]
- Bogorad, R.L.; Yin, H.; Zeigerer, A.; Nonaka, H.; Ruda, V.M.; Zerial, M.; Anderson, D.G.; Koteliansky, V. Nanoparticle-Formulated SiRNA Targeting Integrins Inhibits Hepatocellular Carcinoma Progression in Mice. Nat. Commun. 2014, 5, 3869. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, C.; Willenbring, H. A Reproducible and Well-Tolerated Method for 2/3 Partial Hepatectomy in Mice. Nat. Protoc. 2008, 3, 1167–1170. [Google Scholar] [CrossRef]
- Ferreira, T.; Rasband, W. ImageJ User Guide. 1998. Available online: http://imagej.nih.gov/ij/docs/guide (accessed on 15 July 2021).
- PANTHER. Available online: http://www.pantherdb.org/ (accessed on 11 May 2022).
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, Encapsulated Proteomic-Sample Processing Applied to Copy-Number Estimation in Eukaryotic Cells. Nat. Methods 2014, 11, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Kovalchuk, S.I.; Jensen, O.N.; Rogowska-Wrzesinska, A. FlashPack: Fast and Simple Preparation of Ultrahigh-Performance Capillary Columns for LC-MS*. Mol. Cell. Proteom. 2019, 18, 383–390. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant Computational Platform for Mass Spectrometry-Based Shotgun Proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus Computational Platform for Comprehensive Analysis of (Prote)Omics Data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Goracci, L.; Tortorella, S.; Tiberi, P.; Pellegrino, R.M.; Di Veroli, A.; Valeri, A.; Cruciani, G. Lipostar, a Comprehensive Platform-Neutral Cheminformatics Tool for Lipidomics. Anal. Chem. 2017, 89, 6257–6264. [Google Scholar] [CrossRef]
- Korotkevich, G.; Sukhov, V.; Sergushichev, A. Fast Gene Set Enrichment Analysis. bioRxiv 2019. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Frolkis, A.; Knox, C.; Lim, E.; Jewison, T.; Law, V.; Hau, D.D.; Liu, P.; Gautam, B.; Ly, S.; Guo, A.C.; et al. SMPDB: The Small Molecule Pathway Database. Nucleic Acids Res. 2010, 38, D480–D487. [Google Scholar] [CrossRef]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A Web Server for Metabolomic Data Analysis and Interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Hastie, T.; Tibshirani, R. Sparse Principal Component Analysis. J. Comput. Graph. Stat. 2006, 15, 265–286. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. MixOmics: An R Package for ’omics Feature Selection and Multiple Data Integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: https://bioinfogp.cnb.csic.es/tools/venny/ (accessed on 11 May 2022).
- Bolha, L.; Ravnik-Glavač, M.; Glavač, D. Long Noncoding RNAs as Biomarkers in Cancer. Dis. Markers 2017, 2017, 7243968. [Google Scholar] [CrossRef]
- Sergeeva, O.; Sviridov, E.; Zatsepin, T. Noncoding RNA in Liver Regeneration—From Molecular Mechanisms to Clinical Implications. Semin. Liver Dis. 2020, 40, 70–83. [Google Scholar] [CrossRef]
- Fausto, N.; Campbell, J.S. The Role of Hepatocytes and Oval Cells in Liver Regeneration and Repopulation. Mech. Dev. 2003, 120, 117–130. [Google Scholar] [CrossRef]
- Mehlem, A.; Hagberg, C.E.; Muhl, L.; Eriksson, U.; Falkevall, A. Imaging of Neutral Lipids by Oil Red O for Analyzing the Metabolic Status in Health and Disease. Nat. Protoc. 2013, 8, 1149–1154. [Google Scholar] [CrossRef]
- Chen, L.; Chen, X.-W.; Huang, X.; Song, B.-L.; Wang, Y.; Wang, Y. Regulation of Glucose and Lipid Metabolism in Health and Disease. Sci. China Life Sci. 2019, 62, 1420–1458. [Google Scholar] [CrossRef] [PubMed]
- Norton, L.; Shannon, C.; Gastaldelli, A.; DeFronzo, R.A. Insulin: The Master Regulator of Glucose Metabolism. Metabolism 2022, 129, 155142. [Google Scholar] [CrossRef]
- Gallagher, E.J.; LeRoith, D. The Proliferating Role of Insulin and Insulin-like Growth Factors in Cancer. Trends Endocrinol. Metab. 2010, 21, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Ge, D.; Dauchy, R.T.; Liu, S.; Zhang, Q.; Mao, L.; Dauchy, E.M.; Blask, D.E.; Hill, S.M.; Rowan, B.G.; Brainard, G.C.; et al. Insulin and IGF1 Enhance IL-17-Induced Chemokine Expression through a GSK3B-Dependent Mechanism: A New Target for Melatonin’s Anti-Inflammatory Action. J. Pineal Res. 2013, 55, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Manokawinchoke, J.; Nattasit, P.; Thongngam, T.; Pavasant, P.; Tompkins, K.A.; Egusa, H.; Osathanon, T. Indirect Immobilized Jagged1 Suppresses Cell Cycle Progression and Induces Odonto/Osteogenic Differentiation in Human Dental Pulp Cells. Sci. Rep. 2017, 7, 10124. [Google Scholar] [CrossRef]
- Pancewicz, J.; Taylor, J.M.; Datta, A.; Baydoun, H.H.; Waldmann, T.A.; Hermine, O.; Nicot, C. Notch Signaling Contributes to Proliferation and Tumor Formation of Human T-Cell Leukemia Virus Type 1–Associated Adult T-Cell Leukemia. Proc. Natl. Acad. Sci. USA 2010, 107, 16619–16624. [Google Scholar] [CrossRef]
- Luedde, T.; Schwabe, R.F. NF-ΚB in the Liver—Linking Injury, Fibrosis and Hepatocellular Carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, O.; Chang, H.Y. Long Noncoding RNAs and Human Disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef]
- Du, Z.; Fei, T.; Verhaak, R.G.W.; Su, Z.; Zhang, Y.; Brown, M.; Chen, Y.; Liu, X.S. Integrative Genomic Analyses Reveal Clinically Relevant Long Noncoding RNAs in Human Cancer. Nat. Struct. Mol. Biol. 2013, 20, 908–913. [Google Scholar] [CrossRef]
- Chen, L.-L. Linking Long Noncoding RNA Localization and Function. Trends Biochem. Sci. 2016, 41, 761–772. [Google Scholar] [CrossRef]
- Zhang, H.-F.; Li, W.; Han, Y.-D. LINC00261 Suppresses Cell Proliferation, Invasion and Notch Signaling Pathway in Hepatocellular Carcinoma. Cancer Biomark. 2018, 21, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Sui, J.; Miao, Y.; Han, J.; Nan, H.; Shen, B.; Zhang, X.; Zhang, Y.; Wu, Y.; Wu, W.; Liu, T.; et al. Systematic Analyses of a Novel LncRNA-Associated Signature as the Prognostic Biomarker for Hepatocellular Carcinoma. Cancer Med. 2018, 7, 3240–3256. [Google Scholar] [CrossRef]
- Niu, J.-Z.; Liang, X.-C.; Xu, Z.-W.; Li, Z.-H.; Li, J.; Meng, Y.; Sun, Z.-W. Long non-coding RNA Linc00261 as a novel potential diagnostic and prognostic biomarker for gallbladder cancer. Transl. Cancer Res. 2020, 9, 6078–6085. [Google Scholar] [CrossRef] [PubMed]
- Debacker, A.J.; Voutila, J.; Catley, M.; Blakey, D.; Habib, N. Delivery of Oligonucleotides to the Liver with GalNAc: From Research to Registered Therapeutic Drug. Mol. Ther. 2020, 28, 1759–1771. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Liu, Y.; Liu, R.; Zhang, K.; Zhang, Y. The LncRNA DEANR1 Facilitates Human Endoderm Differentiation by Activating FOXA2 Expression. Cell Rep. 2015, 11, 137–148. [Google Scholar] [CrossRef]
- Shahabi, S.; Kumaran, V.; Castillo, J.; Cong, Z.; Nandagopal, G.; Mullen, D.J.; Alvarado, A.; Correa, M.R.; Saizan, A.; Goel, R.; et al. LINC00261 Is an Epigenetically Regulated Tumor Suppressor Essential for Activation of the DNA Damage Response. Cancer Res. 2019, 79, 3050–3062. [Google Scholar] [CrossRef]
- Zhang, T.-N.; Wang, W.; Yang, N.; Huang, X.-M.; Liu, C.-F. Regulation of Glucose and Lipid Metabolism by Long Non-Coding RNAs: Facts and Research Progress. Front. Endocrinol. 2020, 11, 457. [Google Scholar] [CrossRef]
- Liu, C.; Yang, Z.; Wu, J.; Zhang, L.; Lee, S.; Shin, D.-J.; Tran, M.; Wang, L. Long Noncoding RNA H19 Interacts with Polypyrimidine Tract-Binding Protein 1 to Reprogram Hepatic Lipid Homeostasis. Hepatology 2018, 67, 1768–1783. [Google Scholar] [CrossRef]
- Meng, X.-D.; Yao, H.-H.; Wang, L.-M.; Yu, M.; Shi, S.; Yuan, Z.-X.; Liu, J. Knockdown of GAS5 Inhibits Atherosclerosis Progression via Reducing EZH2-Mediated ABCA1 Transcription in ApoE−/− Mice. Mol. Ther. Nucleic Acids 2020, 19, 84–96. [Google Scholar] [CrossRef]
- Li, P.; Ruan, X.; Yang, L.; Kiesewetter, K.; Zhao, Y.; Luo, H.; Chen, Y.; Gucek, M.; Zhu, J.; Cao, H. A Liver-Enriched Long Non-Coding RNA, LncLSTR, Regulates Systemic Lipid Metabolism in Mice. Cell Metab. 2015, 21, 455–467. [Google Scholar] [CrossRef]
- Parhofer, K.G. Interaction between Glucose and Lipid Metabolism: More than Diabetic Dyslipidemia. Diabetes Metab. J. 2015, 39, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, H.; Wei, X. Long Noncoding RNA LINC00261 Suppresses Prostate Cancer Tumorigenesis through Upregulation of GATA6-Mediated DKK3. Cancer Cell Int. 2020, 20, 474. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xue, K.; Guan, Y.; Jin, Y.; Liu, S.; Wang, Y.; Liu, S.; Wang, L.; Han, L. Long Noncoding RNA LINC00261 Suppresses Cell Proliferation and Invasion and Promotes Cell Apoptosis in Human Choriocarcinoma. Oncol. Res. 2017, 25, 733–742. [Google Scholar] [CrossRef]
- Chen, Z.; Xiang, L.; Huang, Y.; Fang, Y.; Li, X.; Yang, D. Expression of long noncoding RNA linc00261 in hepatocellular carcinoma and its association with postoperative outcomes. Nan Fang Yi Ke Da Xue Xue Bao 2018, 38, 1179–1186. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.-Y.; Chuang, W.-L. Genes Responsible for the Characteristics of Primary Cultured Invasive Phenotype Hepatocellular Carcinoma Cells. Biomed. Pharmacother. 2012, 66, 454–458. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE Database Resources in 2022: A Hub for Mass Spectrometry-Based Proteomics Evidences. Nucleic Acids Res. 2021, 50, D543–D552. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shcherbinina, E.; Abakumova, T.; Bobrovskiy, D.; Kurochkin, I.; Deinichenko, K.; Stekolshchikova, E.; Anikanov, N.; Ziganshin, R.; Melnikov, P.; Khrameeva, E.; et al. Murine Falcor/LL35 lncRNA Contributes to Glucose and Lipid Metabolism In Vitro and In Vivo. Biomedicines 2022, 10, 1397. https://doi.org/10.3390/biomedicines10061397
Shcherbinina E, Abakumova T, Bobrovskiy D, Kurochkin I, Deinichenko K, Stekolshchikova E, Anikanov N, Ziganshin R, Melnikov P, Khrameeva E, et al. Murine Falcor/LL35 lncRNA Contributes to Glucose and Lipid Metabolism In Vitro and In Vivo. Biomedicines. 2022; 10(6):1397. https://doi.org/10.3390/biomedicines10061397
Chicago/Turabian StyleShcherbinina, Evgeniya, Tatiana Abakumova, Daniil Bobrovskiy, Ilia Kurochkin, Ksenia Deinichenko, Elena Stekolshchikova, Nickolay Anikanov, Rustam Ziganshin, Pavel Melnikov, Ekaterina Khrameeva, and et al. 2022. "Murine Falcor/LL35 lncRNA Contributes to Glucose and Lipid Metabolism In Vitro and In Vivo" Biomedicines 10, no. 6: 1397. https://doi.org/10.3390/biomedicines10061397
APA StyleShcherbinina, E., Abakumova, T., Bobrovskiy, D., Kurochkin, I., Deinichenko, K., Stekolshchikova, E., Anikanov, N., Ziganshin, R., Melnikov, P., Khrameeva, E., Logacheva, M., Zatsepin, T., & Sergeeva, O. (2022). Murine Falcor/LL35 lncRNA Contributes to Glucose and Lipid Metabolism In Vitro and In Vivo. Biomedicines, 10(6), 1397. https://doi.org/10.3390/biomedicines10061397







