High-Frequency Irreversible Electroporation (H-FIRE) Induced Blood–Brain Barrier Disruption Is Mediated by Cytoskeletal Remodeling and Changes in Tight Junction Protein Regulation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Assurances and High-Frequency Irreversible Electroporation Delivery
2.2. Immunoprecipitation and Western Blotting
2.2.1. Tight Junction Proteins
2.2.2. Ubiquitin
2.2.3. Cytoskeleton
2.3. Gene Expression Profiling and Pathway Analysis
2.4. Immunofluorescent Imaging
2.4.1. Tight Junction Proteins
2.4.2. Cytoskeleton
2.5. Statistical Analysis
3. Results
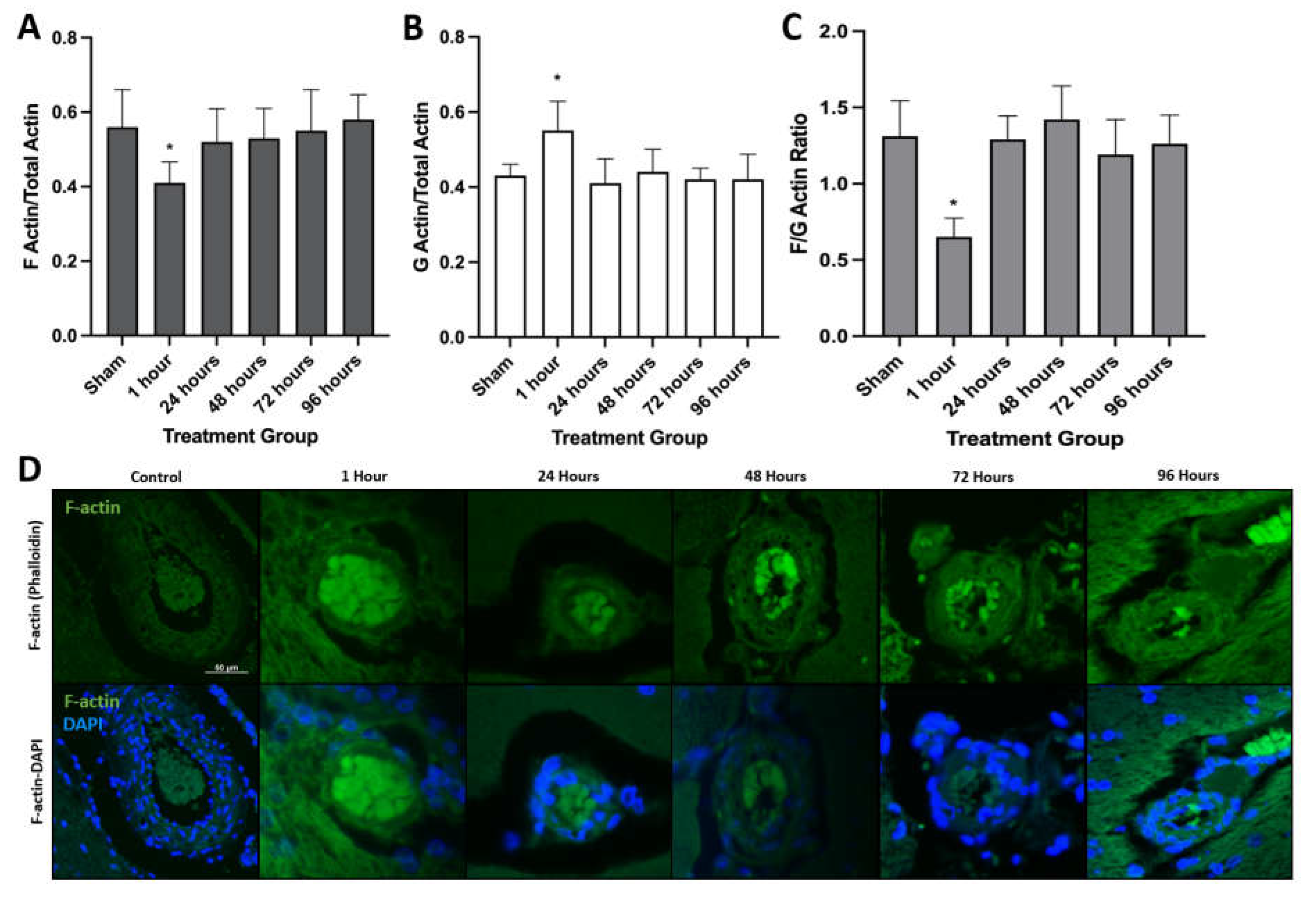
3.1. H-FIRE-Induced BBBD Is Mediated by Cytoskeletal Remodeling
3.2. H-FIRE-Mediated BBBD Is a Transient Process Mediated by Decreases in Tight Junction Protein Concentrations
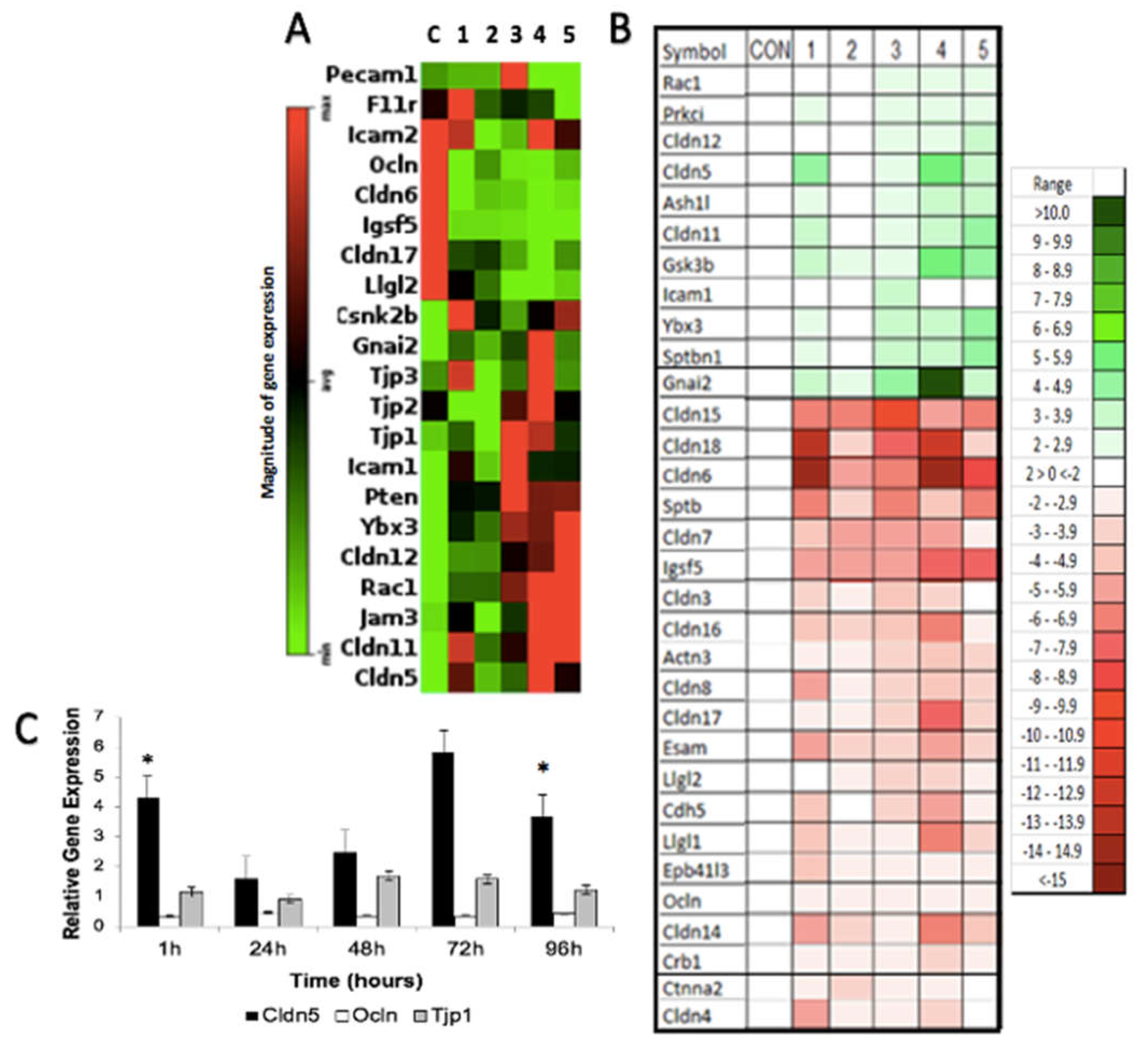
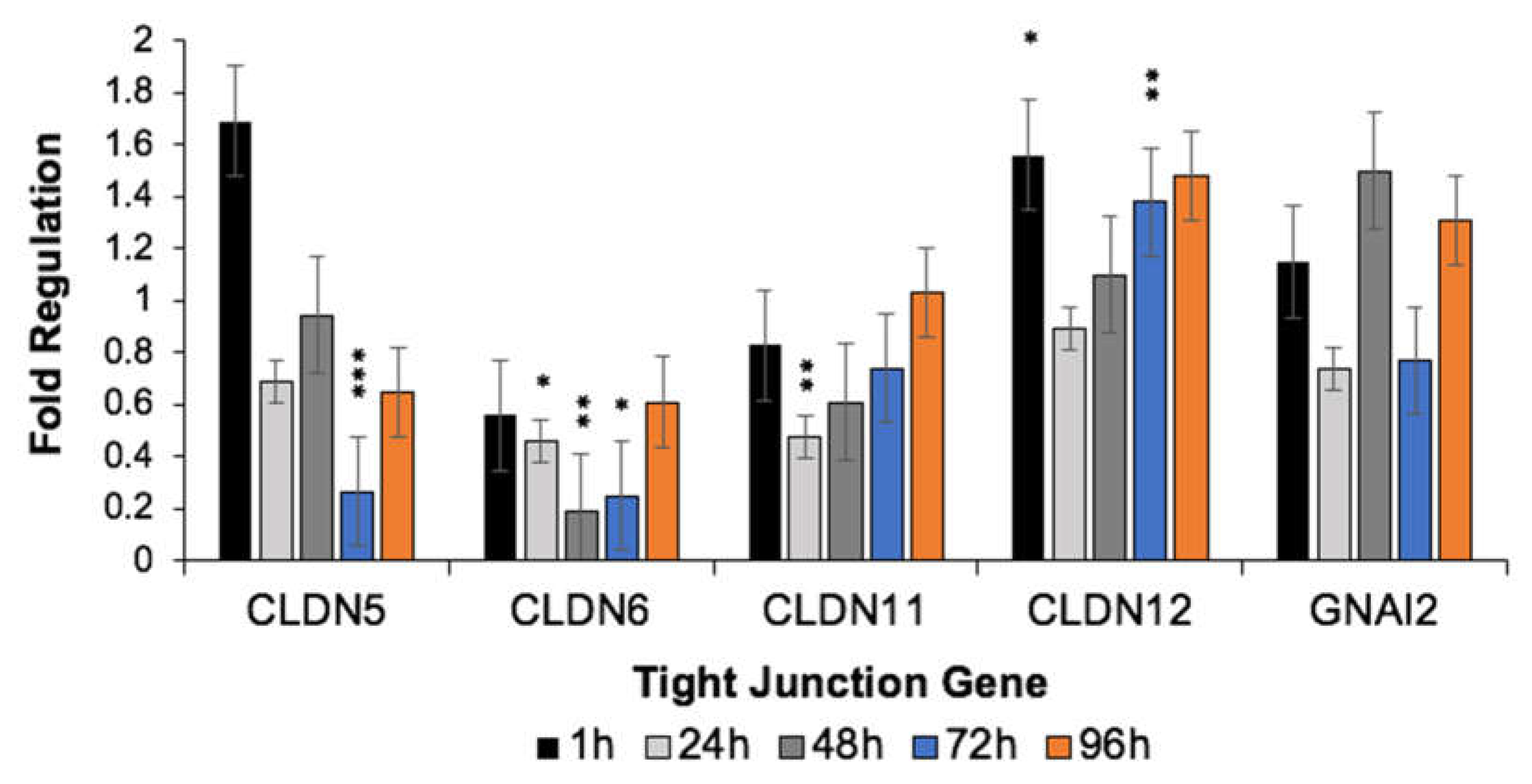
3.3. H-FIRE-Induced Transient BBBD Is Mediated by Alterations in Tight Junction Gene Expression
3.4. H-FIRE-Induced BBBD Is Mediated by Post-Translational Modifications to Tight Junction Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abbott, N.J.; Patabendige, A.A.; Dolman, D.E.; Yusof, S.R.; Begley, D.J. Structure and function of the blood-brain barrier. Neurobiol. Dis. 2010, 37, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Furuse, M.; Hirase, T.; Itoh, M.; Nagafuchi, A.; Yonemura, S.; Tsukita, S.; Tsukita, S. Occludin: A novel integral membrane protein localizing at tight junctions. J. Cell Biol. 1993, 123, 1777–1788. [Google Scholar] [CrossRef] [PubMed]
- Abbott, N.J.; Romero, I.A. Transporting therapeutics across the blood-brain barrier. Mol. Med. Today 1996, 2, 106–113. [Google Scholar] [CrossRef]
- Pardridge, W.M. Molecular biology of the blood-brain barrier. Mol. Biotechnol. 2005, 30, 57–70. [Google Scholar] [CrossRef]
- Aryal, M.; Arvanitis, C.D.; Alexander, P.M.; McDannold, N. Ultrasound-mediated blood-brain barrier disruption for targeted drug delivery in the central nervous system. Adv. Drug Deliv. Rev. 2014, 72, 94–109. [Google Scholar] [CrossRef]
- Barar, J.; Rafi, M.A.; Pourseif, M.M.; Omidi, Y. Blood-brain barrier transport machineries and targeted therapy of brain diseases. Bioimpacts 2016, 6, 225–248. [Google Scholar] [CrossRef]
- Regina, A.; Demeule, M.; Laplante, A.; Jodoin, J.; Dagenais, C.; Berthelet, F.; Moghrabi, A.; Beliveau, R. Multidrug resistance in brain tumors: Roles of the blood-brain barrier. Cancer Metast. Rev. 2001, 20, 13–25. [Google Scholar] [CrossRef]
- Rodriguez, A.; Tatter, S.B.; Debinski, W. Neurosurgical Techniques for Disruption of the Blood-Brain Barrier for Glioblastoma Treatment. Pharmaceutics 2015, 7, 175–187. [Google Scholar] [CrossRef]
- Rossmeisl, J.H., Jr.; Garcia, P.A.; Roberston, J.L.; Ellis, T.L.; Davalos, R.V. Pathology of non-thermal irreversible electroporation (N-TIRE)-induced ablation of the canine brain. J. Vet. Sci. 2013, 14, 433–440. [Google Scholar] [CrossRef]
- Partridge, B.; Rossmeisl, J.H.; Kaloss, A.M.; Basso, E.K.G.; Theus, M.H. Novel ablation methods for treatment of gliomas. J. Neurosci. Methods 2020, 336, 108630. [Google Scholar] [CrossRef]
- Partridge, B.; Lorenzo, M.F.; Dervisis, N.; Davalos, R.V.; Rossmeisl, J.H. Irreversible Electroporation Applications. In Electroporation in Veterinary Oncology Practice: Electrochemotherapy and Gene Electrotransfer for Immunotherapy; Impellizeri, J.A., Ed.; Springer International Publishing: Cham, Switzerland, 2021; pp. 165–204. [Google Scholar] [CrossRef]
- Van den Bos, W.; de Bruin, D.M.; Jurhill, R.R.; Savci-Heijink, C.D.; Muller, B.G.; Varkarakis, I.M.; Skolarikos, A.; Zondervan, P.J.; Laguna-Pes, M.P.; Wijkstra, H.; et al. The correlation between the electrode configuration and histopathology of irreversible electroporation ablations in prostate cancer patients. World J. Urol. 2016, 34, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Niessen, C.; Thumann, S.; Beyer, L.; Pregler, B.; Kramer, J.; Lang, S.; Teufel, A.; Jung, E.M.; Stroszczynski, C.; Wiggermann, P. Percutaneous Irreversible Electroporation: Long-term survival analysis of 71 patients with inoperable malignant hepatic tumors. Sci. Rep. 2017, 7, 43687. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.C., 2nd; Durham, A.N.; Besselink, M.G.; Iannitti, D.; Weiss, M.J.; Wolfgang, C.L.; Huang, K.W. Irreversible electroporation in locally advanced pancreatic cancer: A call for standardization of energy delivery. J. Surg. Oncol. 2016, 114, 865–871. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.C., 2nd; Kwon, D.; Chalikonda, S.; Sellers, M.; Kotz, E.; Scoggins, C.; McMasters, K.M.; Watkins, K. Treatment of 200 locally advanced (stage III) pancreatic adenocarcinoma patients with irreversible electroporation: Safety and efficacy. Ann. Surg. 2015, 262, 486–494; discussion 492–494. [Google Scholar] [CrossRef]
- Dong, S.; Wang, H.; Zhao, Y.; Sun, Y.; Yao, C. First Human Trial of High-Frequency Irreversible Electroporation Therapy for Prostate Cancer. Technol. Cancer Res. Treat 2018, 17, 1533033818789692. [Google Scholar] [CrossRef]
- Bhonsle, S.P.; Arena, C.B.; Sweeney, D.C.; Davalos, R.V. Mitigation of impedance changes due to electroporation therapy using bursts of high-frequency bipolar pulses. Biomed. Eng. Online 2015, 14 (Suppl. 3). [Google Scholar] [CrossRef]
- Lorenzo, M.F.; Thomas, S.C.; Kani, Y.; Hinckley, J.; Lee, M.; Adler, J.; Verbridge, S.S.; Hsu, F.C.; Robertson, J.L.; Davalos, R.V.; et al. Temporal Characterization of Blood-Brain Barrier Disruption with High-Frequency Electroporation. Cancers 2019, 11, 1850. [Google Scholar] [CrossRef]
- Lai, C.H.; Kuo, K.H.; Leo, J.M. Critical role of actin in modulating BBB permeability. Brain. Res. Brain. Res. Rev. 2005, 50, 7–13. [Google Scholar] [CrossRef]
- Graybill, P.M.; Davalos, R.V. Cytoskeletal Disruption after Electroporation and Its Significance to Pulsed Electric Field Therapies. Cancers 2020, 12, 1132. [Google Scholar] [CrossRef]
- Lynn, K.S.; Peterson, R.J.; Koval, M. Ruffles and spikes: Control of tight junction morphology and permeability by claudins. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183339. [Google Scholar] [CrossRef]
- Fanning, A.S.; Ma, T.Y.; Anderson, J.M. Isolation and functional characterization of the actin binding region in the tight junction protein ZO-1. FASEB J. 2002, 16, 1835–1837. [Google Scholar] [CrossRef] [PubMed]
- Luissint, A.C.; Artus, C.; Glacial, F.; Ganeshamoorthy, K.; Couraud, P.O. Tight junctions at the blood brain barrier: Physiological architecture and disease-associated dysregulation. Fluids Barriers CNS 2012, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Pehlivanova, V.N.; Tsoneva, I.H.; Tzoneva, R.D. Multiple effects of electroporation on the adhesive behaviour of breast cancer cells and fibroblasts. Cancer Cell Int. 2012, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Kanthou, C.; Kranjc, S.; Sersa, G.; Tozer, G.; Zupanic, A.; Cemazar, M. The endothelial cytoskeleton as a target of electroporation-based therapies. Mol. Cancer Ther. 2006, 5, 3145–3152. [Google Scholar] [CrossRef] [PubMed]
- Chopinet, L.; Roduit, C.; Rols, M.P.; Dague, E. Destabilization induced by electropermeabilization analyzed by atomic force microscopy. Biochim. Biophys. Acta 2013, 1828, 2223–2229. [Google Scholar] [CrossRef] [PubMed]
- Jaepel, J.; Blum, R. Capturing ER calcium dynamics. Eur. J. Cell Biol. 2011, 90, 613–619. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, E.T.; Salmon, E.D.; Erickson, H.P. How calcium causes microtubule depolymerization. Cell Motil Cytoskelet. 1997, 36, 125–135. [Google Scholar] [CrossRef]
- Tsai, F.C.; Kuo, G.H.; Chang, S.W.; Tsai, P.J. Ca2+ signaling in cytoskeletal reorganization, cell migration, and cancer metastasis. Biomed. Res. Int. 2015, 2015, 409245. [Google Scholar] [CrossRef]
- Atkinson, S.J.; Hosford, M.A.; Molitoris, B.A. Mechanism of actin polymerization in cellular ATP depletion. J. Biol. Chem. 2004, 279, 5194–5199. [Google Scholar] [CrossRef]
- Shen, L.; Turner, J.R. Actin depolymerization disrupts tight junctions via caveolae-mediated endocytosis. Mol. Biol. Cell 2005, 16, 3919–3936. [Google Scholar] [CrossRef]
- Reiche, J.; Huber, O. Post-translational modifications of tight junction transmembrane proteins and their direct effect on barrier function. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183330. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Felinski, E.A.; Antonetti, D.A. Occludin phosphorylation and ubiquitination regulate tight junction trafficking and vascular endothelial growth factor-induced permeability. J. Biol. Chem. 2009, 284, 21036–21046. [Google Scholar] [CrossRef] [PubMed]
- Bonifacino, J.S.; Weissman, A.M. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Ann. Rev. Cell Dev. Biol. 1998, 14, 19–57. [Google Scholar] [CrossRef] [PubMed]
- Tracz, M.; Bialek, W. Beyond K48 and K63: Non-canonical protein ubiquitination. Cell Mol. Biol. Lett. 2021, 26, 1. [Google Scholar] [CrossRef]
- Sadowski, M.; Sarcevic, B. Mechanisms of mono- and poly-ubiquitination: Ubiquitination specificity depends on compatibility between the E2 catalytic core and amino acid residues proximal to the lysine. Cell Div. 2010, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Lochhead, J.J.; Yang, J.; Ronaldson, P.T.; Davis, T.P. Structure, Function, and Regulation of the Blood-Brain Barrier Tight Junction in Central Nervous System Disorders. Front. Physiol. 2020, 11, 914. [Google Scholar] [CrossRef]
- Cai, J.; Culley, M.K.; Zhao, Y.; Zhao, J. The role of ubiquitination and deubiquitination in the regulation of cell junctions. Protein Cell 2018, 9, 754–769. [Google Scholar] [CrossRef]
- Bauer, H.C.; Krizbai, I.A.; Bauer, H.; Traweger, A. “You Shall Not Pass”-tight junctions of the blood brain barrier. Front. Neurosci. 2014, 8, 392. [Google Scholar] [CrossRef]
- Krause, G.; Winkler, L.; Mueller, S.L.; Haseloff, R.F.; Piontek, J.; Blasig, I.E. Structure and function of claudins. Biochim. Biophys. Acta 2008, 1778, 631–645. [Google Scholar] [CrossRef]
- Gunzel, D.; Yu, A.S. Claudins and the modulation of tight junction permeability. Physiol. Rev. 2013, 93, 525–569. [Google Scholar] [CrossRef]
- Sugimoto, K.; Ichikawa-Tomikawa, N.; Kashiwagi, K.; Endo, C.; Tanaka, S.; Sawada, N.; Watabe, T.; Higashi, T.; Chiba, H. Cell adhesion signals regulate the nuclear receptor activity. Proc. Natl. Acad. Sci. USA 2019, 116, 24600–24609. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Guo, C.; Li, Y.; Liu, K.; Zhao, Q.; Ouyang, L. Identification of Claudin-6 as a Molecular Biomarker in Pan-Cancer Through Multiple Omics Integrative Analysis. Front. Cell Dev. Biol. 2021, 9, 726656. [Google Scholar] [CrossRef] [PubMed]
- Kojima, M.; Sugimoto, K.; Tanaka, M.; Endo, Y.; Kato, H.; Honda, T.; Furukawa, S.; Nishiyama, H.; Watanabe, T.; Soeda, S.; et al. Prognostic Significance of Aberrant Claudin-6 Expression in Endometrial Cancer. Cancers 2020, 12, 2748. [Google Scholar] [CrossRef] [PubMed]
- Kohmoto, T.; Masuda, K.; Shoda, K.; Takahashi, R.; Ujiro, S.; Tange, S.; Ichikawa, D.; Otsuji, E.; Imoto, I. Claudin-6 is a single prognostic marker and functions as a tumor-promoting gene in a subgroup of intestinal type gastric cancer. Gastric Cancer 2020, 23, 403–417. [Google Scholar] [CrossRef]
- Liu, Y.; Jin, X.; Li, Y.; Ruan, Y.; Lu, Y.; Yang, M.; Lin, D.; Song, P.; Guo, Y.; Zhao, S.; et al. DNA methylation of claudin-6 promotes breast cancer cell migration and invasion by recruiting MeCP2 and deacetylating H3Ac and H4Ac. J. Exp. Clin. Cancer Res. 2016, 35, 120. [Google Scholar] [CrossRef]
- Kratzer, I.; Vasiljevic, A.; Rey, C.; Fevre-Montange, M.; Saunders, N.; Strazielle, N.; Ghersi-Egea, J.F. Complexity and developmental changes in the expression pattern of claudins at the blood-CSF barrier. Histochem Cell Biol. 2012, 138, 861–879. [Google Scholar] [CrossRef]
- Pires, F.; Ferreira, Q.; Rodrigues, C.A.; Morgado, J.; Ferreira, F.C. Neural stem cell differentiation by electrical stimulation using a cross-linked PEDOT substrate: Expanding the use of biocompatible conjugated conductive polymers for neural tissue engineering. Biochim. Biophys. Acta 2015, 1850, 1158–1168. [Google Scholar] [CrossRef]
- Greene, C.; Hanley, N.; Campbell, M. Claudin-5: Gatekeeper of neurological function. Fluids Barriers CNS 2019, 16, 3. [Google Scholar] [CrossRef]
- Krug, S.M.; Gunzel, D.; Conrad, M.P.; Rosenthal, R.; Fromm, A.; Amasheh, S.; Schulzke, J.D.; Fromm, M. Claudin-17 forms tight junction channels with distinct anion selectivity. Cell Mol. Life Sci. 2012, 69, 2765–2778. [Google Scholar] [CrossRef]
- Frische, S.; Alexander, R.T.; Ferreira, P.; Tan, R.S.G.; Wang, W.; Svenningsen, P.; Skjodt, K.; Dimke, H. Localization and regulation of claudin-14 in experimental models of hypercalcemia. Am. J. Physiol. Renal Physiol. 2021, 320, F74–F86. [Google Scholar] [CrossRef]
- Samanta, P.; Wang, Y.; Fuladi, S.; Zou, J.; Li, Y.; Shen, L.; Weber, C.; Khalili-Araghi, F. Molecular determination of claudin-15 organization and channel selectivity. J. Gen. Physiol. 2018, 150, 949–968. [Google Scholar] [CrossRef] [PubMed]
- Linares, G.R.; Brommage, R.; Powell, D.R.; Xing, W.; Chen, S.T.; Alshbool, F.Z.; Lau, K.H.; Wergedal, J.E.; Mohan, S. Claudin 18 is a novel negative regulator of bone resorption and osteoclast differentiation. J. Bone Miner. Res. 2012, 27, 1553–1565. [Google Scholar] [CrossRef] [PubMed]
- Luissint, A.C.; Federici, C.; Guillonneau, F.; Chretien, F.; Camoin, L.; Glacial, F.; Ganeshamoorthy, K.; Couraud, P.O. Guanine nucleotide-binding protein Galphai2: A new partner of claudin-5 that regulates tight junction integrity in human brain endothelial cells. J. Cereb. Blood Flow Metab. 2012, 32, 860–873. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, S.; Tajima, M.; Yao, I.; Nishimura, W.; Mori, H.; Hata, Y. JAM4, a junctional cell adhesion molecule interacting with a tight junction protein, MAGI-1. Mol. Cell Biol. 2003, 23, 4267–4282. [Google Scholar] [CrossRef] [PubMed]
- Jossin, Y.; Lee, M.; Klezovitch, O.; Kon, E.; Cossard, A.; Lien, W.H.; Fernandez, T.E.; Cooper, J.A.; Vasioukhin, V. Llgl1 Connects Cell Polarity with Cell-Cell Adhesion in Embryonic Neural Stem Cells. Dev. Cell 2017, 41, 481–495.e485. [Google Scholar] [CrossRef]
- Leng, S.; Xie, F.; Liu, J.; Shen, J.; Quan, G.; Wen, T. LLGL2 Increases Ca(2+) Influx and Exerts Oncogenic Activities via PI3K/AKT Signaling Pathway in Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 683629. [Google Scholar] [CrossRef]
- Fan, X.; Xie, X.; Yang, M.; Wang, Y.; Wu, H.; Deng, T.; Weng, X.; Wen, W.; Nie, G. YBX3 Mediates the Metastasis of Nasopharyngeal Carcinoma via PI3K/AKT Signaling. Front. Oncol. 2021, 11, 617621. [Google Scholar] [CrossRef]
- Muller, N. The Role of Intercellular Adhesion Molecule-1 in the Pathogenesis of Psychiatric Disorders. Front. Pharmacol. 2019, 10, 1251. [Google Scholar] [CrossRef]
- Sasaki, S.; Takeda, K.; Ouhara, K.; Shirawachi, S.; Kajiya, M.; Matsuda, S.; Kono, S.; Shiba, H.; Kurihara, H.; Mizuno, N. Involvement of Rac1 in macrophage activation by brain-derived neurotrophic factor. Mol. Biol. Rep. 2021, 48, 5249–5257. [Google Scholar] [CrossRef]
- Sawada, N.; Kim, H.H.; Moskowitz, M.A.; Liao, J.K. Rac1 is a critical mediator of endothelium-derived neurotrophic activity. Sci Signal 2009, 2, ra10. [Google Scholar] [CrossRef]
- Cole, A.R. GSK3 as a Sensor Determining Cell Fate in the Brain. Front. Mol. Neurosci. 2012, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Cemazar, M.; Parkins, C.S.; Holder, A.L.; Chaplin, D.J.; Tozer, G.M.; Sersa, G. Electroporation of human microvascular endothelial cells: Evidence for an anti-vascular mechanism of electrochemotherapy. Br. J. Cancer 2001, 84, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Sersa, G.; Jarm, T.; Kotnik, T.; Coer, A.; Podkrajsek, M.; Sentjurc, M.; Miklavcic, D.; Kadivec, M.; Kranjc, S.; Secerov, A.; et al. Vascular disrupting action of electroporation and electrochemotherapy with bleomycin in murine sarcoma. Br. J. Cancer 2008, 98, 388–398. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Partridge, B.R.; Kani, Y.; Lorenzo, M.F.; Campelo, S.N.; Allen, I.C.; Hinckley, J.; Hsu, F.-C.; Verbridge, S.S.; Robertson, J.L.; Davalos, R.V.; et al. High-Frequency Irreversible Electroporation (H-FIRE) Induced Blood–Brain Barrier Disruption Is Mediated by Cytoskeletal Remodeling and Changes in Tight Junction Protein Regulation. Biomedicines 2022, 10, 1384. https://doi.org/10.3390/biomedicines10061384
Partridge BR, Kani Y, Lorenzo MF, Campelo SN, Allen IC, Hinckley J, Hsu F-C, Verbridge SS, Robertson JL, Davalos RV, et al. High-Frequency Irreversible Electroporation (H-FIRE) Induced Blood–Brain Barrier Disruption Is Mediated by Cytoskeletal Remodeling and Changes in Tight Junction Protein Regulation. Biomedicines. 2022; 10(6):1384. https://doi.org/10.3390/biomedicines10061384
Chicago/Turabian StylePartridge, Brittanie R., Yukitaka Kani, Melvin F. Lorenzo, Sabrina N. Campelo, Irving C. Allen, Jonathan Hinckley, Fang-Chi Hsu, Scott S. Verbridge, John L. Robertson, Rafael V. Davalos, and et al. 2022. "High-Frequency Irreversible Electroporation (H-FIRE) Induced Blood–Brain Barrier Disruption Is Mediated by Cytoskeletal Remodeling and Changes in Tight Junction Protein Regulation" Biomedicines 10, no. 6: 1384. https://doi.org/10.3390/biomedicines10061384
APA StylePartridge, B. R., Kani, Y., Lorenzo, M. F., Campelo, S. N., Allen, I. C., Hinckley, J., Hsu, F.-C., Verbridge, S. S., Robertson, J. L., Davalos, R. V., & Rossmeisl, J. H. (2022). High-Frequency Irreversible Electroporation (H-FIRE) Induced Blood–Brain Barrier Disruption Is Mediated by Cytoskeletal Remodeling and Changes in Tight Junction Protein Regulation. Biomedicines, 10(6), 1384. https://doi.org/10.3390/biomedicines10061384










