Gut Microbiota and Associated Mucosal Immune Response in Eosinophilic Granulomatosis with Polyangiitis (EGPA)
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Setting
2.2. Clinical Assessment
2.3. Microbiota Characterization
2.4. Short- and Medium-Chain Fatty Acids Analysis
2.5. Generation and Characterization of T-Cell Clones from Intestinal Biopsies
2.6. Statistical Analysis
2.7. Patient and Public Involvement Statement
3. Results
3.1. Study Population
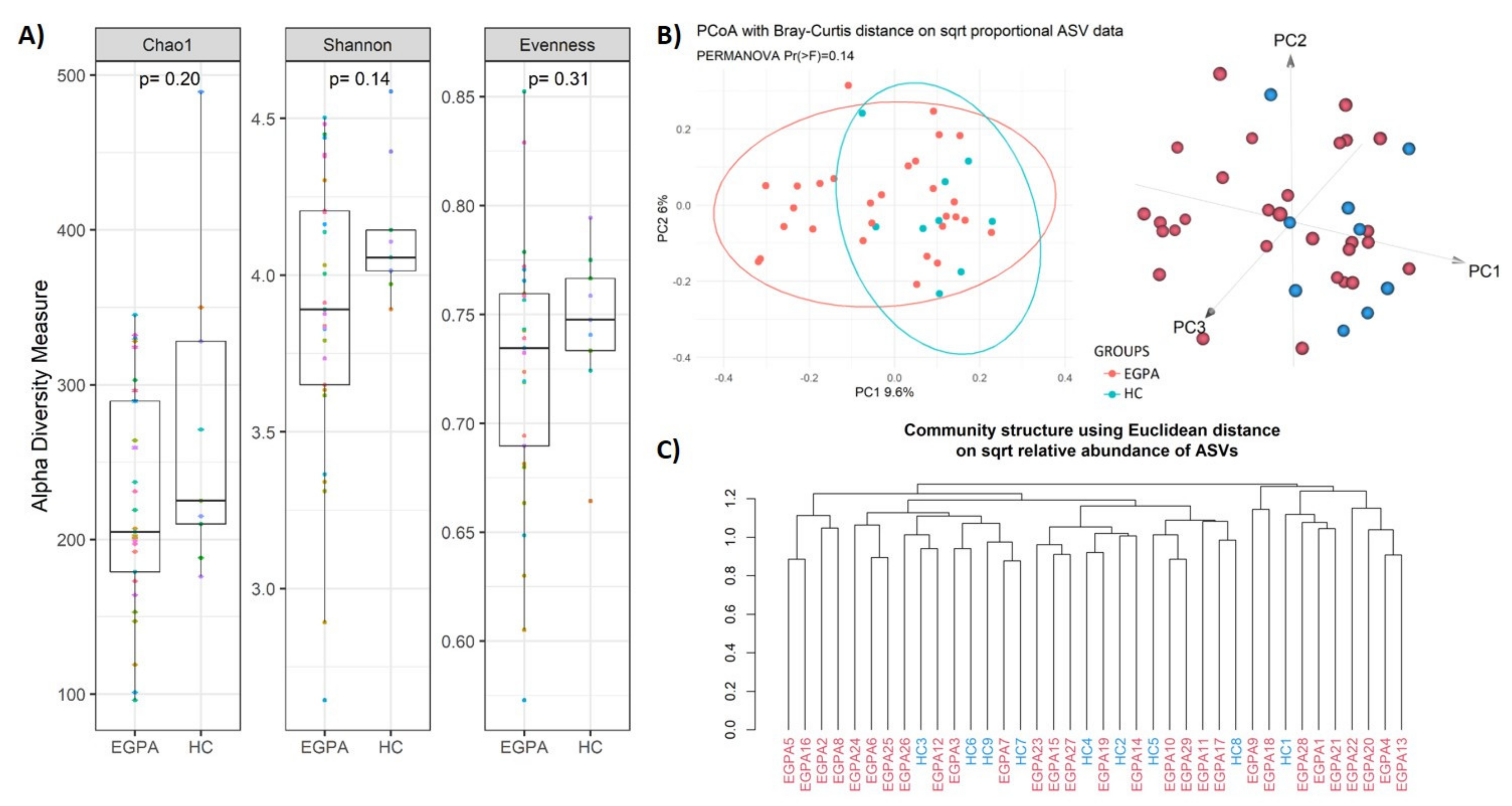
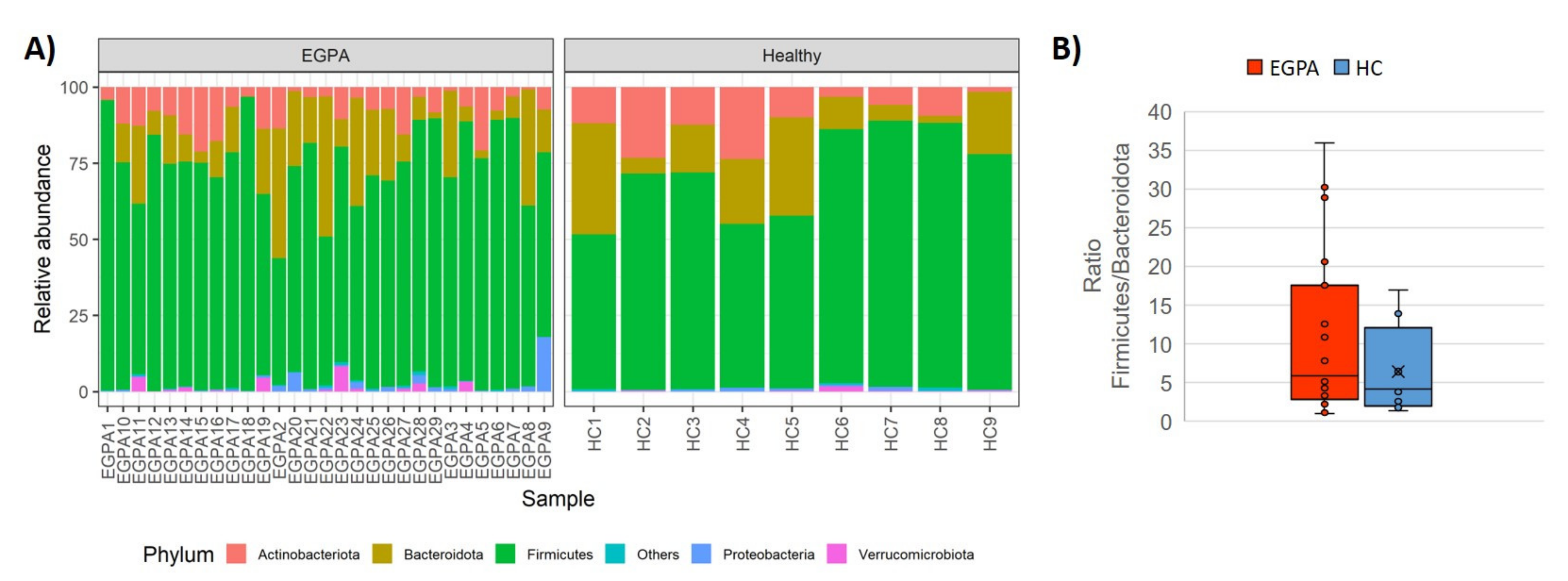
3.2. Faecal Microbiota Composition
3.3. Microbiota-Derived Metabolites
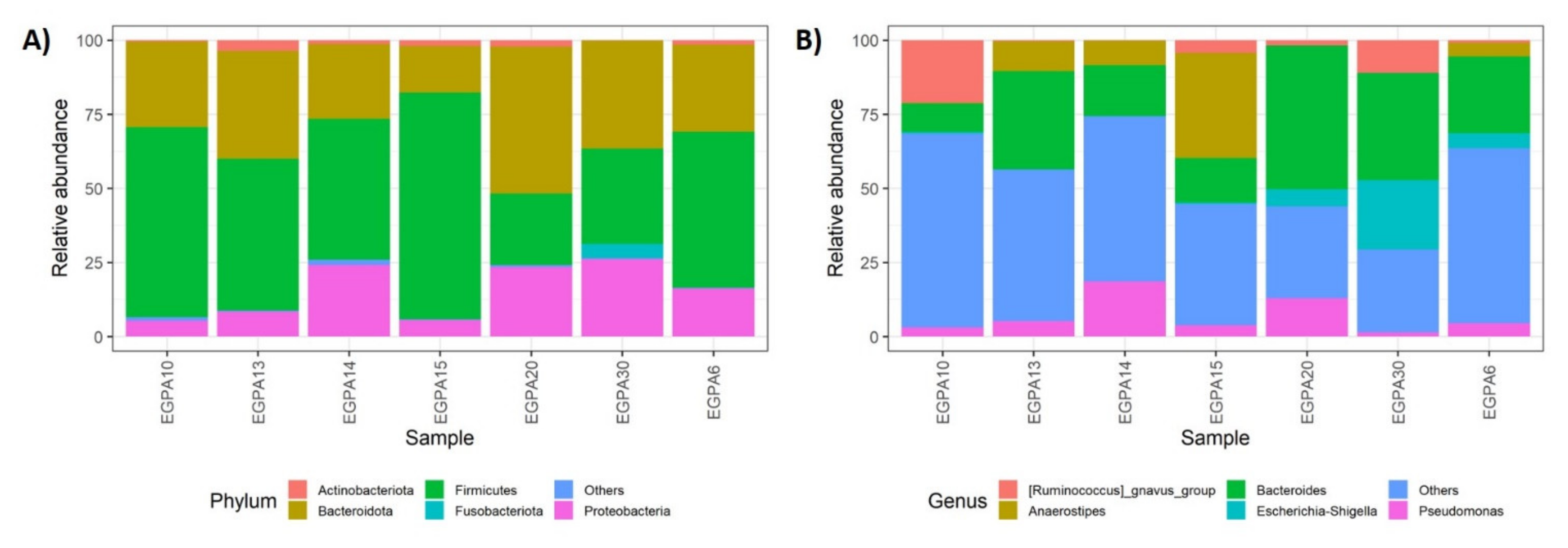
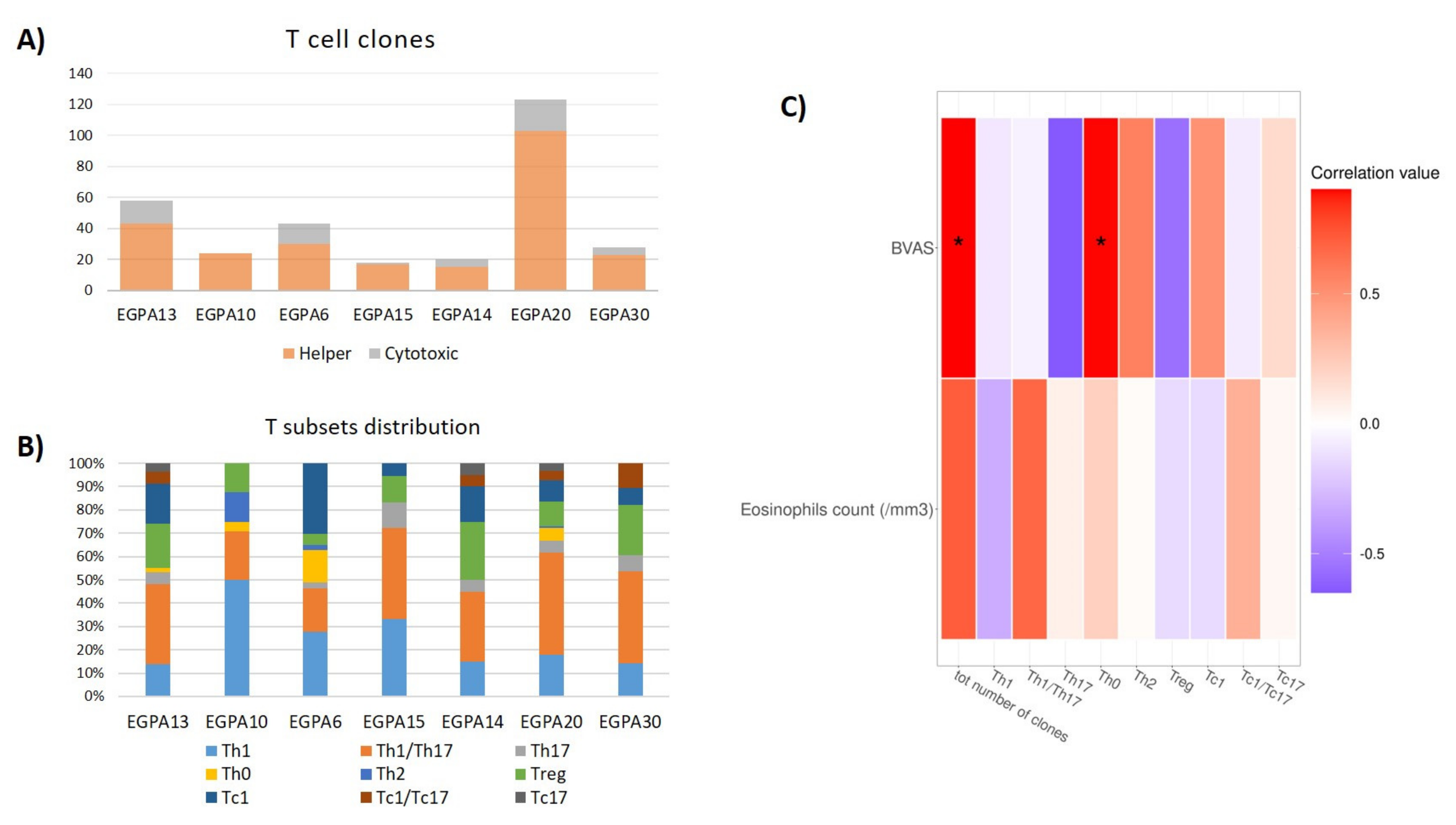
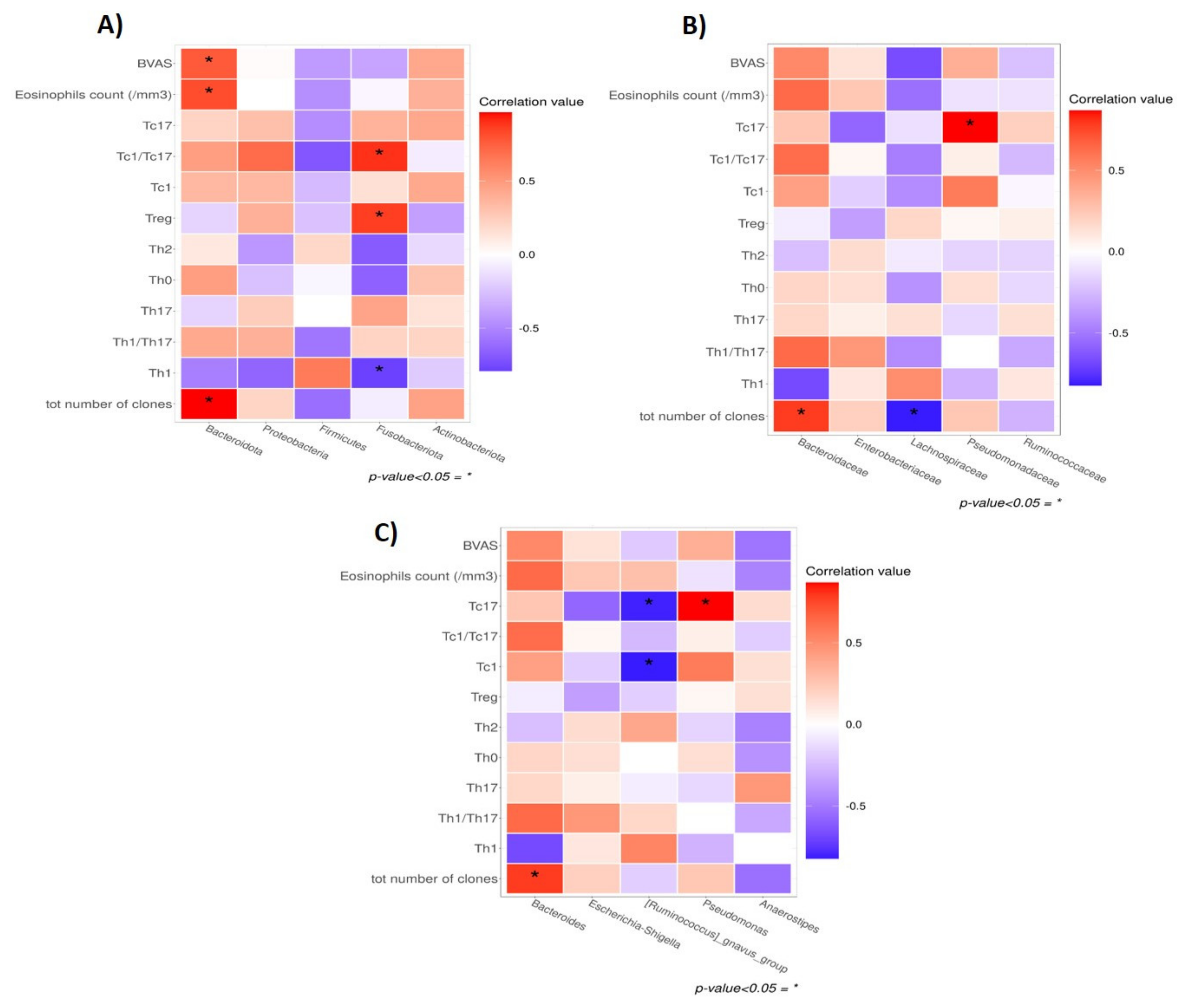
3.4. Gut Microbiota and Mucosal T-Cell Response in EGPA Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Trivioli, G.; Terrier, B.; Vaglio, A. Eosinophilic granulomatosis with polyangiitis: Understanding the disease and its management. Rheumatology 2020, 59, iii84–iii94. [Google Scholar] [CrossRef] [PubMed]
- Moiseev, S.; Bossuyt, X.; Arimura, Y.; Blockmans, D.; Csernok, E.; Damoiseaux, J.; Emmi, G.; Flores-Suarez, L.F.; Hellmich, B.; Jayne, D.; et al. International Consensus on ANCA Testing in Eosinophilic Granulomatosis with Polyangiitis. Am. J. Respir. Crit. Care Med. 2020, 202, 1360–1372. [Google Scholar] [CrossRef] [PubMed]
- Papo, M.; Sinico, R.A.; Teixeira, V.; Venhoff, N.; Urban, M.L.; Iudici, M.; Mahrhold, J.; Locatelli, F.; Cassone, G.; Schiavon, F.; et al. Significance of PR3-ANCA positivity in eosinophilic granulomatosis with polyangiitis (Churg-Strauss). Rheumatology 2021, 60, 4355–4360. [Google Scholar] [CrossRef] [PubMed]
- Fagni, F.; Bello, F.; Emmi, G. Eosinophilic Granulomatosis with Polyangiitis: Dissecting the Pathophysiology. Front. Med. 2021, 8, 627776. [Google Scholar] [CrossRef]
- Vaglio, A.; Martorana, D.; Maggiore, U.; Grasselli, C.; Zanetti, A.; Pesci, A.; Garini, G.; Manganelli, P.; Bottero, P.; Tumiati, B.; et al. HLA-DRB4 as a genetic risk factor for Churg-Strauss syndrome. Arthritis Rheum. Off. J. Am. Coll. Rheumatol. 2007, 56, 3159–3166. [Google Scholar] [CrossRef]
- Maritati, F.; Peyronel, F.; Fenaroli, P.; Pegoraro, F.; Lastrucci, V.; Benigno, G.D.; Palmisano, A.; Rossi, G.M.; Urban, M.L.; Alberici, F.; et al. Occupational Exposures and Smoking in Eosinophilic Granulomatosis with Polyangiitis: A Case-Control Study. Arthritis Rheumatol. 2021, 73, 1694–1702. [Google Scholar] [CrossRef]
- Lyons, P.A.; Peters, J.E.; Alberici, F.; Liley, J.; Coulson, R.M.R.; Astle, W.; Baldini, C.; Bonatti, F.; Cid, M.C.; Elding, H.; et al. Genome-wide association study of eosinophilic granulomatosis with polyangiitis reveals genomic loci stratified by ANCA status. Nat. Commun. 2019, 10, 5120. [Google Scholar] [CrossRef]
- Candela, M.; Biagi, E.; Turroni, S.; Maccaferri, S.; Figini, P.; Brigidi, P. Dynamic efficiency of the human intestinal microbiota. Crit. Rev. Microbiol. 2015, 41, 165–171. [Google Scholar] [CrossRef]
- Centanni, M.; Turroni, S.; Consolandi, C.; Rampelli, S.; Peano, C.; Severgnini, M.; Biagi, E.; Caredda, G.; De Bellis, G.; Brigidi, P.; et al. The enterocyte-associated intestinal microbiota of breast-fed infants and adults responds differently to a TNF-alpha-mediated pro-inflammatory stimulus. PLoS ONE 2013, 8, e81762. [Google Scholar] [CrossRef][Green Version]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef]
- Niccolai, E.; Boem, F.; Emmi, G.; Amedei, A. The link “Cancer and autoimmune diseases” in the light of microbiota: Evidence of a potential culprit. Immunol. Lett. 2020, 222, 12–28. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Niccolai, E.; Di Pilato, V.; Nannini, G.; Baldi, S.; Russo, E.; Zucchi, E.; Martinelli, I.; Menicatti, M.; Bartolucci, G.; Mandrioli, J.; et al. The Gut Microbiota-Immunity Axis in ALS: A Role in Deciphering Disease Heterogeneity? Biomedicines 2021, 9, 753. [Google Scholar] [CrossRef] [PubMed]
- D’Amato, A.; Di Cesare Mannelli, L.; Lucarini, E.; Man, A.L.; Le Gall, G.; Branca, J.J.V.; Ghelardini, C.; Amedei, A.; Bertelli, E.; Regoli, M.; et al. Faecal microbiota transplant from aged donor mice affects spatial learning and memory via modulating hippocampal synaptic plasticity- and neurotransmission-related proteins in young recipients. Microbiome 2020, 8, 140. [Google Scholar] [CrossRef] [PubMed]
- Tariq, S.; Clifford, A.H. An update on the microbiome in vasculitis. Curr. Opin. Rheumatol. 2021, 33, 15–23. [Google Scholar] [CrossRef]
- Consolandi, C.; Turroni, S.; Emmi, G.; Severgnini, M.; Fiori, J.; Peano, C.; Biagi, E.; Grassi, A.; Rampelli, S.; Silvestri, E.; et al. Behcet’s syndrome patients exhibit specific microbiome signature. Autoimmun. Rev. 2015, 14, 269–276. [Google Scholar] [CrossRef]
- Emmi, G.; Bettiol, A.; Niccolai, E.; Ramazzotti, M.; Amedei, A.; Pagliai, G.; Taddei, N.; Sofi, F.; Fiorillo, C.; Prisco, D.; et al. Butyrate-Rich Diets Improve Redox Status and Fibrin Lysis in Behcet’s Syndrome. Circ. Res. 2021, 128, 278–280. [Google Scholar] [CrossRef]
- Rhee, R.L.; Sreih, A.G.; Najem, C.E.; Grayson, P.C.; Zhao, C.; Bittinger, K.; Collman, R.G.; Merkel, P.A. Characterisation of the nasal microbiota in granulomatosis with polyangiitis. Ann. Rheum. Dis. 2018, 77, 1448–1453. [Google Scholar] [CrossRef]
- Rhee, R.L.; Lu, J.; Bittinger, K.; Lee, J.J.; Mattei, L.M.; Sreih, A.G.; Chou, S.; Miner, J.J.; Cohen, N.A.; Kelly, B.J.; et al. Dynamic Changes in the Nasal Microbiome Associated With Disease Activity in Patients With Granulomatosis with Polyangiitis. Arthritis Rheumatol. 2021, 73, 1703–1712. [Google Scholar] [CrossRef]
- Kronbichler, A.; Harrison, E.M.; Wagner, J. Nasal microbiome research in ANCA-associated vasculitis: Strengths, limitations, and future directions. Comput. Struct. Biotechnol. J. 2021, 19, 415–423. [Google Scholar] [CrossRef]
- Wagner, J.; Harrison, E.M.; Martinez Del Pero, M.; Blane, B.; Mayer, G.; Leierer, J.; Gopaluni, S.; Holmes, M.A.; Parkhill, J.; Peacock, S.J.; et al. The composition and functional protein subsystems of the human nasal microbiome in granulomatosis with polyangiitis: A pilot study. Microbiome 2019, 7, 137. [Google Scholar] [CrossRef] [PubMed]
- Lamprecht, P.; Fischer, N.; Huang, J.; Burkhardt, L.; Lutgehetmann, M.; Arndt, F.; Rolfs, I.; Kerstein, A.; Iking-Konert, C.; Laudien, M. Changes in the composition of the upper respiratory tract microbial community in granulomatosis with polyangiitis. J. Autoimmun. 2019, 97, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Masi, A.T.; Hunder, G.G.; Lie, J.T.; Michel, B.A.; Bloch, D.A.; Arend, W.P.; Calabrese, L.H.; Edworthy, S.M.; Fauci, A.S.; Leavitt, R.Y.; et al. The American College of Rheumatology 1990 criteria for the classification of Churg-Strauss syndrome (allergic granulomatosis and angiitis). Arthritis Rheum. 1990, 33, 1094–1100. [Google Scholar] [CrossRef] [PubMed]
- Wechsler, M.E.; Akuthota, P.; Jayne, D.; Khoury, P.; Klion, A.; Langford, C.A.; Merkel, P.A.; Moosig, F.; Specks, U.; Cid, M.C.; et al. Mepolizumab or Placebo for Eosinophilic Granulomatosis with Polyangiitis. N. Engl. J. Med. 2017, 376, 1921–1932. [Google Scholar] [CrossRef]
- Mukhtyar, C.; Lee, R.; Brown, D.; Carruthers, D.; Dasgupta, B.; Dubey, S.; Flossmann, O.; Hall, C.; Hollywood, J.; Jayne, D.; et al. Modification and validation of the Birmingham Vasculitis Activity Score (version 3). Ann. Rheum. Dis. 2009, 68, 1827–1832. [Google Scholar] [CrossRef]
- Niccolai, E.; Baldi, S.; Ricci, F.; Russo, E.; Nannini, G.; Menicatti, M.; Poli, G.; Taddei, A.; Bartolucci, G.; Calabro, A.S.; et al. Evaluation and comparison of short chain fatty acids composition in gut diseases. World J. Gastroenterol. 2019, 25, 5543–5558. [Google Scholar] [CrossRef]
- Amedei, A.; Della Bella, C.; Niccolai, E.; Stanflin, N.; Benagiano, M.; Duranti, R.; Del Prete, G.; Murphy, T.F.; D’Elios, M.M. Moraxella catarrhalis-specific Th1 cells in BAL fluids of chronic obstructive pulmonary disease patients. Int. J. Immunopathol. Pharmacol. 2009, 22, 979–990. [Google Scholar] [CrossRef]
- Niccolai, E.; Ricci, F.; Russo, E.; Nannini, G.; Emmi, G.; Taddei, A.; Ringressi, M.N.; Melli, F.; Miloeva, M.; Cianchi, F.; et al. The Different Functional Distribution of “Not Effector” T Cells (Treg/Tnull) in Colorectal Cancer. Front. Immunol. 2017, 8, 1900. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, 2.6–2. 2022. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 20 March 2022).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Harrell, F.E.; Dupont, C. Hmisc: Harrell Miscellaneous, 4.7-0. 2022. Available online: https://cran.r-project.org/web/packages/Hmisc/Hmisc.pdf (accessed on 20 March 2022).
- Weiner, J. Three Dimensional PCA Plots. 2020. Available online: https://cran.r-project.org/web/packages/pca3d/pca3d.pdf (accessed on 20 March 2022).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 12. [Google Scholar] [CrossRef]
- Edgar, R. Taxonomy annotation and guide tree errors in 16S rRNA databases. PeerJ 2018, 6, e5030. [Google Scholar] [CrossRef] [PubMed]
- Darfeuille-Michaud, A.; Neut, C.; Barnich, N.; Lederman, E.; Di Martino, P.; Desreumaux, P.; Gambiez, L.; Joly, B.; Cortot, A.; Colombel, J.F. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology 1998, 115, 1405–1413. [Google Scholar] [CrossRef]
- Lupp, C.; Robertson, M.L.; Wickham, M.E.; Sekirov, I.; Champion, O.L.; Gaynor, E.C.; Finlay, B.B. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2007, 2, 204. [Google Scholar] [CrossRef]
- Mukhopadhya, I.; Hansen, R.; El-Omar, E.M.; Hold, G.L. IBD-what role do Proteobacteria play? Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 219–230. [Google Scholar] [CrossRef]
- Getz, T.M.; Hoffman, G.S.; Padmanabhan, R.; Villa-Forte, A.; Roselli, E.E.; Blackstone, E.; Johnston, D.; Pettersson, G.; Soltesz, E.; Svensson, L.G.; et al. Microbiomes of Inflammatory Thoracic Aortic Aneurysms Due to Giant Cell Arteritis and Clinically Isolated Aortitis Differ From Those of Non-Inflammatory Aneurysms. Pathog. Immun. 2019, 4, 105–123. [Google Scholar] [CrossRef][Green Version]
- Saad, R.; Tsoi, K.; Onac, I.A.; Davies, K.A.; Hajela, V.K.; Tacu, C. Streptococcus-associated vasculitis: A role for antibiotic therapy? IDCases 2021, 24, e01071. [Google Scholar] [CrossRef]
- Kinumaki, A.; Sekizuka, T.; Hamada, H.; Kato, K.; Yamashita, A.; Kuroda, M. Characterization of the gut microbiota of Kawasaki disease patients by metagenomic analysis. Front. Microbiol. 2015, 6, 824. [Google Scholar] [CrossRef]
- Chow, J.; Tang, H.; Mazmanian, S.K. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr. Opin. Immunol. 2011, 23, 473–480. [Google Scholar] [CrossRef]
- Bhaskaran, N.; Quigley, C.; Paw, C.; Butala, S.; Schneider, E.; Pandiyan, P. Role of Short Chain Fatty Acids in Controlling Tregs and Immunopathology During Mucosal Infection. Front. Microbiol. 2018, 9, 1995. [Google Scholar] [CrossRef]
- Dallos, T.; Heiland, G.R.; Strehl, J.; Karonitsch, T.; Gross, W.L.; Moosig, F.; Holl-Ulrich, C.; Distler, J.H.; Manger, B.; Schett, G.; et al. CCL17/thymus and activation-related chemokine in Churg-Strauss syndrome. Arthritis Rheum. 2010, 62, 3496–3503. [Google Scholar] [CrossRef] [PubMed]
- Kiene, M.; Csernok, E.; Muller, A.; Metzler, C.; Trabandt, A.; Gross, W.L. Elevated interleukin-4 and interleukin-13 production by T cell lines from patients with Churg-Strauss syndrome. Arthritis Rheum. 2001, 44, 469–473. [Google Scholar] [CrossRef]
- Jakiela, B.; Sanak, M.; Szczeklik, W.; Sokolowska, B.; Plutecka, H.; Mastalerz, L.; Musial, J.; Szczeklik, A. Both Th2 and Th17 responses are involved in the pathogenesis of Churg-Strauss syndrome. Clin. Exp. Rheumatol. 2011, 29, S23–S34. [Google Scholar] [PubMed]
- Tsurikisawa, N.; Oshikata, C.; Tsuburai, T.; Sugano, S.; Nakamura, Y.; Shimoda, T.; Tamama, S.; Adachi, K.; Horita, A.; Saito, I.; et al. Th17 cells reflect colon submucosal pathologic changes in active eosinophilic granulomatosis with polyangiitis. BMC Immunol. 2015, 16, 75. [Google Scholar] [CrossRef]
- Free, M.E.; Bunch, D.O.; McGregor, J.A.; Jones, B.E.; Berg, E.A.; Hogan, S.L.; Hu, Y.; Preston, G.A.; Jennette, J.C.; Falk, R.J.; et al. Patients with antineutrophil cytoplasmic antibody-associated vasculitis have defective Treg cell function exacerbated by the presence of a suppression-resistant effector cell population. Arthritis Rheum. 2013, 65, 1922–1933. [Google Scholar] [CrossRef]
- Emmi, G.; Silvestri, E.; Bella, C.D.; Grassi, A.; Benagiano, M.; Cianchi, F.; Squatrito, D.; Cantarini, L.; Emmi, L.; Selmi, C.; et al. Cytotoxic Th1 and Th17 cells infiltrate the intestinal mucosa of Behcet patients and exhibit high levels of TNF-alpha in early phases of the disease. Medicine 2016, 95, e5516. [Google Scholar] [CrossRef]
- Belkaid, Y.; Bouladoux, N.; Hand, T.W. Effector and memory T cell responses to commensal bacteria. Trends Immunol. 2013, 34, 299–306. [Google Scholar] [CrossRef]
- Van Damme, N.; De Vos, M.; Baeten, D.; Demetter, P.; Mielants, H.; Verbruggen, G.; Cuvelier, C.; Veys, E.M.; De Keyser, F. Flow cytometric analysis of gut mucosal lymphocytes supports an impaired Th1 cytokine profile in spondyloarthropathy. Ann. Rheum. Dis. 2001, 60, 495–499. [Google Scholar] [CrossRef]
- Annunziato, F.; Cosmi, L.; Santarlasci, V.; Maggi, L.; Liotta, F.; Mazzinghi, B.; Parente, E.; Fili, L.; Ferri, S.; Frosali, F.; et al. Phenotypic and functional features of human Th17 cells. J. Exp. Med. 2007, 204, 1849–1861. [Google Scholar] [CrossRef]

| Participants’ Characteristics at Time of Sampling | EGPA, n (% Out of 29) | Controls, n (% Out of 9) | p-Value |
|---|---|---|---|
| Age (years; median (IQR)) | 58 (55) | 62 (30) | 0.868 |
| Female sex | 16 (55%) | 4 (44%) | 0.980 |
| ANCA positivity | 10 (34%) | - | |
| Disease manifestations in the medical history | |||
| Pulmonary | 28 (97%) | - | |
| ENT | 24 (83%) | - | |
| Neurological | 18 (62%) | - | |
| General | 17 (59%) | - | |
| Cutaneous | 11 (38%) | - | |
| Cardiac | 8 (28%) | - | |
| Gastrointestinal | 7 (24%) | - | |
| Renal | 1 (3%) | - | |
| Current active disease | 22 (76%) | - | |
| Eosinophilia (>500 cell/mm3) | 10 (35%) | - | |
| Ongoing pharmacological treatments | |||
| Systemic glucocorticoid | 28 (96%) | - | |
| DMARDs | 22 (76%) | - | |
| Mycophenolate mofetil | 5 (17%) | - | |
| Cyclosporine | 3 (10%) | - | |
| Azathioprine | 2 (7%) | - | |
| Cyclophosphamide | 1 (3%) | - | |
| Rituximab | 1 (3%) | - | |
| Intravenous Immunoglobulin | 2 (7%) | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niccolai, E.; Bettiol, A.; Baldi, S.; Silvestri, E.; Di Gloria, L.; Bello, F.; Nannini, G.; Ricci, F.; Nicastro, M.; Ramazzotti, M.; et al. Gut Microbiota and Associated Mucosal Immune Response in Eosinophilic Granulomatosis with Polyangiitis (EGPA). Biomedicines 2022, 10, 1227. https://doi.org/10.3390/biomedicines10061227
Niccolai E, Bettiol A, Baldi S, Silvestri E, Di Gloria L, Bello F, Nannini G, Ricci F, Nicastro M, Ramazzotti M, et al. Gut Microbiota and Associated Mucosal Immune Response in Eosinophilic Granulomatosis with Polyangiitis (EGPA). Biomedicines. 2022; 10(6):1227. https://doi.org/10.3390/biomedicines10061227
Chicago/Turabian StyleNiccolai, Elena, Alessandra Bettiol, Simone Baldi, Elena Silvestri, Leandro Di Gloria, Federica Bello, Giulia Nannini, Federica Ricci, Maria Nicastro, Matteo Ramazzotti, and et al. 2022. "Gut Microbiota and Associated Mucosal Immune Response in Eosinophilic Granulomatosis with Polyangiitis (EGPA)" Biomedicines 10, no. 6: 1227. https://doi.org/10.3390/biomedicines10061227
APA StyleNiccolai, E., Bettiol, A., Baldi, S., Silvestri, E., Di Gloria, L., Bello, F., Nannini, G., Ricci, F., Nicastro, M., Ramazzotti, M., Vaglio, A., Bartolucci, G., Emmi, G., Amedei, A., & Prisco, D. (2022). Gut Microbiota and Associated Mucosal Immune Response in Eosinophilic Granulomatosis with Polyangiitis (EGPA). Biomedicines, 10(6), 1227. https://doi.org/10.3390/biomedicines10061227








