Frequency of the Dopamine Receptor D3 (rs6280) vs. Opioid Receptor µ1 (rs1799971) Polymorphic Risk Alleles in Patients with Opioid Use Disorder: A Preponderance of Dopaminergic Mechanisms?
Abstract
1. Introduction
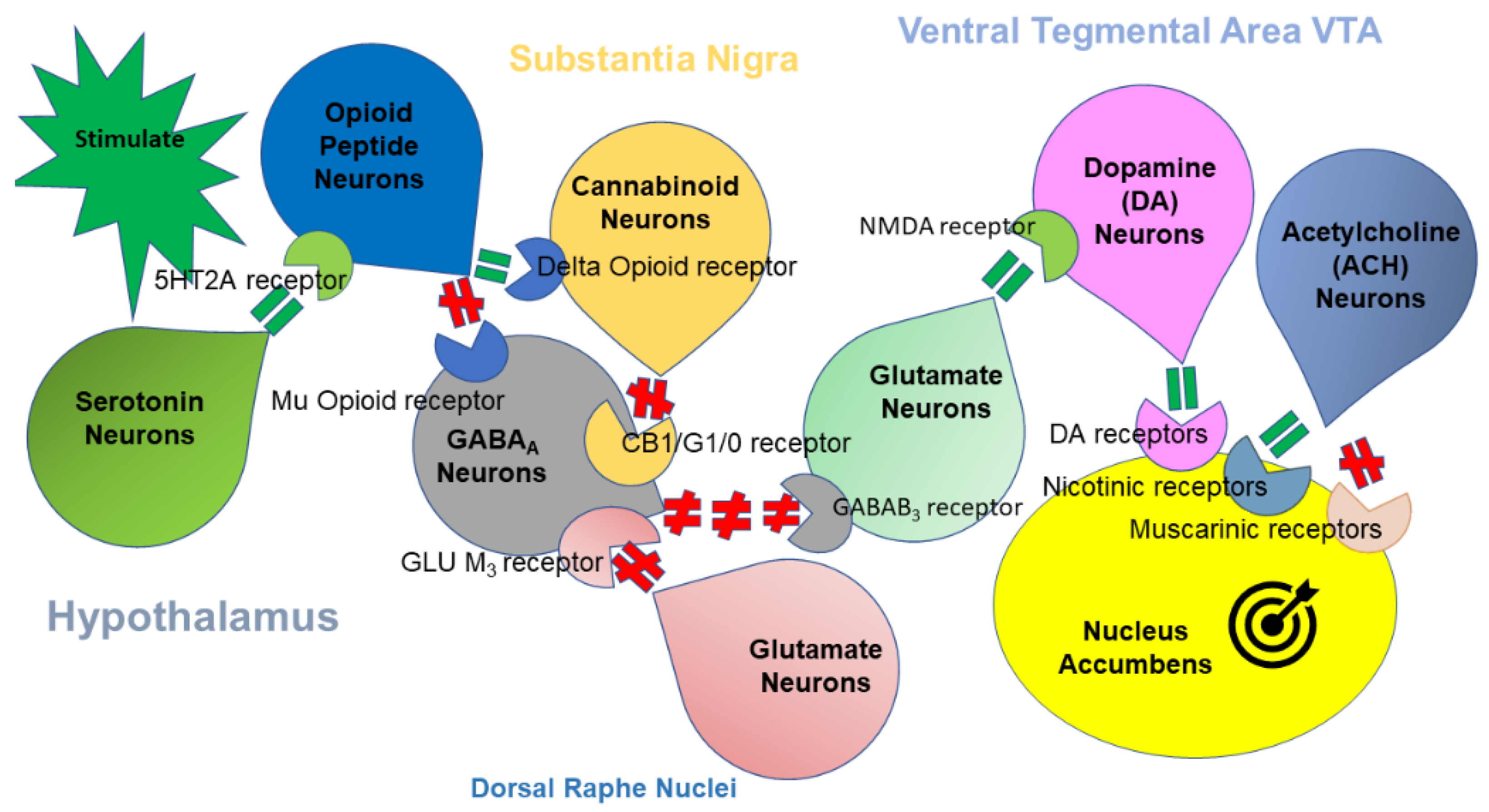
1.1. Brain Reward Function
1.2. Dopaminergic Aspects of Addictive Behvaior
1.3. Dopamine D3 Receptor Function and Addiction Vulnerability
1.4. Neurogenetics of Dopamine D3 Receptor in Opioid Use Disorder
1.5. Neurogenetics of the Opioid Receptor µ1 in Opioid Use Disorder
1.6. Genetic Addiction Risk Severity
1.7. Different Allelic Frequencies of the Dopamine D3 Receptor and the Opioid Receptor in Opioid Use Disorder
1.7.1. Study 1: Opioid Use Disorder Patients in Pain Clinics
1.7.2. Study 2: Opioid Use Disorder Patients at Howard University
1.7.3. Combined Analyses of Pain OUD Study #1 and HU- OUD Study #2
2. Discussion
3. Summary
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Azadfard, M.; Huecker, M.R.; Leaming, J.M. Opioid Addiction. Available online: https://pubmed.ncbi.nlm.nih.gov/28846246/ (accessed on 20 March 2022).
- WISQARS (Web-Based Injury Statistics Query and Reporting System)/Injury Center/Cdc. Available online: http://www.cdc.gov/injury/wisqars/index.html (accessed on 19 March 2022).
- Lopez, G. A Rising Death Toll. Available online: https://www.nytimes.com/2022/02/13/briefing/opioids-drug-overdose-death-toll.html (accessed on 20 March 2022).
- Wilcox, H.C.; Conner, K.R.; Caine, E.D. Association of Alcohol and Drug Use Disorders and Completed Suicide: An Empirical Review of Cohort Studies. Drug Alcohol Depend. 2004, 76, S11–S19. [Google Scholar] [CrossRef] [PubMed]
- Harper, S.; Riddell, C.A.; King, N.B. Declining Life Expectancy in the United States: Missing the Trees for the Forest. Annu. Rev. Public Health 2021, 42, 381–403. [Google Scholar] [PubMed]
- Bech, A.B.; Clausen, T.; Waal, H.; Šaltytė Benth, J.; Skeie, I. Mortality and Causes of Death among Patients with Opioid Use Disorder Receiving Opioid Agonist Treatment: A National Register Study. BMC Health Serv. Res. 2019, 19, 1–10. [Google Scholar]
- Mason, M.; Soliman, R.; Kim, H.S.; Post, L.A. Disparities by Sex and Race and Ethnicity in Death Rates Due to Opioid Overdose among Adults 55 Years or Older, 1999 to 2019. Jama Netw. Open 2022, 5, e2142982. [Google Scholar] [PubMed]
- Larochelle, M.R.; Slavova, S.; Root, E.D.; Feaster, D.J.; Ward, P.J.; Selk, S.C.; Knott, C.; Villani, J.; Samet, J.H. Disparities in Opioid Overdose Death Trends by Race/Ethnicity, 2018–2019, from the Healing Communities Study. Am. J. Public Health 2021, 111, 1851–1854. [Google Scholar] [CrossRef]
- Bohnert, K.M.; Ilgen, M.A.; Louzon, S.; McCarthy, J.F.; Katz, I.R. Substance Use Disorders and the Risk of Suicide Mortality among Men and Women in the US Veterans Health Administration. Addiction 2017, 112, 1193–1201. [Google Scholar] [CrossRef]
- Volkow, N.D.; Collins, F.S. The Role of Science in Addressing the Opioid Crisis. New Engl. J. Med. 2017, 377, 391–394. [Google Scholar] [CrossRef]
- Volkow, N.D.; Morales, M. The Brain on Drugs: From Reward to Addiction. Cell 2015, 162, 712–725. [Google Scholar] [CrossRef]
- Mohebi, A.; Pettibone, J.R.; Hamid, A.A.; Wong, J.-M.T.; Vinson, L.T.; Patriarchi, T.; Tian, L.; Kennedy, R.T.; Berke, J.D. Dissociable Dopamine Dynamics for Learning and Motivation. Nature 2019, 570, 65–70. [Google Scholar]
- Berridge, K.C.; Robinson, T.E.; Aldridge, J.W. Dissecting Components of Reward: ‘Liking’, ‘Wanting’, and Learning. Curr. Opin. Pharmacol. 2009, 9, 65–73. [Google Scholar] [CrossRef]
- Elman, I.; Borsook, D.; Volkow, N.D. Pain and Suicidality: Insights from Reward and Addiction Neuroscience. Prog. Neurobiol. 2013, 109, 1–27. [Google Scholar] [CrossRef]
- Elman, I.; Zubieta, J.-K.; Borsook, D. The Missing P in Psychiatric Training. Arch. Gen. Psychiatry 2011, 68, 12. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, J.M.; Rauscher, N.A.; Cheer, J.F.; Oleson, E.B. A Role for Phasic Dopamine Release within the Nucleus Accumbens in Encoding Aversion: A Review of the Neurochemical Literature. ACS Chem. Neurosci. 2014, 6, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Olsen, C.M. Natural Rewards, Neuroplasticity, and Non-Drug Addictions. Neuropharmacology 2011, 61, 1109–1122. [Google Scholar] [CrossRef] [PubMed]
- Elman, I.; Borsook, D. Common Brain Mechanisms of Chronic Pain and Addiction. Neuron 2016, 89, 11–36. [Google Scholar] [CrossRef]
- Biliński, P.; Wojtyła, A.; Kapka-Skrzypczak, L.; Chwedorowicz, R.; Cyranka, M.; Studziński, T. Epigenetic Regulation in Drug Addiction. Ann. Agric. Environ. Med. 2012, 19, 491–496. [Google Scholar]
- Blum, K.; Baron, D.; McLaughlin, T.; Gold, M.S. Molecular Neurological Correlates of Endorphinergic/Dopaminergic Mechanisms in Reward Circuitry Linked to Endorphinergic Deficiency Syndrome (EDS). J. Neurol. Sci. 2020, 411, 116733. [Google Scholar] [CrossRef]
- Elman, I.; Borsook, D. The Failing Cascade: Comorbid Post Traumatic Stress-and Opioid Use Disorders. Neurosci. Biobehav. Rev. 2019, 103, 374–383. [Google Scholar] [CrossRef]
- Blum, K. Allelic Association of Human Dopamine D2 Receptor Gene in Alcoholism. JAMA 1990, 263, 2055. [Google Scholar] [CrossRef]
- Cadet, J.L.; Jayanthi, S. Epigenetics of Addiction. In Encyclopedia of Behavioral Neuroscience, 2nd ed.; Elsevier: New York, NY, USA, 2022; pp. 383–389. [Google Scholar]
- Robison, A.J.; Nestler, E.J. Transcriptional and Epigenetic Mechanisms of Addiction. Nat. Rev. Neurosci. 2011, 12, 623–637. [Google Scholar] [CrossRef]
- Numan, S.; Lane-Ladd, S.B.; Zhang, L.; Lundgren, K.H.; Russell, D.S.; Seroogy, K.B.; Nestler, E.J. Differential Regulation of Neurotrophin Andtrkreceptor Mrnas in Catecholaminergic Nuclei during Chronic Opiate Treatment and Withdrawal. J. Neurosci. 1998, 18, 10700–10708. [Google Scholar] [CrossRef] [PubMed]
- Gaffney, R.J.; O’Dwyer, T.P.; Maguire, A.J. Bezold’s Abscess. J. Laryngol. Otol. 1991, 105, 765–766. [Google Scholar] [CrossRef] [PubMed]
- Volkow, N.D. Addiction Should Be Treated, Not Penalized. Neuropsychopharmacology 2021, 46, 2048–2050. [Google Scholar] [CrossRef] [PubMed]
- Blum, K. Genetic Addiction Risk Score Gars Trade a Predictor of Vulnerability to Opioid Dependence. Front. Biosci. 2018, 10, 175–196. [Google Scholar] [CrossRef]
- Gardner, E.L. Addiction and Brain Reward and Antireward Pathways. Chronic Pain Addict. 2011, 30, 22–60. [Google Scholar]
- Borsook, D.; Linnman, C.; Faria, V.; Strassman, A.M.; Becerra, L.; Elman, I. Reward Deficiency and Anti-Reward in Pain Chronification. Neurosci. Biobehav. Rev. 2016, 68, 282–297. [Google Scholar] [CrossRef]
- Elman, I.; Lowen, S.; Frederick, B.B.; Chi, W.; Becerra, L.; Pitman, R.K. Functional Neuroimaging of Reward Circuitry Responsivity to Monetary Gains and Losses in Posttraumatic Stress Disorder. Biol. Psychiatry 2009, 66, 1083–1090. [Google Scholar] [CrossRef]
- Elman, I.; Borsook, D.; Lukas, S.E. Food Intake and Reward Mechanisms in Patients with Schizophrenia: Implications for Metabolic Disturbances and Treatment with Second-Generation Antipsychotic Agents. Neuropsychopharmacology 2006, 31, 2091–2120. [Google Scholar] [CrossRef]
- Elman, I.; Tschibelu, E.; Borsook, D. Psychosocial Stress and Its Relationship to Gambling Urges in Individuals with Pathological Gambling. Am. J. Addict. 2010, 19, 332–339. [Google Scholar] [CrossRef]
- Volkow, N.D.; Koob, G. Brain Disease Model of Addiction: Why Is It so Controversial? Lancet Psychiatry 2015, 2, 677–679. [Google Scholar] [CrossRef]
- Hancock, D.B.; Markunas, C.A.; Bierut, L.J.; Johnson, E.O. Human Genetics of Addiction: New Insights and Future Directions. Curr. Psychiatry Rep. 2018, 20, 8. [Google Scholar] [CrossRef] [PubMed]
- Blum, K.; Oscar-Berman, M.; Demetrovics, Z.; Barh, D.; Gold, M.S. Genetic Addiction Risk Score (GARS): Molecular Neurogenetic Evidence for Predisposition to Reward Deficiency Syndrome (RDS). Mol. Neurobiol. 2014, 50, 765–796. [Google Scholar] [CrossRef] [PubMed]
- Blum, K.; Oscar-Berman, M.; Barh, D.; Giordano, J.; Gold, M. Dopamine Genetics and Function in Food and Substance Abuse. J. Genet. Syndr. Gene Ther. 2013, 4, 1000121. [Google Scholar] [PubMed]
- Chavez, A.; Scheiman, J.; Vora, S.; Pruitt, B.W.; Tuttle, M.; PR Iyer, E.; Lin, S.; Kiani, S.; Guzman, C.D.; Wiegand, D.J.; et al. Highly Efficient Cas9-Mediated Transcriptional Programming. Nat. Methods 2015, 12, 326–328. [Google Scholar] [CrossRef] [PubMed]
- Prieto, G.A. Abnormalities of Dopamine D3 Receptor Signaling in the Diseased Brain. J. Cent. Nerv. Syst. Dis. 2017, 9, 117957351772633. [Google Scholar] [CrossRef]
- Shao, D.; Cao, Z.; Fu, Y.; Yang, H.; Gao, P.; Zheng, P.; Lai, B. Projection from the Basolateral Amygdala to the Anterior Cingulate Cortex Facilitates the Consolidation of Long-Term Withdrawal Memory. Addict. Biol. 2021, 26, e13048. [Google Scholar] [CrossRef]
- Wang, S. Historical Review: Opiate Addiction and Opioid Receptors. Cell Transpl. 2018, 28, 233–238. [Google Scholar] [CrossRef]
- Rosen, L.G.; Rushlow, W.J.; Laviolette, S.R. Opiate Exposure State Controls Dopamine D3 Receptor and CDK5/Calcineurin Signaling in the Basolateral Amygdala during Reward and Withdrawal Aversion Memory Formation. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2017, 79, 59–66. [Google Scholar] [CrossRef]
- Rice, O.V.; Ashby, C.R.; Dixon, C.; Laurenzo, W.; Hayden, J.; Song, R.; Li, J.; Tiwari, A.K.; Gardner, E.L. Selective Dopamine D3 Receptor Antagonism Significantly Attenuates Stress-Induced Immobility in a Rat Model of Post-Traumatic Stress Disorder. Synapse 2018, 72, e22035. [Google Scholar] [CrossRef]
- Heidbreder, C.A.; Gardner, E.L.; Xi, Z.-X.; Thanos, P.K.; Mugnaini, M.; Hagan, J.J.; Ashby, C.R. The Role of Central Dopamine D3 Receptors in Drug Addiction: A Review of Pharmacological Evidence. Brain Res. Rev. 2005, 49, 77–105. [Google Scholar] [CrossRef]
- Vorel, S.R.; Ashby, C.R.; Paul, M.; Liu, X.; Hayes, R.; Hagan, J.J.; Middlemiss, D.N.; Stemp, G.; Gardner, E.L. Dopamine D3 receptor Antagonism Inhibits Cocaine-Seeking and Cocaine-Enhanced Brain Reward in Rats. J. Neurosci. 2002, 22, 9595–9603. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.G.; Newman, A.H.; Gardner, E.L.; Ashby, C.R.; Heidbreder, C.A.; Pak, A.C.; Peng, X.-Q.; Xi, Z.-X. Acute Administration of SB-277011A, NGB 2904, or BP 897 Inhibits Cocaine Cue-Induced Reinstatement of Drug-Seeking Behavior in Rats: Role of Dopamine D3 Receptors. Synapse 2005, 57, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Heidbreder, C.A.; Andreoli, M.; Marcon, C.; Thanos, P.K.; Ashby, C.R.; Gardner, E.L. Role of Dopamine D3 Receptors in the Addictive Properties of Ethanol. Drugs Today 2004, 40, 355. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xi, Z.-X.; Gilbert, J.; Campos, A.C.; Kline, N.; Ashby, C.R.; Hagan, J.J.; Heidbreder, C.A.; Gardner, E.L. Blockade of Mesolimbic Dopamine D3 Receptors Inhibits Stress-Induced Reinstatement of Cocaine-Seeking in Rats. Psychopharmacology 2004, 176, 57–65. [Google Scholar] [CrossRef]
- Zhan, J.; Jordan, C.J.; Bi, G.-h.; He, X.-h.; Gardner, E.L.; Wang, Y.-L.; Xi, Z.-X. Genetic Deletion of the Dopamine D3 Receptor Increases Vulnerability to Heroin in Mice. Neuropharmacology 2018, 141, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Jordan, C.J.; Humburg, B.; Rice, M.; Bi, G.-H.; You, Z.-B.; Shaik, A.B.; Cao, J.; Bonifazi, A.; Gadiano, A.; Rais, R.; et al. The Highly Selective Dopamine D R Antagonist, R-VK4-40 Attenuates Oxycodone Reward and Augments Analgesia in Rodents. Neuropharmacology 2019, 158, 107597. [Google Scholar] [CrossRef] [PubMed]
- Ragia, G.; Veresies, I.; Veresie, L.; Veresies, K.; Manolopoulos, V.G. Association Study of DRD2 A2/A1, DRD3 ser9gly, DβH −1021c>T, oprm1 A118G and GRIK1 RS2832407C>a Polymorphisms with Alcohol Dependence. Drug Metab. Pers. Ther. 2016, 31, 143–150. [Google Scholar] [CrossRef]
- Sadat-Shirazi, M.-S.; Zarrindast, M.-R.; Daneshparvar, H.; Ziaie, A.; Fekri, M.; Abbasnezhad, E.; Ashabi, G.; Khalifeh, S.; Vousooghi, N. Alteration of Dopamine Receptors Subtypes in the Brain of Opioid Abusers: A Postmortem Study in Iran. Neurosci. Lett. 2018, 687, 169–176. [Google Scholar] [CrossRef]
- Liang, J.; Zheng, X.; Chen, J.; Li, Y.; Xing, X.; Bai, Y.; Li, Y. Roles of BDNF, Dopamine D3 Receptors, and Their Interactions in the Expression of Morphine-Induced Context-Specific Locomotor Sensitization. Eur. Neuropsychopharmacol. 2011, 21, 825–834. [Google Scholar] [CrossRef]
- Abijo, T.; Blum, K.; Gondré-Lewis, M.C. Neuropharmacological and Neurogenetic Correlates of Opioid Use Disorder (OUD) as a Function of Ethnicity: Relevance to Precision Addiction Medicine. Curr. Neuropharmacol. 2020, 18, 578–595. [Google Scholar] [CrossRef]
- Kuo, S.C.; Yeh, Y.W.; Chen, C.Y.; Huang, C.C.; Chang, H.A.; Yen, C.H.; Ho, P.S.; Liang, C.S.; Chou, H.W.; Lu, R.B.; et al. DRD3 Variation Associates with Early-Onset Heroin Dependence, but Not Specific Personality Traits. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2014, 51, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Levran, O.; Randesi, M.; da Rosa, J.C.; Ott, J.; Rotrosen, J.; Adelson, M.; Kreek, M.J. Overlapping Dopaminergic Pathway Genetic Susceptibility to Heroin and Cocaine Addictions in African Americans. Ann. Hum. Genet. 2015, 79, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Chen, S.L.; Chen, S.H.; Chu, C.H.; Chang, Y.H.; Lin, S.H.; Huang, S.Y.; Tzeng, N.S.; Kuo, P.H.; Lee, I.H.; et al. Interaction of the DRD3 and BDNF Gene Variants in Subtyped Bipolar Disorder. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2012, 39, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Chen, S.L.; Chen, S.H.; Huang, S.Y.; Tzeng, N.S.; Chang, Y.H.; Wang, C.L.; Lee, I.H.; Yeh, T.L.; Yang, Y.K.; et al. TheCOMTandDRD3genes Interacted in Bipolar I but Not Bipolar II Disorder. World J. Biol. Psychiatry 2010, 12, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kuzhikandathil, E.V. Molecular Characterization of Individual D3 Dopamine Receptor-Expressing Cells Isolated from Multiple Brain Regions of a Novel Mouse Model. Brain Struct. Function 2012, 217, 809–833. [Google Scholar] [CrossRef]
- Vengeliene, V.; Leonardi-Essmann, F.; Perreau-Lenz, S.; Gebicke-Haerter, P.; Drescher, K.; Gross, G.; Spanagel, R. The Dopamine D3 Receptor Plays an Essential Role in Alcohol-Seeking and Relapse. FASEB J. 2006, 20, 2223–2233. [Google Scholar] [CrossRef]
- Mulert, C.; Juckel, G.; Giegling, I.; Pogarell, O.; Leicht, G.; Karch, S.; Mavrogiorgou, P.; Möller, H.-J.; Hegerl, U.; Rujescu, D. A ser9gly Polymorphism in the Dopamine D3 Receptor Gene (DRD3) and Event-Related P300 Potentials. Neuropsychopharmacology 2006, 31, 1335–1344. [Google Scholar] [CrossRef]
- Limosin, F.; Romo, L.; Batel, P.; Adès, J.; Boni, C.; Gorwood, P. Association between Dopamine Receptor D3 Gene BALI Polymorphism and Cognitive Impulsiveness in Alcohol-Dependent Men. Eur. Psychiatry 2005, 20, 304–306. [Google Scholar] [CrossRef]
- Duaux, E.; Gorwood, P.; Griffon, N.; Bourdel, M.-C.; Sautel, F.; Sokoloff, P.; Schwartz, J.-C.; Ades, J.; Lôo, H.; Poirier, M.-F. Homozygosity at the Dopamine D3 Receptor Gene Is Associated with Opiate Dependence. Mol. Psychiatry 1998, 3, 333–336. [Google Scholar] [CrossRef][Green Version]
- Spangler, R.; Goddard, N.L.; Avena, N.M.; Hoebel, B.G.; Leibowitz, S.F. Elevated D3 Dopamine Receptor Mrna in Dopaminergic and Dopaminoceptive Regions of the Rat Brain in Response to Morphine. Mol. Brain Res. 2003, 111, 74–83. [Google Scholar] [CrossRef]
- Comings, D.E.; Gonzalez, N.; Wu, S.; Saucier, G.; Johnson, P.; Verde, R.; MacMurray, J.P. Homozygosity at the Dopamine DRD3 Receptor Gene in Cocaine Dependence. Mol. Psychiatry 1999, 4, 484–487. [Google Scholar] [CrossRef] [PubMed]
- Raggio, M.; González, R.; Hohl, D.; Glesmann, L.; Catanesi, C. Genetic Variations of OPRM1, OPRK1, and COMT Genes and Their Possible Associations with Oral Pain in a Population from Argentina. J. Oral Facial Pain Headache 2018, 32, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Clarke, T.-K.; Crist, R.C.; Kampman, K.M.; Dackis, C.A.; Pettinati, H.M.; O’Brien, C.P.; Oslin, D.W.; Ferraro, T.N.; Lohoff, F.W.; Berrettini, W.H. Low Frequency Genetic Variants in the μ-Opioid Receptor (OPRM1) Affect Risk for Addiction to Heroin and Cocaine. Neurosci. Lett. 2013, 542, 71–75. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hirasawa-Fujita, M.; Bly, M.J.; Ellingrod, V.L.; Dalack, G.W.; Domino, E.F. Genetic Variation of the Mu Opioid Receptor (OPRM1) and Dopamine D2 Receptor (DRD2) Is Related to Smoking Differences in Patients with Schizophrenia but Not Bipolar Disorder. Clin. Schizophr. Relat. Psychoses 2017, 11, 39–48. [Google Scholar] [CrossRef]
- Bond, C.; LaForge, K.S.; Tian, M.; Melia, D.; Zhang, S.; Borg, L.; Gong, J.; Schluger, J.; Strong, J.A.; Leal, S.M.; et al. Single-Nucleotide Polymorphism in the Human Mu Opioid Receptor Gene Alters β-Endorphin Binding and Activity: Possible Implications for Opiate Addiction. Proc. Natl. Acad. Sci. USA 1998, 95, 9608–9613. [Google Scholar] [CrossRef]
- Beer, B.; Erb, R.; Pavlic, M.; Ulmer, H.; Giacomuzzi, S.; Riemer, Y.; Oberacher, H. Association of Polymorphisms in Pharmacogenetic Candidate Genes (oprd1, Gal, ABCB1, OPRM1) with Opioid Dependence in European Population: A Case-Control Study. PLoS ONE 2013, 8, e75359. [Google Scholar] [CrossRef]
- Luo, X.; Kranzler, H.R.; Zhao, H.; Gelernter, J. Haplotypes at the Oprm1 Locus Are Associated with Susceptibility to Substance Dependence in European-Americans. Am. J. Med. Genet. 2003, 120B, 97–108. [Google Scholar] [CrossRef]
- Crowley, J.J.; Oslin, D.W.; Patkar, A.A.; Gottheil, E.; DeMaria, P.A.; O’Brien, C.P.; Berrettini, W.H.; Grice, D.E. A Genetic Association Study of the Mu Opioid Receptor and Severe Opioid Dependence. Psychiatr. Genet. 2003, 13, 169–173. [Google Scholar] [CrossRef]
- Szeto, C.Y.; Tang, N.L.; Lee, D.T.; Stadlin, A. Association between Mu Opioid Receptor Gene Polymorphisms and Chinese Heroin Addicts. Neuroreport 2001, 12, 1103–1106. [Google Scholar] [CrossRef]
- Wachman, E.M.; Hayes, M.J.; Brown, M.S.; Paul, J.; Harvey-Wilkes, K.; Terrin, N.; Huggins, G.S.; Aranda, J.V.; Davis, J.M. Association of OPRM1 and COMT Single-Nucleotide Polymorphisms with Hospital Length of Stay and Treatment of Neonatal Abstinence Syndrome. JAMA 2013, 309, 1821. [Google Scholar] [CrossRef]
- Shi, J.; Hui, L.; Xu, Y.; Wang, F.; Huang, W.; Hu, G. Sequence Variations in the Mu-Opioid Receptor Gene (OPRM1) Associated with Human Addiction to Heroin. Hum. Mutat. 2002, 19, 459–460. [Google Scholar] [CrossRef] [PubMed]
- Crist, R.C.; Doyle, G.A.; Nelson, E.C.; Degenhardt, L.; Martin, N.G.; Montgomery, G.W.; Saxon, A.J.; Ling, W.; Berrettini, W.H. A Polymorphism in the Oprm1 3′-Untranslated Region Is Associated with Methadone Efficacy in Treating Opioid Dependence. Pharm. J. 2016, 18, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Ray, R.; Ruparel, K.; Newberg, A.; Wileyto, E.P.; Loughead, J.W.; Divgi, C.; Blendy, J.A.; Logan, J.; Zubieta, J.-K.; Lerman, C. Human Mu Opioid Receptor ( oprm1 A118G) Polymorphism Is Associated with Brain Mu-Opioid Receptor Binding Potential in Smokers. Proc. Natl. Acad. Sci. USA 2011, 108, 9268–9273. [Google Scholar] [CrossRef] [PubMed]
- Bart, G.; Kreek, M.J.; Ott, J.; LaForge, K.S.; Proudnikov, D.; Pollak, L.; Heilig, M. Increased Attributable Risk Related to a Functional μ-Opioid Receptor Gene Polymorphism in Association with Alcohol Dependence in Central Sweden. Neuropsychopharmacology 2004, 30, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Hall, F.S.; Sora, I.; Uhl, G.R. Ethanol Consumption and Reward Are Decreased in µ-Opiate Receptor Knockout Mice. Psychopharmacology 2001, 154, 43–49. [Google Scholar] [CrossRef]
- Blum, K.; Modestino, E.J.; Gondre-Lewis, M.; Chapman, E.J.; Neary, J.; Siwicki, D.; Baron, D.; Hauser, M.; Smith, D.E.; Kenison Roy, A.; et al. The Benefits of Genetic Addiction Risk Score (GARS™) Testing in Substance Use Disorder (SUD). Int. J. Genom. Data Min. 2018, 2018, 115. [Google Scholar] [CrossRef]
- Haerian, B.S.; Haerian, M.S. Oprm1 rs1799971 Polymorphism and Opioid Dependence: Evidence from a Meta-Analysis. Pharmacogenomics 2013, 14, 813–824. [Google Scholar] [CrossRef]
- Pattullo, G.G. Opioids in acute pain: Towards getting the right balance. Anaesth. Intensive Care 2022, 50, 68–80. [Google Scholar] [CrossRef]
- Fried, L.; Modestino, E.J.; Siwicki, D.; Lott, L.; Thanos, P.K.; Baron, D.; Badgaiyan, R.D.; Ponce, J.V.; Giordano, J.; Downs, W.B.; et al. Hypodopaminergia and “Precision Behavioral Management” (PBM): It Is a Generational Family Affair. Curr. Pharm. Biotechnol. 2020, 21, 528–541. [Google Scholar] [CrossRef]
- dbGaP/Database of Genotypes and Phenotypes/National Center for Biotechnology Information, National Library of Medicine (NCBI/NLM). Available online: https://www.ncbi.nlm.nih.gov/gap (accessed on 20 March 2022).
- RS1799971 RefSNP Report-dbSNP-NCBI. Available online: https://www.ncbi.nlm.nih.gov/snp/rs1799971?horizontal_tab=true (accessed on 20 March 2022).
- RS6280 RefSNP Report-dbSNP-NCBI. Available online: https://www.ncbi.nlm.nih.gov/snp/rs6280?horizontal_tab=true (accessed on 20 March 2022).
- Francès, H.; Le Foll, B.; Diaz, J.; Smirnova, M.; Sokoloff, P. Role of DRD3 in Morphine-Induced Conditioned Place Preference Using DRD3-Knockout Mice. NeuroReport 2004, 15, 2245–2249. [Google Scholar] [CrossRef]
- You, Z.-B.; Gao, J.-T.; Bi, G.-H.; He, Y.; Boateng, C.; Cao, J.; Gardner, E.L.; Newman, A.H.; Xi, Z.-X. The Novel Dopamine D3 Receptor Antagonists/Partial Agonists CAB2-015 and BAK4-54 Inhibit Oxycodone-Taking and Oxycodone-Seeking Behavior in Rats. Neuropharmacology 2017, 126, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Elman, I.; Howard, M.; Borodovsky, J.T.; Mysels, D.; Rott, D.; Borsook, D.; Albanese, M. Metabolic and Addiction Indices in Patients on Opioid Agonist Medication-Assisted Treatment: A Comparison of Buprenorphine and Methadone. Sci. Rep. 2020, 10, 5617. [Google Scholar] [CrossRef] [PubMed]
- Meehl, P.E. Hedonic Capacity: Some Conjectures. Bull. Menn. Clin. 1975, 39, 295–307. [Google Scholar]
- Elman, I.; Upadhyay, J.; Langleben, D.D.; Albanese, M.; Becerra, L.; Borsook, D. Reward and Aversion Processing in Patients with Post-Traumatic Stress Disorder: Functional Neuroimaging with Visual and Thermal Stimuli. Transl. Psychiatry 2018, 8, 240. [Google Scholar] [CrossRef] [PubMed]
- Blum, K.; Chen, A.L.; Oscar-Berman, M.; Chen, T.J.; Lubar, J.; White, N.; Lubar, J.; Bowirrat, A.; Braverman, E.; Schoolfield, J.; et al. Generational Association Studies of DOPAMINERGIC Genes in Reward Deficiency Syndrome (RDS) Subjects: Selecting Appropriate Phenotypes for Reward Dependence Behaviors. Int. J. Environ. Res. Public Health 2011, 8, 4425–4459. [Google Scholar] [CrossRef] [PubMed]
- Volkow, N.; Czernin, J. A Conversation between Nora Volkow and Johannes Czernin. J. Nucl. Med. 2019, 60, 717–720. [Google Scholar] [CrossRef] [PubMed]
- Ettienne, E.B.; Ofoegbu, A.; Maneno, M.K.; Briggs, J.; Ezeude, G.; Williams, S.; Walker, C.; Chapman, E. Pharmacogenomics and Opioid Use Disorder: Clinical Decision Support in an African American Cohort. J. Natl. Med. Assoc. 2019, 111, 674–681. [Google Scholar] [CrossRef]
- Hyman, S.E. The Daunting Polygenicity of Mental Illness: Making a New Map. Philos. Trans. R. Soc. B: Biol. Sci. 2018, 373, 20170031. [Google Scholar] [CrossRef]
- Hyman, S.E.; Malenka, R.C.; Nestler, E.J. Neural Mechanisms of Addiction: The Role of Reward-Related Learning and Memory. Annu. Rev. Neurosci. 2006, 29, 565–598. [Google Scholar] [CrossRef]
- Everitt, B.J.; Robbins, T.W. Neural Systems of Reinforcement for Drug Addiction: From Actions to Habits to Compulsion. Nat. Neurosci. 2005, 8, 1481–1489. [Google Scholar] [CrossRef]
- Niehaus, J.L.; Murali, M.; Kauer, J.A. Drugs of Abuse and Stress Impair LTP at Inhibitory Synapses in the Ventral Tegmental Area. Eur. J. Neurosci. 2010, 32, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Chen, A.L.C.; Stokes, S.D.; Silverman, S.; Bowirrat, A.; Manka, M.; Manka, D.; Miller, D.K.; Perrine, K.; Chen, T.J.H.; et al. Early Intervention of Intravenous KB220IV-Neuroadaptagen Amino-Acid Therapy (NAAT)™ Improves Behavioral Outcomes in a Residential Addiction Treatment Program: A Pilot Study. J. Psychoact. Drugs 2012, 44, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Wurtman, R.J. Daily Rhythms in Tyrosine Transaminase and Other Hepatic Enzymes That Metabolize Amino Acids: Mechanisms and Possible Consequences. Life Sci. 1974, 15, 827–847. [Google Scholar] [CrossRef]
- Wang, Y.; Yin, F.; Guo, H.; Zhang, J.; Yan, P.; Lai, J. The Role of Dopamine D1 and D3 Receptors in N-Methyl-D-Aspartate (Nmda)/Glycineb Site-Regulated Complex Cognitive Behaviors Following Repeated Morphine Administration. Int. J. Neuropsychopharmacol. 2017, 20, 562–574. [Google Scholar] [CrossRef] [PubMed]
- Gingrich, J.A.; Caron, M.G. Recent Advances in the Molecular Biology of Dopamine Receptors. Annu. Rev. Neurosci. 1993, 16, 299–321. [Google Scholar] [CrossRef]
- Li, Y.; Xia, B.; Li, R.; Yin, D.; Liang, W. Changes in Expression of Dopamine, Its Receptor, and Transporter in Nucleus Accumbens of Heroin-Addicted Rats with Brain-Derived Neurotrophic Factor (BDNF) Overexpression. Med. Sci. Monit. 2017, 23, 2805–2815. [Google Scholar] [CrossRef]
- Von Deneen, K.M.; Qin, W.; Liu, P.; Dong, M.; Chen, P.; Xie, H.; Zhang, Y.; Gold, M.S.; Liu, Y.; Tian, J. Connectivity Study of the Neuromechanism of Acute Acupuncture Needling during Fmri in “Overweight” Subjects. Evid. -Based Complement. Altern. Med. 2015, 2015, 384389. [Google Scholar] [CrossRef]
- Johnson, R.J.; Gold, M.S.; Johnson, D.R.; Ishimoto, T.; Lanaspa, M.A.; Zahniser, N.R.; Avena, N.M. Attention-Deficit/Hyperactivity Disorder: Is It Time to Reappraise the Role of Sugar Consumption? Postgrad. Med. 2011, 123, 39–49. [Google Scholar] [CrossRef]
- Blum, K.; Oscar-Berman, M.; Braverman, E.R.; Febo, M.; Li, M.; Gold, M.S. Enhancing Brain Pregnenolone May Protect Cannabis Intoxication but Should Not Be Considered as an Anti-Addiction Therapeutic: Hypothesizing Dopaminergic Blockade and Promoting Anti- Reward. J. Reward Defic. Syndr. 2015, 1, 20–23. [Google Scholar] [CrossRef]
- Dackis, C.A.; Gold, M.S. New Concepts in Cocaine Addiction: The Dopamine Depletion Hypothesis. Neurosci. Biobehav. Rev. 1985, 9, 469–477. [Google Scholar] [CrossRef]
- Dackis, C.A.; Gold, M.S. Addictiveness of Central Stimulants. Adv. Alcohol Subst. Abus. 1990, 9, 9–26. [Google Scholar] [CrossRef]
- Krasnova, I.N.; Chiflikyan, M.; Justinova, Z.; McCoy, M.T.; Ladenheim, B.; Jayanthi, S.; Quintero, C.; Brannock, C.; Barnes, C.; Adair, J.E.; et al. CREB Phosphorylation Regulates Striatal Transcriptional Responses in the Self-Administration Model of Methamphetamine Addiction in the Rat. Neurobiol. Dis. 2013, 58, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Gold, M.S.; Dackis, C.A. New Insights and Treatments: Opiate Withdrawal and Cocaine Addiction. Clin. Ther. 1984, 7, 6–21. [Google Scholar]
- Dackis, C.; Gold, M.S. Neurotransmitter and Neuroendocrine Abnormalities Associated with Cocaine Use. Psychiatr. Med. 1985, 3, 461–483. [Google Scholar] [PubMed]
- Blum, K.; Thompson, B.; Demotrovics, Z.; Femino, J.; Giordano, J.; Oscar-Berman, M.; Teitelbaum, S.; Smith, D.E.; Roy, A.K.; Agan, G.; et al. The Molecular Neurobiology of Twelve Steps Program & Fellowship: Connecting the Dots for Recovery. J. Reward Defic. Syndr. 2015, 1, 46. [Google Scholar] [PubMed]

| Gene | Polymorphism | Study Findings | Reference |
|---|---|---|---|
| DRD3 OMIM 126451 | rs6280 | Significant interaction for BDNF Val66Met Val/Val genotype with, both DRD3 Ser9Gly Ser/Ser and Ser/Gly SNPs in bipolar-II patients (p = 0.027 and 0.006, respectively). | Lee et al. [57,58] |
| DRD3 KO mice | DRD3 knockout mice (DRD3 KO): hypoalgesia, lower morphine-induced tolerance and attenuated withdrawal signs compared with the wild type mice. | Li et al. [59] | |
| rs6280 | Upregulation of DRD3 in the striatum of alcohol preferring (P) and high alcohol drinking (HAD)rats through DNA microarrarys, confirmed by qRT-polymerase chain reaction. | Vengeliene et al. [60] | |
| rs6280 | Respectively decreased and increased parietal and frontal P300 amplitudes in Gly9 homozygotes versus Ser9 carriers. | Mulert et al. [61] | |
| BalI | Impulsive alcohol dependent patients were more frequently heterozygous for DRD3 BalI in comparison to both, alcohol-dependent patients with lower impulsivity ratings (OR = 2.51, p < 0.02) and to healthy controls (OR = 2.32, p < 0.03). | Limosin et al. [62] | |
| High sensation-seeking score was more frequent in homozygous for both alleles than those with a low sensation-seeking score under 24 (p < 0.04) or controls (p = 0.03). | Duaux et al. [63] | ||
| Binging on a sucrose solution increased the expression of DRD3 gene (NAc > caudate, putamen) and decreases that of the DRD2 and of the preproenkephalin and preprotachykinin genes. | Spangler et al. [64] | ||
| MscI/BalI | Increased homozygosity in cocaine dependence (29.8%) vs. controls (46.9%) particularly in those with chronic cocaine consumption for > 10 years (25%) and > 15 years (46.5%). | Comings et al. [65] |
| Gene | Polymorphism (Study Conditions) | Study Findings | Reference |
|---|---|---|---|
| OPRM1 OMIM 610064 | rs1799971 | Independent of session, smokers homozygous for the wild-type OPRM1 A allele exhibited significantly higher levels of non-dominant mu opioid receptor A118G than smokers carrying the G allele in bilateral amygdala, left thalamus, and left anterior cingulate cortex. | Ray et al. [77] |
| rs1799971 | Found a significant association for both A118G and C1031G polymorphisms and opioid dependence. The G allele is more common in the heroin-dependent group (39.5% and 30.8% for A118G and C1031G polymorphisms, respectively) when compared to the controls (29.4% and 21.1% for A118G and C1031G polymorphisms, respectively). *this is the only study reporting C1031G | Szeto et al. [73] | |
| rs1799971 | There was a significant overall association between genotypes with an 118G allele and alcohol dependence (p = 0.0074). The attributable risk for alcohol dependence in subjects with an 118G allele was 11.1% | Bart et al. [78] | |
| OPRM1 KO mice | Wild type mice consumed more alcohol than heterozygous or homozygous MOR KO mice (female KO mice > male KO mice). MOR KO mice exhibited less ethanol reward in a conditioned place preference paradigm (females < males). | Hall et al. [79] |
| Population | All | Male | Female | |
|---|---|---|---|---|
| Number (n) | 121 | 55 (45%) | 66 (55%) | |
| Average Age (n = 121) | 53 | 54 | 53 | |
| Ethnicity | ||||
| Caucasian | 67% | |||
| Hispanic | 17% | |||
| Unknown | 10% | |||
| Black or African American | 4% | |||
| Asian | 2% |
| Population | All | Male | Female | |
|---|---|---|---|---|
| Number (n) | 39 | 28 (72%) | 11(28%) | |
| Ethnicity | ||||
| Black or African American | 97% | |||
| Unknown | 3% |
| Population | All | Male | Female | |
|---|---|---|---|---|
| Number (n) | 160 | 83 (52%) | 76(48%) | |
| Average Age (n = 121) | 53 | 54 | 53 | |
| Ethnicity | ||||
| Caucasian | 51% | |||
| Black or African American | 27% | |||
| Hispanic | 13% | |||
| Unknown | 8% | |||
| Asian | 1% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gondré-Lewis, M.C.; Elman, I.; Alim, T.; Chapman, E.; Settles-Reaves, B.; Galvao, C.; Gold, M.S.; Baron, D.; Kazmi, S.; Gardner, E.; et al. Frequency of the Dopamine Receptor D3 (rs6280) vs. Opioid Receptor µ1 (rs1799971) Polymorphic Risk Alleles in Patients with Opioid Use Disorder: A Preponderance of Dopaminergic Mechanisms? Biomedicines 2022, 10, 870. https://doi.org/10.3390/biomedicines10040870
Gondré-Lewis MC, Elman I, Alim T, Chapman E, Settles-Reaves B, Galvao C, Gold MS, Baron D, Kazmi S, Gardner E, et al. Frequency of the Dopamine Receptor D3 (rs6280) vs. Opioid Receptor µ1 (rs1799971) Polymorphic Risk Alleles in Patients with Opioid Use Disorder: A Preponderance of Dopaminergic Mechanisms? Biomedicines. 2022; 10(4):870. https://doi.org/10.3390/biomedicines10040870
Chicago/Turabian StyleGondré-Lewis, Marjorie C., Igor Elman, Tanya Alim, Edwin Chapman, Beverlyn Settles-Reaves, Carine Galvao, Mark S. Gold, David Baron, Shan Kazmi, Eliot Gardner, and et al. 2022. "Frequency of the Dopamine Receptor D3 (rs6280) vs. Opioid Receptor µ1 (rs1799971) Polymorphic Risk Alleles in Patients with Opioid Use Disorder: A Preponderance of Dopaminergic Mechanisms?" Biomedicines 10, no. 4: 870. https://doi.org/10.3390/biomedicines10040870
APA StyleGondré-Lewis, M. C., Elman, I., Alim, T., Chapman, E., Settles-Reaves, B., Galvao, C., Gold, M. S., Baron, D., Kazmi, S., Gardner, E., Gupta, A., Dennen, C., & Blum, K. (2022). Frequency of the Dopamine Receptor D3 (rs6280) vs. Opioid Receptor µ1 (rs1799971) Polymorphic Risk Alleles in Patients with Opioid Use Disorder: A Preponderance of Dopaminergic Mechanisms? Biomedicines, 10(4), 870. https://doi.org/10.3390/biomedicines10040870









