HREM, RNAseq and Cell Cycle Analyses Reveal the Role of the G2/M-Regulatory Protein, WEE1, on the Survivability of Chicken Embryos during Diapause
Abstract
1. Introduction
2. Materials and Methods
2.1. Egg Collection and Isolation of Blastoderm
2.2. HREM Imaging
2.3. RNAseq Analysis
2.4. Real-Time PCR Analysis
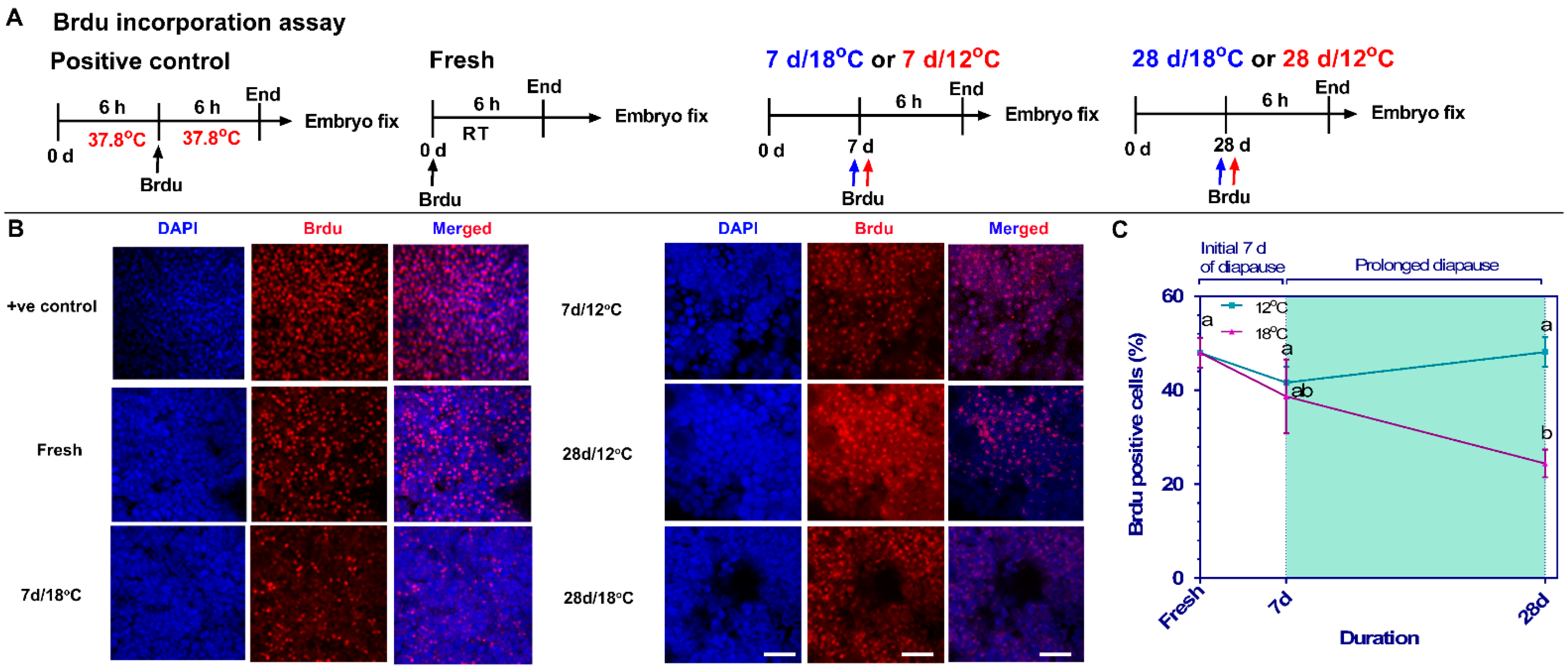
2.5. BrdU Incorporation Assay In-Ovo in Embryos
2.6. Immunohistochemistry
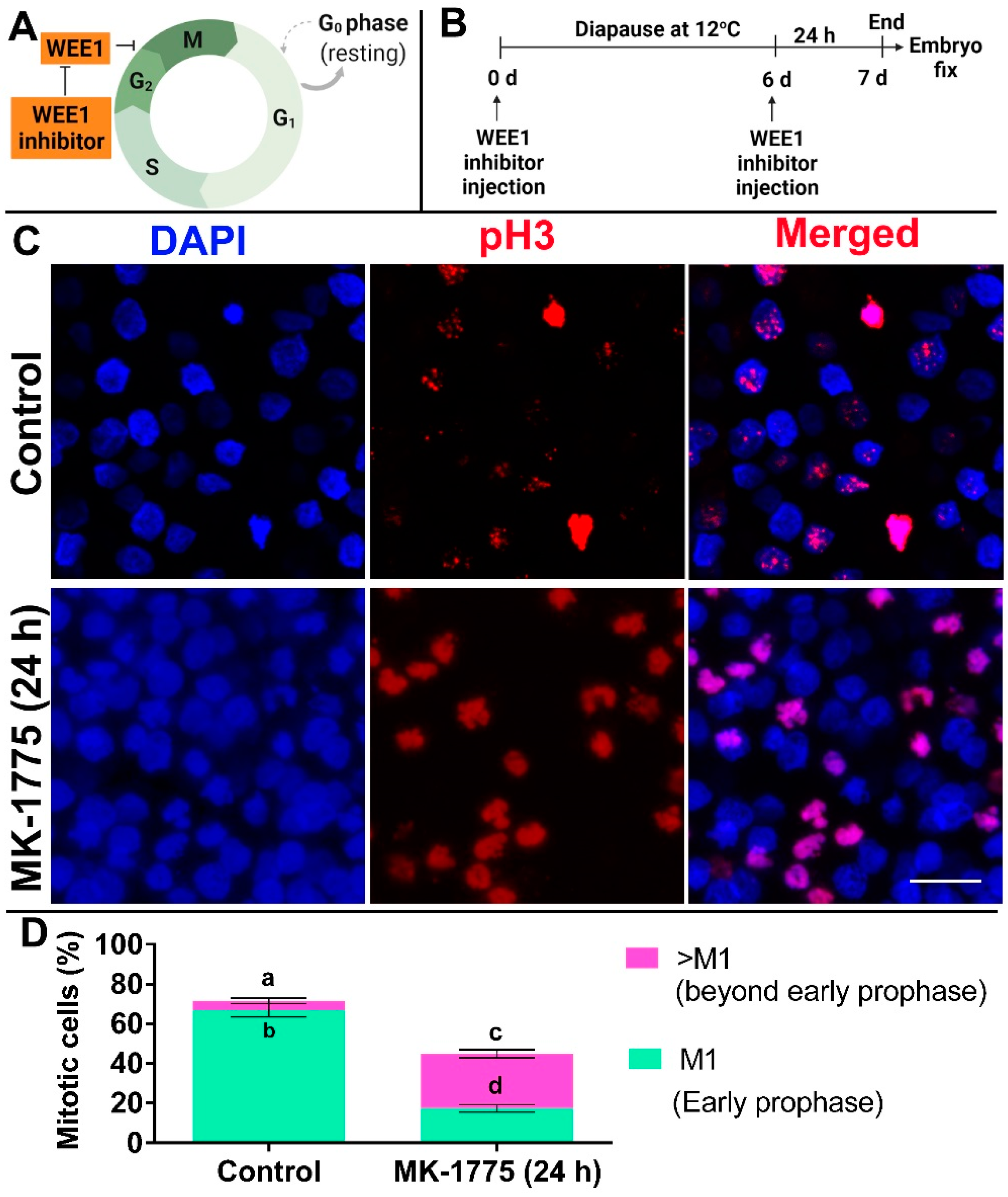
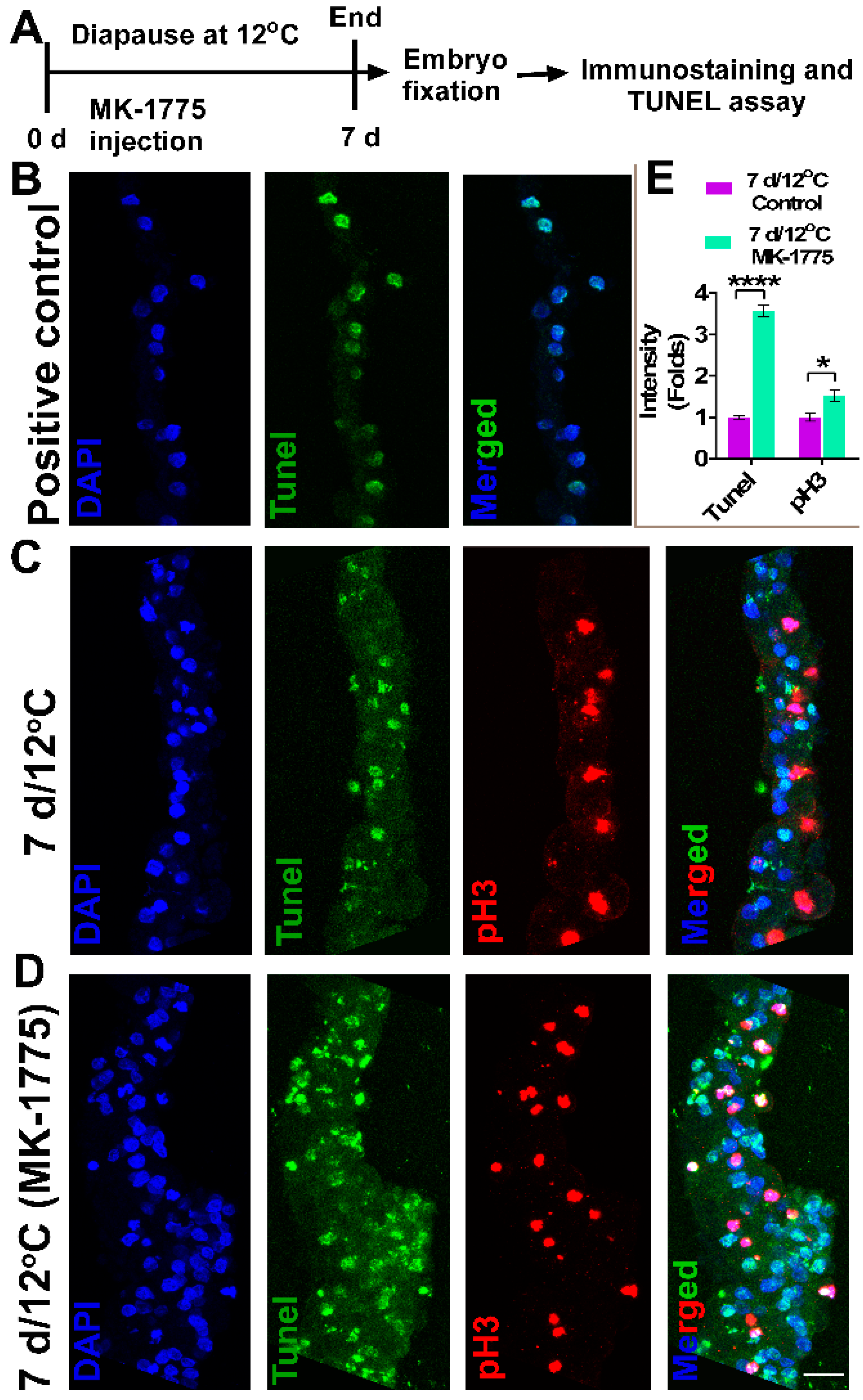
2.7. Embryo Treatment with WEE1 Inhibitor-MK1775
2.8. TUNEL Assay
2.9. Statistical Tests
3. Results
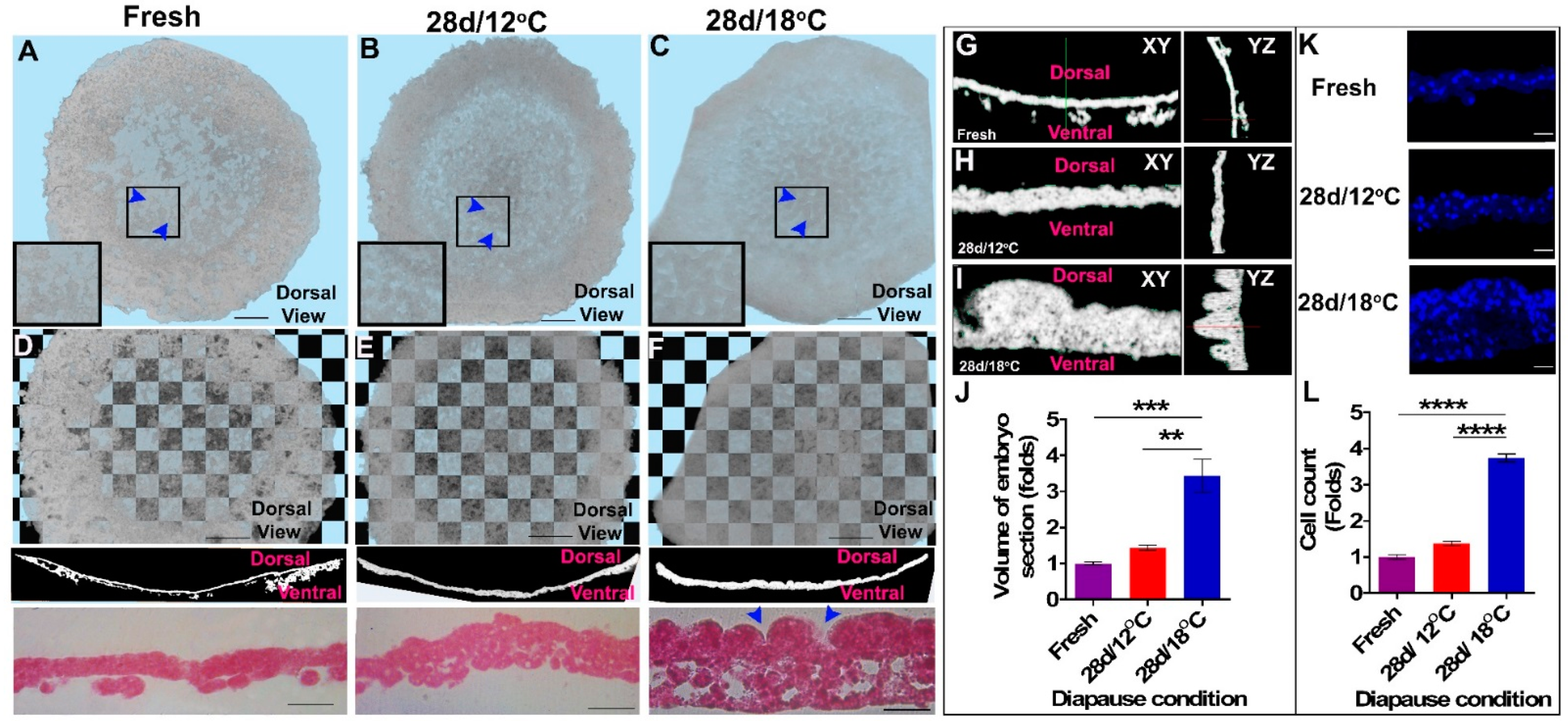
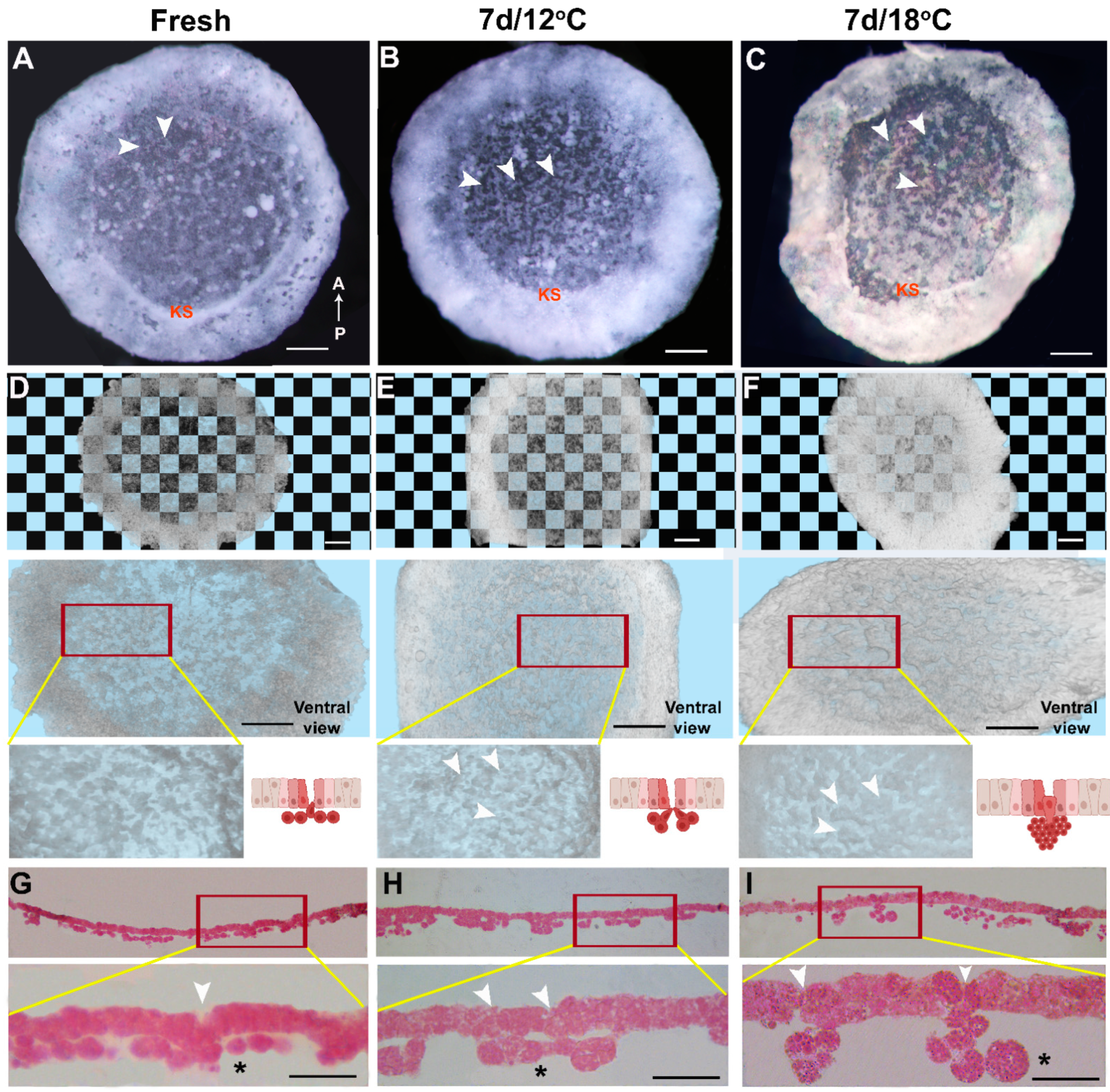
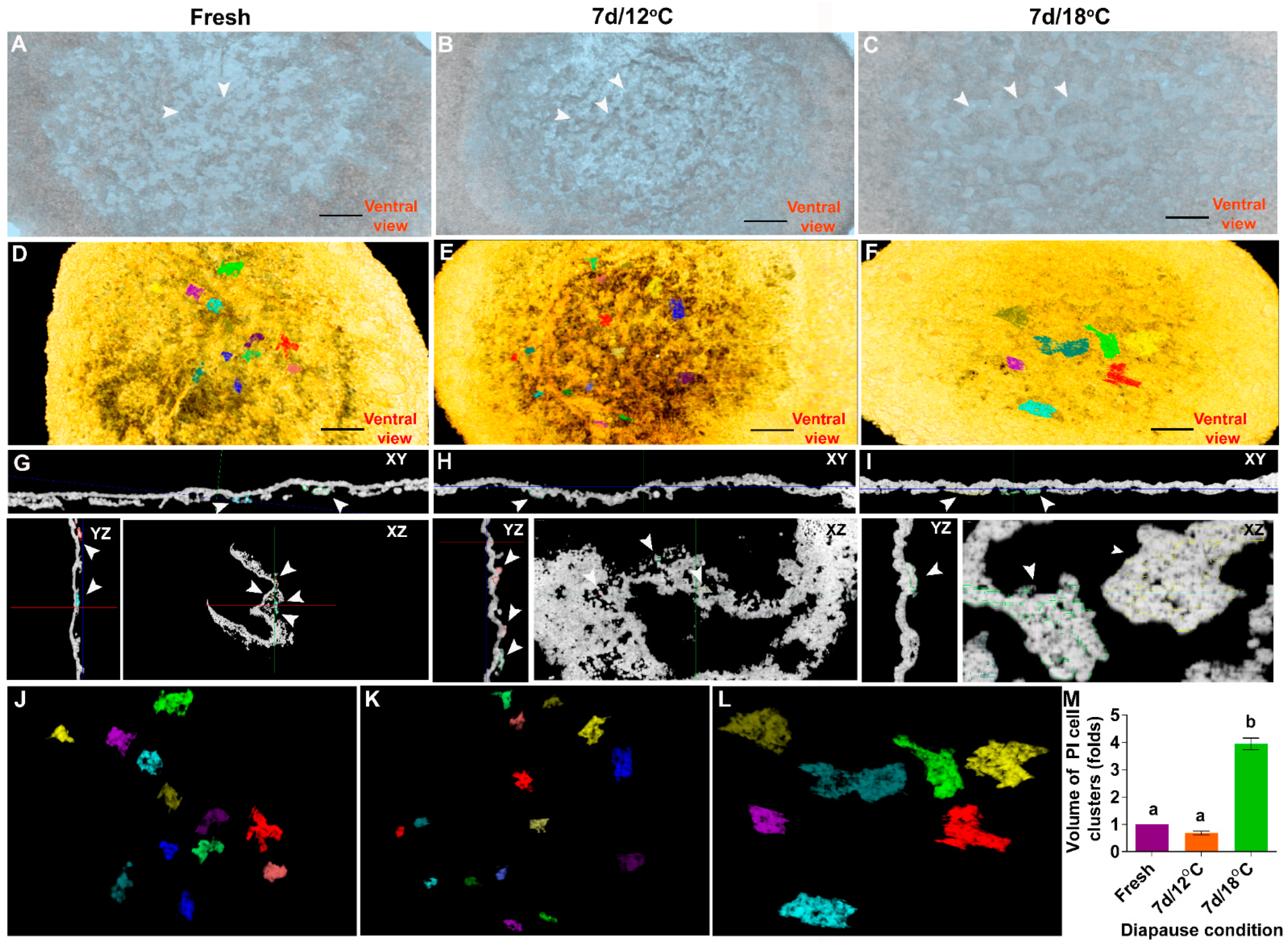
3.1. The Cellular Morphology of the Blastoderm during Diapause
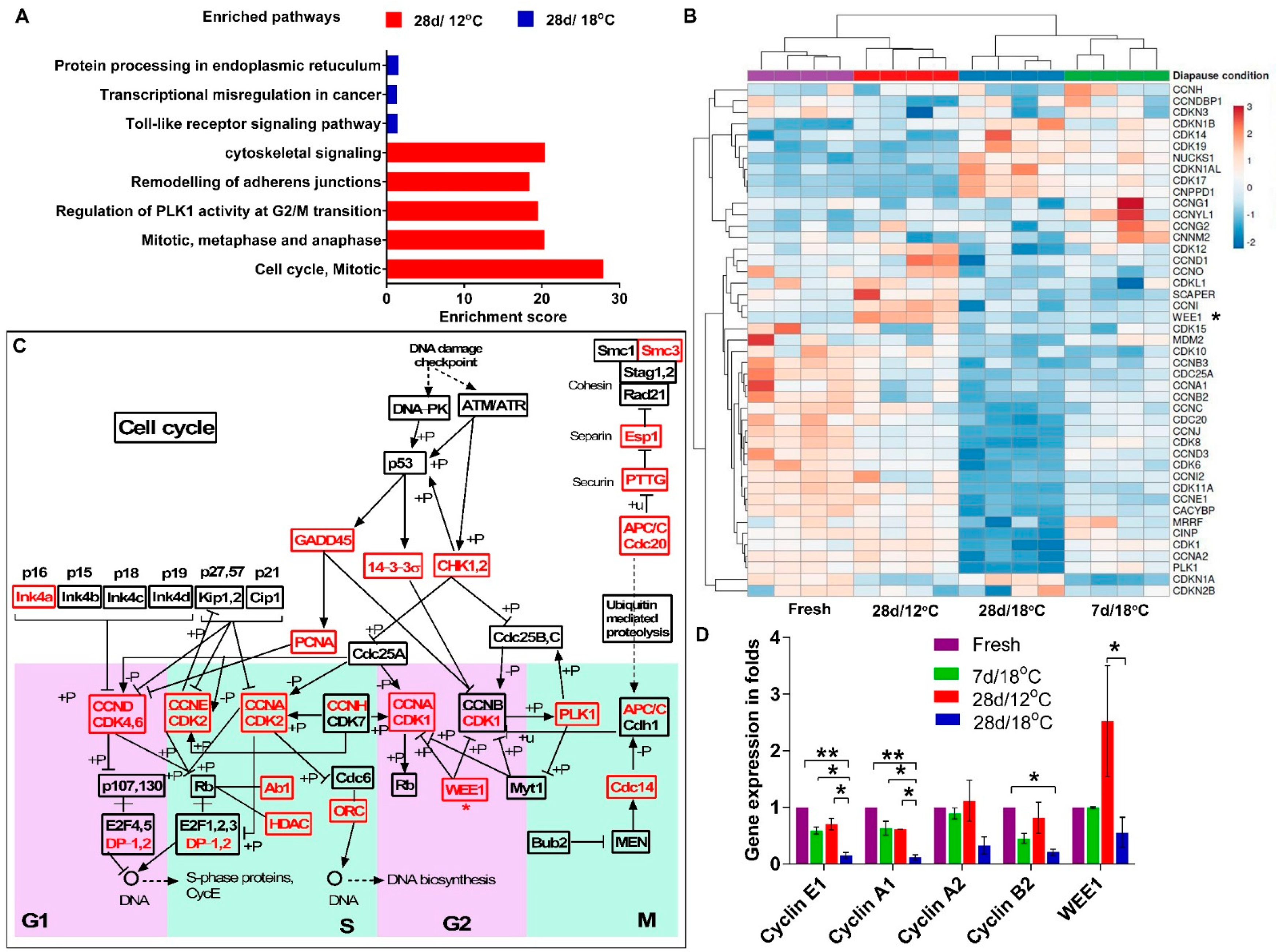
3.2. Transcriptome Profiling Reveals Distinct Regulation in Cell Cycle in Diapaused Embryos at 18 °C and 12 °C
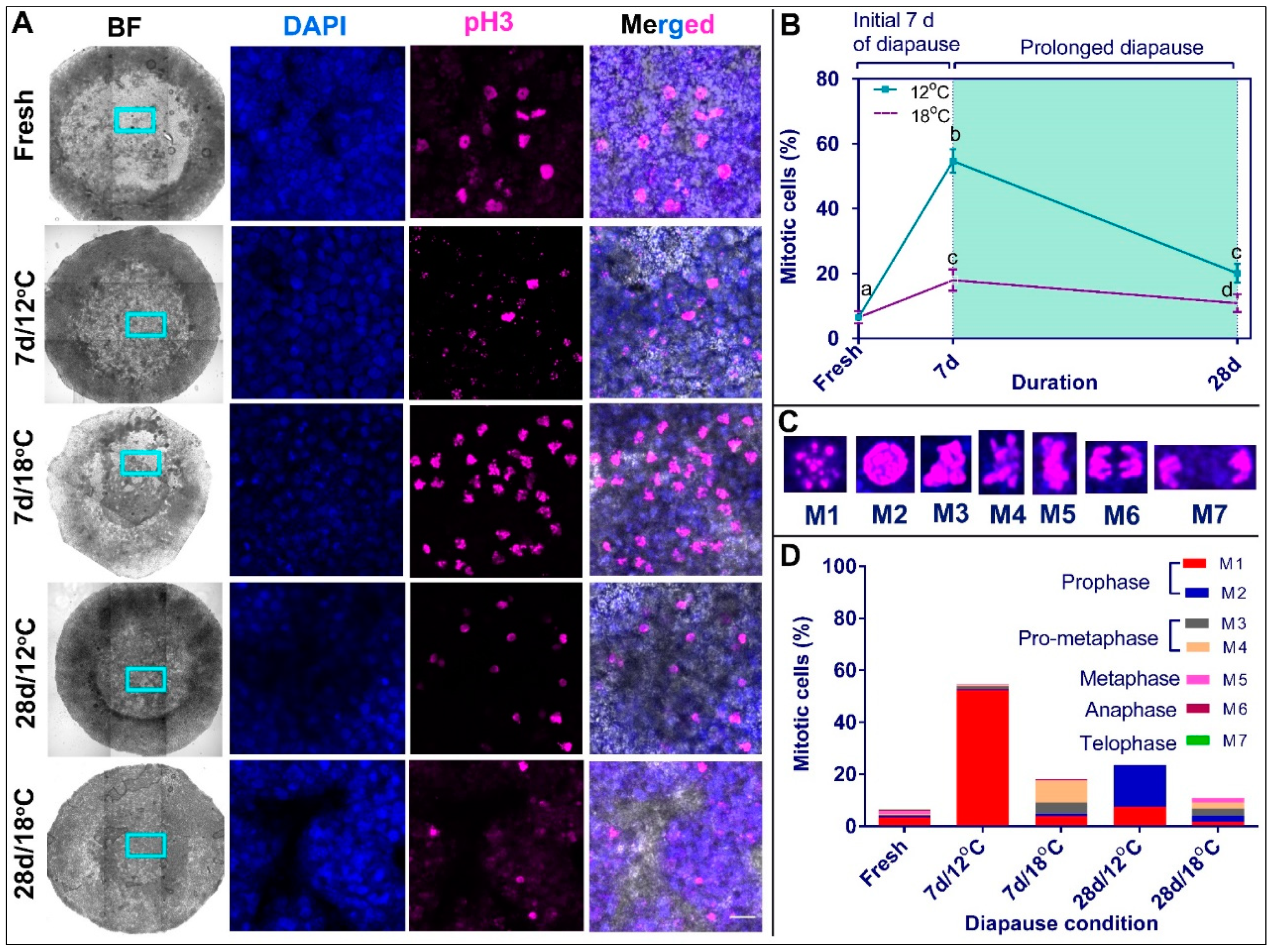
3.3. Mapping the Mitotic Phase of Diapaused Embryos Reveals an Increase in Cell Proliferation at Higher Temperature
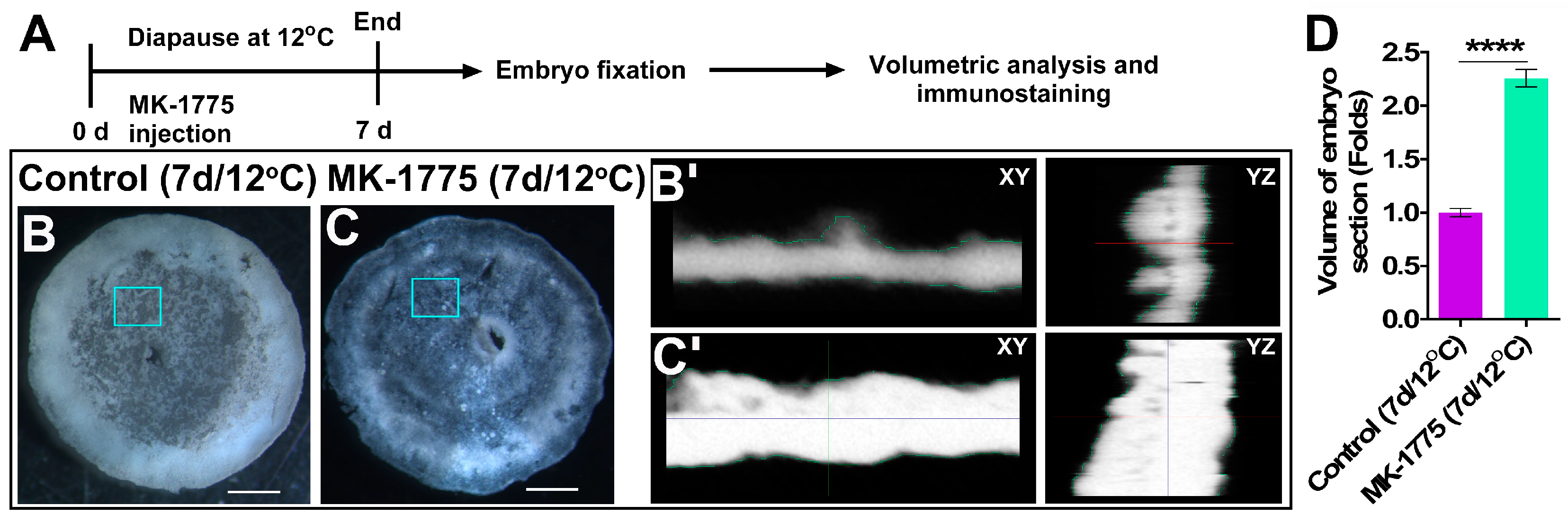
3.4. Inhibition of WEE1 in Embryos Diapaused at 12 °C Results in Cell Cycle Progression beyond Prophase
3.5. Increase in Staining of Cellular Death Marker, TUNEL upon WEE1 Kinase Activity Inhibition in 12 °C Diapaused Embryos
4. Discussion
The Survivability of Embryonic Cells during Diapause Is Linked with Cell Cycle Regulation and Maintaining the Cytoarchitectural Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Stern, C.D.; Downs, K.M. The hypoblast (visceral endoderm): An evo-devo perspective. Development 2012, 139, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- Fasenko, G.M. Egg storage and the embryo. Poult. Sci. 2007, 86, 1020–1024. [Google Scholar] [CrossRef] [PubMed]
- Pokhrel, N.; Cohen, E.B.-T.; Genin, O.; Ruzal, M.; Sela-Donenfeld, D.; Cinnamon, Y. Effects of storage conditions on hatchability, embryonic survival and cytoarchitectural properties in broiler from young and old flocks. Poult. Sci. 2018, 97, 1429–1440. [Google Scholar] [CrossRef] [PubMed]
- Pokhrel, N.; Sela-Donenfeld, D.; Cinnamon, Y. The chick blastoderm during diapause, a landmark for optimization of preincubation storage conditions. Poult. Sci. 2021, 100, 101227. [Google Scholar] [CrossRef] [PubMed]
- Eyal-Giladi, H.; Kochav, S. From cleavage to primitive streak formation: A complementary normal table and a new look at the first stages of the development of the chick. I. General morphology. Dev. Biol. 1976, 49, 321–337. [Google Scholar] [CrossRef]
- Stern, C.D.; Canning, D.R. Origin of cells giving rise to mesoderm and endoderm in chick embryo. Nature 1990, 343, 273–275. [Google Scholar] [CrossRef]
- Karagenc, L.; Sandikci, M. Tissue distribution of cells derived from the area opaca in heterospecific quail-chick blastodermal chimeras. J. Anat. 2010, 216, 16–22. [Google Scholar] [CrossRef]
- George-Weinstein, M.; Gerhart, J.; Reed, R.; Flynn, J.; Callihan, B.; Mattiacci, M.; Miehle, C.; Foti, G.; Lash, J.W.; Weintraub, H. Skeletal Myogenesis: The Preferred Pathway of Chick Embryo Epiblast Cells in Vitro. Dev. Biol. 1996, 173, 279–291. [Google Scholar] [CrossRef]
- Ko, M.H.; Hwang, Y.S.; Rim, J.S.; Han, H.J.; Han, J.Y. Avian blastoderm dormancy arrests cells in G2 and suppresses apoptosis. FASEB J. 2017, 31, 3240–3250. [Google Scholar] [CrossRef]
- Dyson, N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998, 12, 2245–2262. [Google Scholar] [CrossRef]
- Lasorella, A.; Iavarone, A.; Israel, M.A. Id2 specifically alters regulation of the cell cycle by tumor suppressor proteins. Mol. Cell. Biol. 1996, 16, 2570–2578. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zeng, S.; Zhang, Y.; Deng, G.; Qu, Y.; Guo, C.; Yin, L.; Han, Y.; Shen, H. BMP4 enhances hepatocellular carcinoma proliferation by promoting cell cycle progression via ID2/CDKN1B signaling. Mol. Carcinog. 2017, 56, 2279–2289. [Google Scholar] [CrossRef] [PubMed]
- Bertoli, C.; Skotheim, J.M.; De Bruin, R.A.M. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013, 14, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Yam, C.H.; Fung, T.K.; Poon, R.Y.C. Cyclin A in cell cycle control and cancer. Cell. Mol. Life Sci. 2002, 59, 1317–1326. [Google Scholar] [CrossRef]
- Huang, Y.; Sramkoski, R.M.; Jacobberger, J.W. The kinetics of G2 and M transitions regulated by B cyclins. PLoS ONE 2013, 8, 30–35. [Google Scholar] [CrossRef]
- Whittaker, S.R.; Mallinger, A.; Workman, P.; Clarke, P.A. Inhibitors of cyclin-dependent kinases as cancer therapeutics. Pharmacol. Ther. 2017, 173, 83–105. [Google Scholar] [CrossRef]
- Do, K.; Doroshow, J.H.; Kummar, S. Wee1 kinase as a target for cancer therapy. Cell Cycle 2013, 12, 3348–3353. [Google Scholar] [CrossRef]
- Collins, I.; Garrett, M.D. Targeting the cell division cycle in cancer: CDK and cell cycle checkpoint kinase inhibitors. Curr. Opin. Pharmacol. 2005, 5, 366–373. [Google Scholar] [CrossRef]
- Zegerman, P.; Diffley, J.F.X. Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature 2010, 467, 474–478. [Google Scholar] [CrossRef]
- Elbæk, C.R.; Petrosius, V.; Sørensen, C.S. WEE1 kinase limits CDK activities to safeguard DNA replication and mitotic entry. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2020, 819, 111694. [Google Scholar] [CrossRef]
- Takai, H.; Tominaga, K.; Motoyama, N.; Minamishima, Y.A.; Nagahama, H.; Tsukiyama, T.; Ikeda, K.; Nakayama, K.; Nakanishi, M.; Nakayama, K.I. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(−/−) mice. Genes Dev. 2000, 14, 1439–1447. [Google Scholar] [CrossRef] [PubMed]
- Hirai, H.; Iwasawa, Y.; Okada, M.; Arai, T.; Nishibata, T.; Kobayashi, M.; Kimura, T.; Kaneko, N.; Ohtani, J.; Yamanaka, K.; et al. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol. Cancer Ther. 2009, 8, 2992–3000. [Google Scholar] [CrossRef] [PubMed]
- Strauss, B.; Harrison, A.; Coelho, P.A.; Yata, K.; Zernicka-Goetz, M.; Pines, J. Cyclin B1 is essential for mitosis in mouse embryos, and its nuclear export sets the time for mitosis. J. Cell Biol. 2018, 217, 179–193. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, Y.; Li, C.; Wang, R.H.; Deng, C.X. Murine Wee1 plays a critical role in cell cycle regulation and pre-implantation stages of embryonic development. Int. J. Biol. Sci. 2006, 2, 161–170. [Google Scholar] [CrossRef]
- Vassilopoulos, A.; Tominaga, Y.; Kim, H.-S.; Lahusen, T.; Li, B.; Yu, H.; Gius, D.; Deng, C.-X. WEE1 murine deficiency induces hyper-activation of APC/C and results in genomic instability and carcinogenesis. Oncogene 2015, 34, 3023–3035. [Google Scholar] [CrossRef]
- Esposito, F.; Giuffrida, R.; Raciti, G.; Puglisi, C.; Forte, S. Wee1 kinase: A potential target to overcome tumor resistance to therapy. Int. J. Mol. Sci. 2021, 22, 10689. [Google Scholar] [CrossRef]
- Pokhrel, N.; Cohen, E.B.; Genin, O.; Cinnamon, Y. Cellular and morphological characterization of blastoderms from freshly laid broiler eggs. Poult. Sci. 2017, 96, 4399–4408. [Google Scholar] [CrossRef]
- Kohl, A.; Golan, N.; Cinnamon, Y.; Genin, O.; Chefetz, B.; Sela-Donenfeld, D. A proof of concept study demonstrating that environmental levels of carbamazepine impair early stages of chick embryonic development. Environ. Int. 2019, 129, 583–594. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carrera, I.; Frise, E.; Verena, K.; Mark, L.; Tobias, P.; Stephan, P.; Curtis, R.; Stephan, S.; Benjamin, S.; et al. Fiji—An Open platform for biological image analysis. Nat. Methods 2009, 9. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Jin, S.; Ma, C.; Wang, Z.; Fang, Q.; Jiang, R. RNA-Seq reveals seven promising candidate genes affecting the proportion of thick egg albumen in layer-type chickens. Sci. Rep. 2017, 7, 18083. [Google Scholar] [CrossRef]
- Yi, Y.; Fang, Y.; Wu, K.; Liu, Y.; Zhang, W. Comprehensive gene and pathway analysis of cervical cancer progression. Oncol. Lett. 1999, 19, 3316–3332. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef]
- Lunn, J.S.; Fishwick, K.J.; Halley, P.A.; Storey, K.G. A spatial and temporal map of FGF/Erk1/2 activity and response repertoires in the early chick embryo. Dev. Biol. 2007, 302, 536–552. [Google Scholar] [CrossRef]
- Peretz, Y.; Kohl, A.; Slutsky, N.; Komlos, M.; Varshavsky, S.; Sela-Donenfeld, D. Neural stem cells deriving from chick embryonic hindbrain recapitulate hindbrain development in culture. Sci. Rep. 2018, 8, 13920. [Google Scholar] [CrossRef]
- Kalev-Altman, R.; Hanael, E.; Zelinger, E.; Blum, M.; Monsonego-Ornan, E.; Sela-Donenfeld, D. Conserved role of matrix metalloproteases 2 and 9 in promoting the migration of neural crest cells in avian and mammalian embryos. FASEB J. 2020, 34, 5240–5261. [Google Scholar] [CrossRef] [PubMed]
- Slattery, S.D.; Newberg, J.Y.; Szafran, A.T.; Hall, R.M.; Brinkley, B.R.; Mancini, M.A. A framework for image-based classification of mitotic cells in asynchronous populations. Assay Drug Dev. Technol. 2012, 10, 161–178. [Google Scholar] [CrossRef] [PubMed]
- Walczak, C.E.; Cai, S.; Khodjakov, A. Mechanisms of chromosome behaviour during mitosis. Nat. Rev. Mol. Cell Biol. 2010, 11, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Baudoin, N.C.; Cimini, D. A guide to classifying mitotic stages and mitotic defects in fixed cells. Chromosoma 2018, 127, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Mori, S.; Bernardi, R.; Laurent, A.; Resnati, M.; Crippa, A.; Gabrieli, A.; Keough, R.; Gonda, T.J.; Blasi, F. Myb-Binding Protein 1A (MYBBP1A) Is Essential for Early Embryonic Development, Controls Cell Cycle and Mitosis, and Acts as a Tumor Suppressor. PLoS ONE 2012, 7, e39723. [Google Scholar] [CrossRef]
- Hirai, H.; Arai, T.; Okada, M.; Nishibata, T.; Kobayashi, M.; Sakai, N.; Imagaki, K.; Ohtani, J.; Sakai, T.; Yoshizumi, T.; et al. MK-1775, a small molecule Wee1 inhibitor, enhances antitumor efficacy of various DNA-damaging agents, including 5-fluorouracil. Cancer Biol. Ther. 2010, 9, 514–522. [Google Scholar] [CrossRef]
- Kreahling, J.M.; Gemmer, J.Y.; Reed, D.; Letson, D.; Bui, M.; Altiok, S. MK1775, A Selective Wee1 Inhibitor, Shows Single-Agent Antitumor Activity Against Sarcoma Cells. Mol Cancer Ther. 2012, 11, 174–182. [Google Scholar] [CrossRef]
- Noiron, J.; Hoareau, M.; Colin, J.; Guénal, I. Apoptosis quantification in tissue: Development of a semi-automatic protocol and assessment of critical steps of image processing. Biomolecules 2021, 11, 1523. [Google Scholar] [CrossRef]
- Daniel, B.; DeCoster, M.A. Quantification of sPLA2-induced early and late apoptosis changes in neuronal cell cultures using combined TUNEL and DAPI staining. Brain Res. Protoc. 2004, 13, 144–150. [Google Scholar] [CrossRef]
- Weisinger, K.; Kayam, G.; Missulawin-Drillman, T.; Sela-Donenfeld, D. Analysis of expression and function of FGF-MAPK signaling components in the hindbrain reveals a central role for FGF3 in the regulation of Krox20, mediated by Pea3. Dev. Biol. 2010, 344, 881–895. [Google Scholar] [CrossRef][Green Version]
- Weninger, W.J.; Geyer, S.H.; Mohun, T.J.; Rasskin-Gutman, D.; Matsui, T.; Ribeiro, I.; da Costa, F.L.; Izpisúa-Belmonte, J.C.; Müller, G.B. High-resolution episcopic microscopy: A rapid technique for high detailed 3D analysis of gene activity in the context of tissue architecture and morphology. Anat. Embryol. 2006, 211, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Geyer, S.H.; Maurer-Gesek, B.; Reissig, L.F.; Weninger, W.J. High-resolution episcopic microscopy (HREM)—Simple and robust protocols for processing and visualizing organic materials. J. Vis. Exp. 2017, 125, e56071. [Google Scholar] [CrossRef] [PubMed]
- Matthews, H.K.; Bertoli, C.; de Bruin, R.A.M. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2022, 23, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Dominguez, D.; Chen, S.; Fan, J.; Qin, L.; Long, A.; Li, X.; Zhang, Y.; Shi, H.; Zhang, B. WEE1 inhibition by MK1775 as a single-agent therapy inhibits ovarian cancer viability. Oncol. Lett. 2017, 14, 3580–3586. [Google Scholar] [CrossRef]
- Yuan, M.L.; Li, P.; Xing, Z.H.; Di, J.M.; Liu, H.; Yang, A.K.; Lin, X.J.; Jiang, Q.W.; Yang, Y.; Huang, J.R.; et al. Inhibition of WEE1 Suppresses the Tumor Growth in Laryngeal Squamous Cell Carcinoma. Front. Pharmacol. 2018, 9, 1041. [Google Scholar] [CrossRef]
- Ha, D.H.; Min, A.; Kim, S.; Jang, H.; Kim, S.H.; Kim, H.J.; Ryu, H.S.; Ku, J.L.; Lee, K.H.; Im, S.A. Antitumor effect of a WEE1 inhibitor and potentiation of olaparib sensitivity by DNA damage response modulation in triple-negative breast cancer. Sci. Rep. 2020, 10, 9930. [Google Scholar] [CrossRef]
- Lee, Y.Y.; Cho, Y.J.; won Shin, S.; Choi, C.; Ryu, J.Y.; Jeon, H.K.; Choi, J.J.; Hwang, J.R.; Choi, C.H.; Kim, T.J.; et al. Anti-Tumor Effects of Wee1 Kinase Inhibitor with Radiotherapy in Human Cervical Cancer. Sci. Rep. 2019, 9, 15394. [Google Scholar] [CrossRef]
- Denton, D.; Kumar, S. Terminal deoxynucleotidyl transferase (TdT)-mediated dutp nick-end labeling (TUNEL) for detection of apoptotic cells in Drosophila. Cold Spring Harb. Protoc. 2015, 2015, 568–571. [Google Scholar] [CrossRef]
- Feng, X.; Krogh, K.A.; Wu, C.Y.; Lin, Y.W.; Tsai, H.C.; Thayer, S.A.; Wei, L.N. Receptor-interacting protein 140 attenuates endoplasmic reticulum stress in neurons and protects against cell death. Nat. Commun. 2014, 5, 4487. [Google Scholar] [CrossRef]
- Serrano Nájera, G.; Weijer, C.J. Cellular processes driving gastrulation in the avian embryo. Mech. Dev. 2020, 163, 103624. [Google Scholar] [CrossRef]
- Harris, T. Adherens Junctions: From Molecular Mechanisms to Tissue Development and Disease; Springer: Dordrecht, The Netherlands, 2012; ISBN 9789400741850. [Google Scholar]
- Scott, A.E.; Vasilescu, D.M.; Seal, K.A.D.; Keyes, S.D.; Mavrogordato, M.N.; Hogg, J.C.; Sinclair, I.; Warner, J.A.; Hackett, T.L.; Lackie, P.M. Three dimensional imaging of paraffin embedded human lung tissue samples by micro-computed tomography. PLoS ONE 2015, 10, e0126230. [Google Scholar] [CrossRef] [PubMed]
- Ugryumova, N.; Stevens-Smith, J.; Scutt, A.; Matcher, S.J. Local variations in bone mineral density: A comparison of OCT versus x-ray micro-CT. Coherence Domain Opt. Methods Opt. Coherence Tomogr. Biomed. XII 2008, 6847, 684725. [Google Scholar] [CrossRef]
- Varner, V.D.; Voronov, D.A.; Taber, L.A. Mechanics of head fold formation: Investigating tissue-level forces during early development. Development 2010, 137, 3801–3811. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.C.; Lu, H.C.; Turmaine, M.; Oliveira, N.M.M.; Yang, Y.; De Almeida, I.; Stern, C.D. Molecular anatomy of the pre-primitive-streak chick embryo. Open Biol. 2020, 10, 190299. [Google Scholar] [CrossRef] [PubMed]
- Eyal-Giladi, H.; Debby, A.; Harel, N. The posterior section of the chick’s area pellucida and its involvement in hypoblast and primitive streak formation. Development 1992, 116, 819–830. [Google Scholar] [CrossRef]
- Geyer, S.H.; Tinhofer, I.E.; Lumenta, D.B.; Kamolz, L.P.; Branski, L.; Finnerty, C.C.; Herndon, D.N.; Weninger, W.J. High-resolution episcopic microscopy (HREM): A useful technique for research in wound care. Ann. Anat. 2015, 197, 3–10. [Google Scholar] [CrossRef]
- Matheson, C.J.; Backos, D.S.; Reigan, P. Targeting WEE1 Kinase in Cancer. Trends Pharmacol. Sci. 2016, 37, 872–881. [Google Scholar] [CrossRef]
- Kamemizu, C.; Fujimori, T. Distinct dormancy progression depending on embryonic regions during mouse embryonic diapause. Biol. Reprod. 2019, 100, 1204–1214. [Google Scholar] [CrossRef]
- Dolfi, L.; Ripa, R.; Antebi, A.; Valenzano, D.R.; Cellerino, A. Cell cycle dynamics during diapause entry and exit in an annual killifish revealed by FUCCI technology. Evodevo 2019, 10, 29. [Google Scholar] [CrossRef]
- Murakami, M.S.; Moody, S.A.; Daar, I.O.; Morrison, D.K. Morphogenesis during Xenopus gastrulation requires Wee1-mediated inhibition of cell proliferation. Development 2004, 131, 571–580. [Google Scholar] [CrossRef]
- Morison, S.A.; Cramp, R.L.; Alton, L.A.; Franklin, C.E. Cooler temperatures slow the repair of DNA damage in tadpoles exposed to ultraviolet radiation: Implications for amphibian declines at high altitude. Glob. Chang. Biol. 2020, 26, 1225–1234. [Google Scholar] [CrossRef] [PubMed]
- Dieser, M.; Battista, J.R.; Christner, B.C. DNA double-strand break repair at −15 °C. Appl. Environ. Microbiol. 2013, 79, 7662–7668. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.H.; Ye, C.X.; Guo, Z.X.; Wang, A.L. Immune and physiological responses of pufferfish (Takifugu obscurus) under cold stress. Fish Shellfish Immunol. 2017, 64, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Wang, W.N.; Wang, L.J.; Liu, Y.F.; Wang, A.L. Oxidative stress, DNA damage and osmolality in the Pacific white shrimp, Litopenaeus vannamei exposed to acute low temperature stress. Comp. Biochem. Physiol. 2011, 154, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xu, Z.P.; Huang, Y.; Hamrick, H.E.; Duerksen-Hughes, P.J.; Yu, Y.N. ATM and ATR: Sensing DNA damage. World J. Gastroenterol. 2004, 10, 155–160. [Google Scholar] [CrossRef]
- Smith, J.; Mun Tho, L.; Xu, N.; Gillespie, D.A. The ATM-Chk2 and ATR-Chk1 Pathways in DNA Damage Signaling and Cancer, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2010; Volume 108. [Google Scholar]
- Maréchal, A.; Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013, 5, a012716. [Google Scholar] [CrossRef]
- Gorecki, L.; Andrs, M.; Korabecny, J. Clinical candidates targeting the ATR–CHK11–WEE1 axis in cancer. Cancers 2021, 13, 795. [Google Scholar] [CrossRef]
- Smith, H.L.; Southgate, H.; Tweddle, D.A.; Curtin, N.J. DNA damage checkpoint kinases in cancer. Expert Rev. Mol. Med. 2020, 22, e2. [Google Scholar] [CrossRef]
- Hustedt, N.; Durocher, D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 2017, 19, 1–9. [Google Scholar] [CrossRef]
- Dixon, A.F.G.; Guo, Y. Egg and cluster size in lady bird beetles (Coleoptera: Coccinellidae): The direct and indirect effects of aphid abundance. Eur. J. Entomol. 1993, 90, 457–463. [Google Scholar]
- Berrigan, D. The Allometry of Egg Size and Number in Insects. Oikos 1991, 60, 313–321. [Google Scholar] [CrossRef]
- Morrongiello, J.R.; Bond, N.R.; Crook, D.A.; Wong, B.B.M. Spatial variation in egg size and egg number reflects trade-offs and bet-hedging in a freshwater fish. J. Anim. Ecol. 2012, 81, 806–817. [Google Scholar] [CrossRef] [PubMed]
- Christians, J.K. Avian egg size: Variation within species and inflexibility within individuals. Biol. Rev. 2002, 77, 1–26. [Google Scholar] [CrossRef]
- Taborsky, B.; Skubic, E.; Bruintjes, R. Mothers adjust egg size to helper number in a cooperatively breeding cichlid. Behav. Ecol. 2007, 18, 652–657. [Google Scholar] [CrossRef]
- Suetsugu, K.; Funaki, S.; Takahashi, A.; Ito, K.; Yokoyama, T. Potential role of bird predation in the dispersal of otherwise flightless stick insects. Ecology 2018, 99, 1504–1506. [Google Scholar] [CrossRef] [PubMed]
- Cassill, D.L. Extending r/K selection with a maternal risk-management model that classifies animal species into divergent natural selection categories. Sci. Rep. 2019, 9, 6111. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, J.; Bresseel, J.; Constant, J.; Kneubühler, B.; Leubner, F.; Michalik, P.; Bradler, S. Extreme convergence in egg-laying strategy across insect orders. Sci. Rep. 2015, 5, 7825. [Google Scholar] [CrossRef]
- Mansour, N.; Lahnsteiner, F.; Patzner, R.A. Physiological and biochemical investigations on egg stickiness in common carp. Anim. Reprod. Sci. 2009, 114, 256–268. [Google Scholar] [CrossRef]
- Mazzini, M.; Callaini, G.; Mencarelli, C. A comparative analysis of the evolution of the egg envelopes and the origin of the yolk. Bolletino di Zool. 1984, 51, 35–101. [Google Scholar] [CrossRef][Green Version]
- Zhang, C.; Wei, D.; Shi, G.; Huang, X.; Cheng, P.; Liu, G.; Guo, X.; Liu, L.; Wang, H.; Miao, F.; et al. Understanding the regulation of overwintering diapause molecular mechanisms in Culex pipiens pallens through comparative proteomics. Sci. Rep. 2019, 9, 6485. [Google Scholar] [CrossRef]
- Ishizaki, H. Arrest of adult development in debrained pupae of the silkworm, Bombyx mori. J. Insect Physiol. 1972, 18, 1621–1627. [Google Scholar] [CrossRef]
- Rafferty, A.R.; Reina, R.D. Arrested embryonic development: A review of strategies to delay hatching in egg-laying reptiles. Proc. R. Soc. B Biol. Sci. 2012, 279, 2299–2308. [Google Scholar] [CrossRef] [PubMed]
- Bai, G.; Smolka, M.B.; Schimenti, J.C. Chronic DNA Replication Stress Reduces Replicative Lifespan of Cells by TRP53-Dependent, microRNA-Assisted MCM2-7 Downregulation. PLoS Genet. 2016, 12, e1005787. [Google Scholar] [CrossRef] [PubMed]
- Calcinotto, A.; Kohli, J.; Zagato, E.; Pellegrini, L.; Demaria, M.; Alimonti, A. Cellular senescence: Aging, cancer, and injury. Physiol. Rev. 2019, 99, 1047–1078. [Google Scholar] [CrossRef] [PubMed]
- Tomasetti, C.; Poling, J.; Roberts, N.J.; London, N.R.; Pittman, M.E.; Haffner, M.C.; Rizzo, A.; Baras, A.; Karim, B.; Kim, A.; et al. Cell division rates decrease with age, providing a potential explanation for the age-dependent deceleration in cancer incidence. Proc. Natl. Acad. Sci. USA. 2019, 116, 20482–20488. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Hiona, A.; Lee, A.S.; Cao, F.; Huang, M.; Li, Z.; Cherry, A.; Pei, X.; Wu, J.C. Effects of long-term culture on human embryonic stem cell aging. Stem Cells Dev. 2011, 20, 127–138. [Google Scholar] [CrossRef]
- Nayan, M.; Paul, A.; Chen, G.; Chiu, R.C.J.; Prakash, S.; Shum-Tim, D. Superior therapeutic potential of young bone marrow mesenchymal stem cells by direct intramyocardial delivery in aged recipients with acutemyocardial infarction: In Vitro and in Vivo investigation. J. Tissue Eng. 2011, 2011, 741213. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Carruthers, R.D.; Ahmed, S.U.; Ramachandran, S.; Strathdee, K.; Kurian, K.M.; Hedley, A.; Gomez-Roman, N.; Kalna, G.; Neilson, M.; Gilmour, L.; et al. Replication stress drives constitutive activation of the DNA damage response and radioresistance in glioblastoma stem-like cells. Cancer Res. 2018, 78, 5060–5071. [Google Scholar] [CrossRef]
- Ngoi, N.Y.L.; Sundararajan, V.; Tan, D.S.P. Exploiting replicative stress in gynecological cancers as a therapeutic strategy. Int. J. Gynecol. Cancer 2020, 30, 1224–1238. [Google Scholar] [CrossRef]
- Pai, C.C.; Hsu, K.F.; Durley, S.C.; Keszthelyi, A.; Kearsey, S.E.; Rallis, C.; Folkes, L.K.; Deegan, R.; Wilkins, S.E.; Pfister, S.X.; et al. An essential role for dNTP homeostasis following CDK-induced replication stress. J. Cell Sci. 2019, 132, jcs226969. [Google Scholar] [CrossRef] [PubMed]
- Heald, R.; McLoughlin, M.; McKeon, F. Human wee1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated cdc2 kinase. Cell 1993, 74, 463–474. [Google Scholar] [CrossRef]
- Shipley, J.R.; Twining, C.W.; Taff, C.C.; Vitousek, M.N.; Flack, A.; Winkler, D.W. Birds advancing lay dates with warming springs face greater risk of chick mortality. Proc. Natl. Acad. Sci. USA 2020, 117, 25590–25594. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pokhrel, N.; Genin, O.; Sela-Donenfeld, D.; Cinnamon, Y. HREM, RNAseq and Cell Cycle Analyses Reveal the Role of the G2/M-Regulatory Protein, WEE1, on the Survivability of Chicken Embryos during Diapause. Biomedicines 2022, 10, 779. https://doi.org/10.3390/biomedicines10040779
Pokhrel N, Genin O, Sela-Donenfeld D, Cinnamon Y. HREM, RNAseq and Cell Cycle Analyses Reveal the Role of the G2/M-Regulatory Protein, WEE1, on the Survivability of Chicken Embryos during Diapause. Biomedicines. 2022; 10(4):779. https://doi.org/10.3390/biomedicines10040779
Chicago/Turabian StylePokhrel, Narayan, Olga Genin, Dalit Sela-Donenfeld, and Yuval Cinnamon. 2022. "HREM, RNAseq and Cell Cycle Analyses Reveal the Role of the G2/M-Regulatory Protein, WEE1, on the Survivability of Chicken Embryos during Diapause" Biomedicines 10, no. 4: 779. https://doi.org/10.3390/biomedicines10040779
APA StylePokhrel, N., Genin, O., Sela-Donenfeld, D., & Cinnamon, Y. (2022). HREM, RNAseq and Cell Cycle Analyses Reveal the Role of the G2/M-Regulatory Protein, WEE1, on the Survivability of Chicken Embryos during Diapause. Biomedicines, 10(4), 779. https://doi.org/10.3390/biomedicines10040779







