An Integrative Pan-Cancer Analysis of Kinesin Family Member C1 (KIFC1) in Human Tumors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genetic Alteration Analysis
2.2. Gene Expression Analysis
2.3. Survival Analysis
2.4. Protein–Protein Interactions of KIFC1 and Similar Genes in Pan Cancer
2.5. Gene Set Enrichment Analysis of KIFC1 in Pan Cancer
2.6. Immune Reactivity Analysis
2.7. Statistical Analysis
3. Results
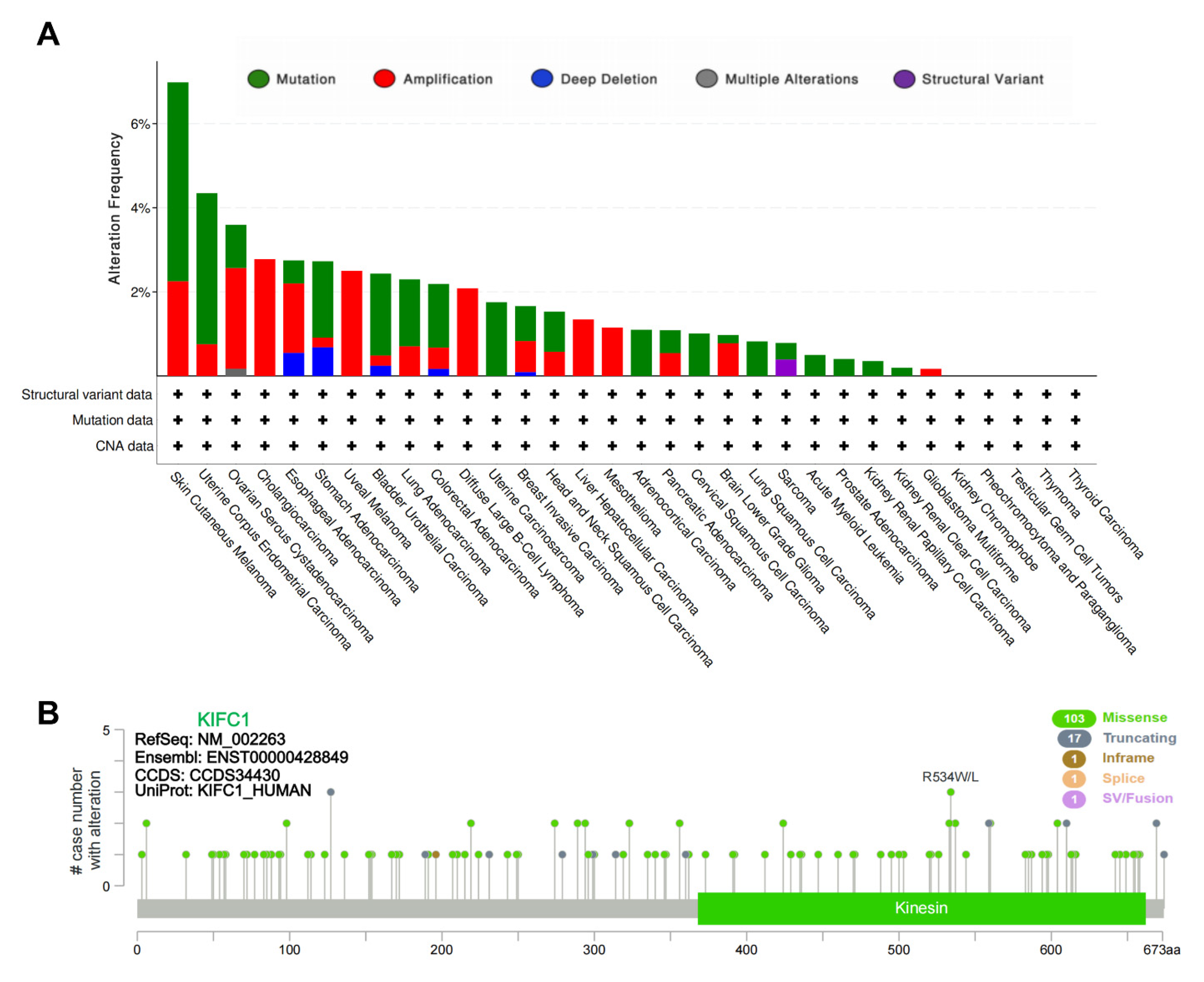
3.1. Genetic Alteration of KIFC1 in Tumors
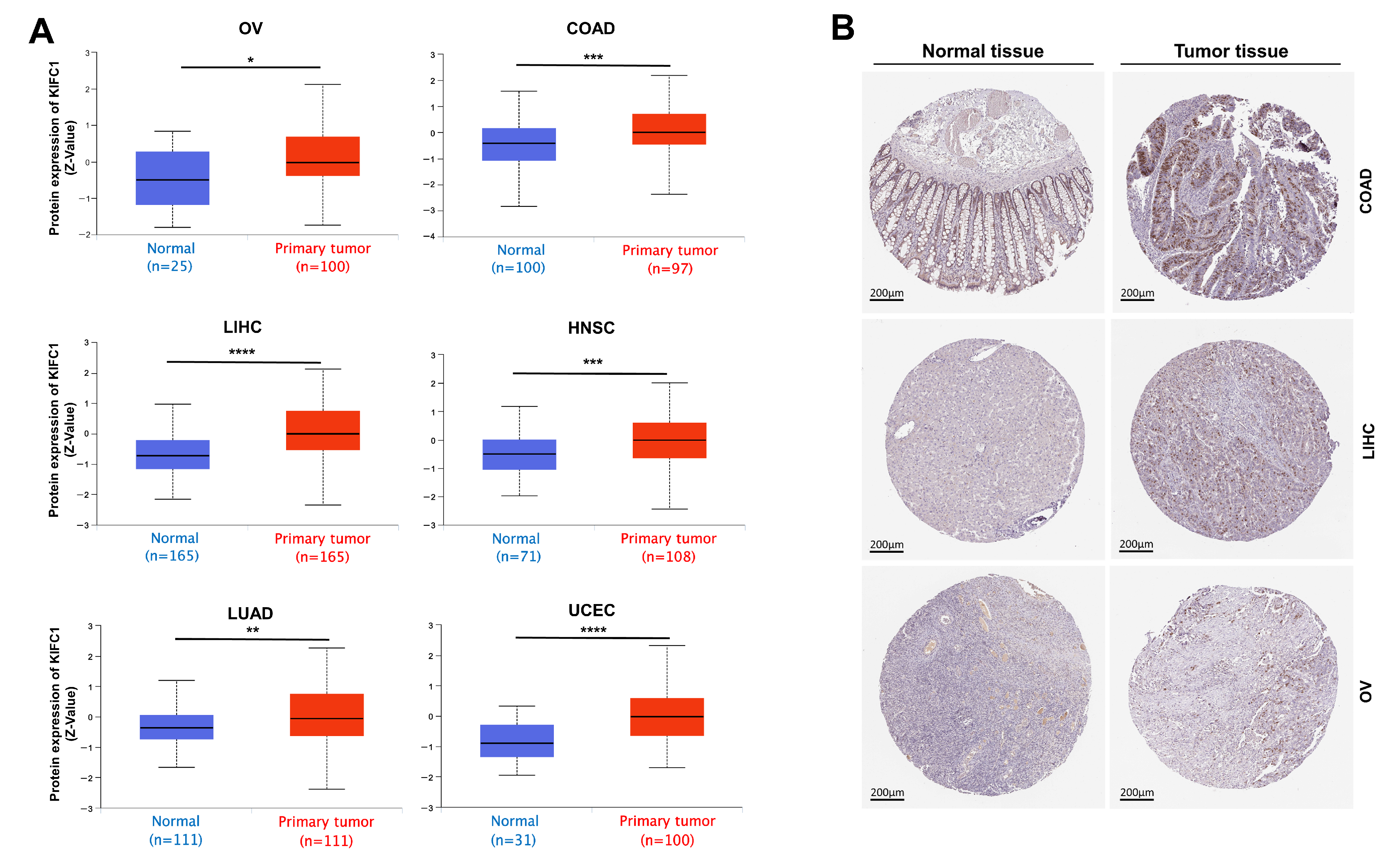
3.2. Gene Expression Analysis Data
3.3. Survival Analysis Data
3.4. Protein–Protein Interactions of KIFC1 and Similar Genes in Pan Cancer
3.5. Gene Set Enrichment Analysis Data
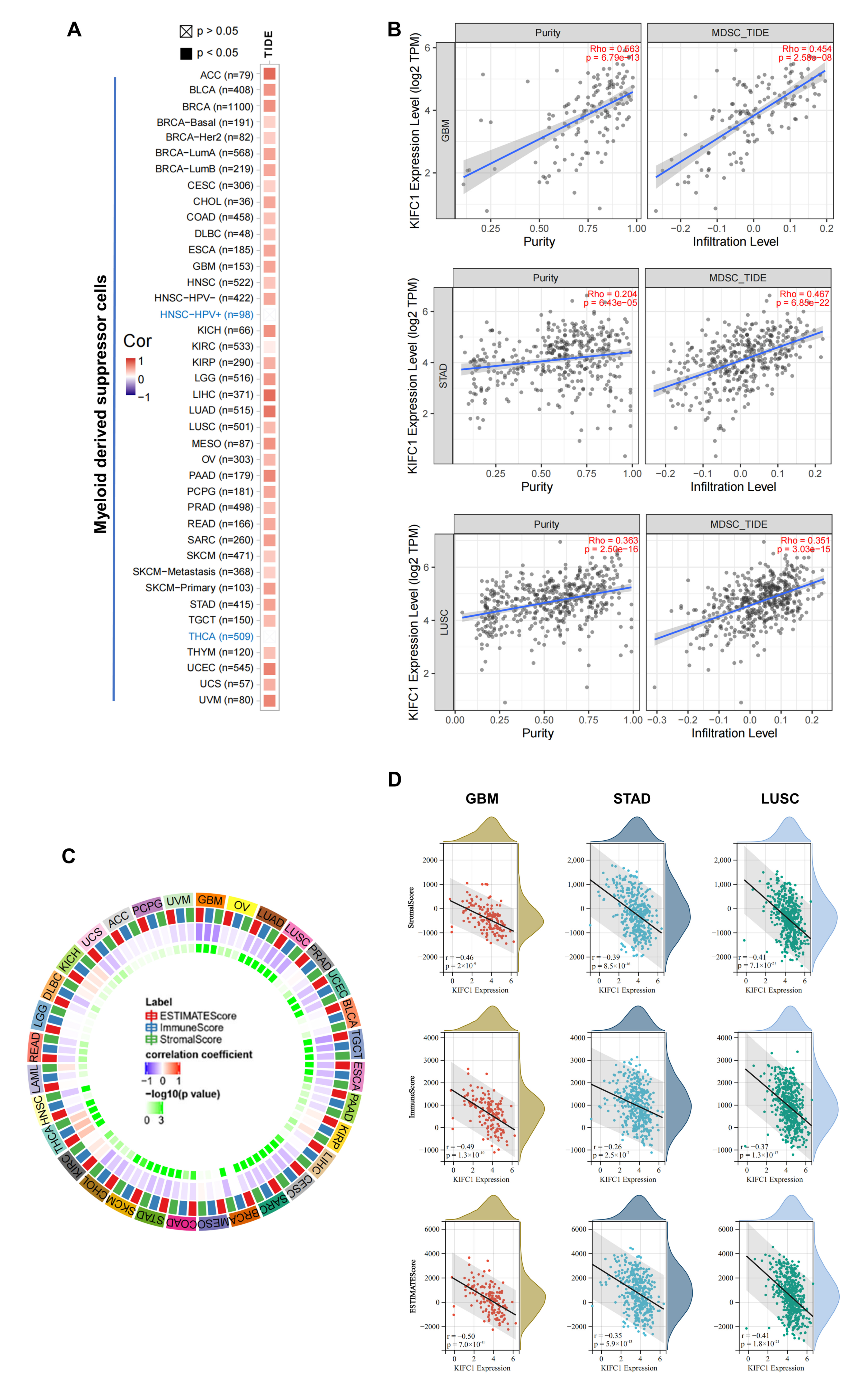
3.6. Immune Reactivity Analysis Data
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Janatipour, M.; Naumov, Y.; Ando, A.; Sugimura, K.; Okamoto, N.; Tsuji, K.; Abe, K.; Inoko, H. Search for MHC-associated genes in human: Five new genes centromeric to HLA-DP with yet unknown functions. Immunogenetics 1992, 35, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mountain, V.; Simerly, C.; Howard, L.; Ando, A.; Schatten, G.; Compton, D. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 1999, 147, 351–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, S.; Weaver, L.; Ems-McClung, S.; Walczak, C. Kinesin-14 family proteins HSET/XCTK2 control spindle length by cross-linking and sliding microtubules. Mol. Biol. Cell 2009, 20, 1348–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.X.; Sperry, A.O. C-terminal kinesin motor KIFC1 participates in acrosome biogenesis and vesicle transport. Biol. Reprod. 2003, 69, 1719–1729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Y.X.; Shen, H.Q.; She, Z.Y.; Sheng, L.; Chen, Q.Q.; Chu, Y.L.; Tan, F.Q.; Yang, W.X. C-terminal kinesin motor KIFC1 participates in facilitating proper cell division of human seminoma. Oncotarget 2017, 8, 61373–61384. [Google Scholar] [CrossRef]
- Muralidharan, H.; Guha, S.; Madugula, K.; Patil, A.; Bennison, S.A.; Sun, X.; Toyo-Oka, K.; Baas, P.W. KIFC1 regulates the trajectory of neuronal migration. J. Neurosci. 2022. [Google Scholar] [CrossRef]
- Ganem, N.; Godinho, S.; Pellman, D. A mechanism linking extra centrosomes to chromosomal instability. Nature 2009, 460, 278–282. [Google Scholar] [CrossRef] [Green Version]
- Kulukian, A.; Holland, A.; Vitre, B.; Naik, S.; Cleveland, D.; Fuchs, E. Epidermal development, growth control, and homeostasis in the face of centrosome amplification. Proc. Natl. Acad. Sci. USA 2015, 112, E6311–E6320. [Google Scholar] [CrossRef] [Green Version]
- Basto, R.; Brunk, K.; Vinadogrova, T.; Peel, N.; Franz, A.; Khodjakov, A.; Raff, J. Centrosome amplification can initiate tumorigenesis in flies. Cell 2008, 133, 1032–1042. [Google Scholar] [CrossRef] [Green Version]
- Levine, M.; Bakker, B.; Boeckx, B.; Moyett, J.; Lu, J.; Vitre, B.; Spierings, D.; Lansdorp, P.; Cleveland, D.; Lambrechts, D.; et al. Centrosome Amplification Is Sufficient to Promote Spontaneous Tumorigenesis in Mammals. Dev. Cell 2017, 40, 313–322.e315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostecka, L.; Olseen, A.; Kang, K.; Torga, G.; Pienta, K.; Amend, S. High KIFC1 expression is associated with poor prognosis in prostate cancer. Med. Oncol. 2021, 38, 47. [Google Scholar] [CrossRef] [PubMed]
- Teng, K.; Wei, S.; Zhang, C.; Chen, J.; Chen, J.; Xiao, K.; Liu, J.; Dai, M.; Guan, X.; Yun, J.; et al. KIFC1 is activated by TCF-4 and promotes hepatocellular carcinoma pathogenesis by regulating HMGA1 transcriptional activity. J. Exp. Clin. Cancer Res. CR 2019, 38, 329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittal, K.; Choi, D.; Klimov, S.; Pawar, S.; Kaur, R.; Mitra, A.; Gupta, M.; Sams, R.; Cantuaria, G.; Rida, P.; et al. A centrosome clustering protein, KIFC1, predicts aggressive disease course in serous ovarian adenocarcinomas. J. Ovarian Res. 2016, 9, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zhan, P.; Zhou, Z.; Xing, Z.; Zhu, S.; Ma, C.; Li, Q.; Zhu, Q.; Miao, Y.; Zhang, J.; et al. The overexpression of KIFC1 was associated with the proliferation and prognosis of non-small cell lung cancer. J. Thorac. Dis. 2016, 8, 2911–2923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Chong, T.; Yang, J.; Li, H.; Chen, H. Kinesin Motor Protein KIFC1 Is a Target Protein of miR-338-3p and Is Associated With Poor Prognosis and Progression of Renal Cell Carcinoma. Oncol. Res. 2018, 27, 125–137. [Google Scholar] [CrossRef]
- Ogden, A.; Garlapati, C.; Li, X.B.; Turaga, R.C.; Oprea-Ilies, G.; Wright, N.; Bhattarai, S.; Mittal, K.; Wetherilt, C.S.; Krishnamurti, U.; et al. Multi-institutional study of nuclear KIFC1 as a biomarker of poor prognosis in African American women with triple-negative breast cancer. Sci. Rep. 2017, 7, 42289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekino, Y.; Oue, N.; Shigematsu, Y.; Ishikawa, A.; Sakamoto, N.; Sentani, K.; Teishima, J.; Matsubara, A.; Yasui, W. KIFC1 induces resistance to docetaxel and is associated with survival of patients with prostate cancer. Urol. Oncol. 2017, 35, 31.e13–31.e20. [Google Scholar] [CrossRef]
- De, S.; Cipriano, R.; Jackson, M.W.; Stark, G.R. Overexpression of kinesins mediates docetaxel resistance in breast cancer cells. Cancer Res. 2009, 69, 8035–8042. [Google Scholar] [CrossRef] [Green Version]
- Fan, G.; Sun, L.; Meng, L.; Hu, C.; Wang, X.; Shi, Z.; Hu, C.; Han, Y.; Yang, Q.; Cao, L.; et al. The ATM and ATR kinases regulate centrosome clustering and tumor recurrence by targeting KIFC1 phosphorylation. Nat. Commun. 2021, 12, 20. [Google Scholar] [CrossRef]
- Kwon, M.; Godinho, S.; Chandhok, N.; Ganem, N.; Azioune, A.; Thery, M.; Pellman, D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008, 22, 2189–2203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Mikule, K.; Wang, W.; Su, N.; Petteruti, P.; Gharahdaghi, F.; Code, E.; Zhu, X.; Jacques, K.; Lai, Z.; et al. Discovery and mechanistic study of a small molecule inhibitor for motor protein KIFC1. ACS Chem. Biol. 2013, 8, 2201–2208. [Google Scholar] [CrossRef] [PubMed]
- Watts, C.A.; Richards, F.M.; Bender, A.; Bond, P.J.; Korb, O.; Kern, O.; Riddick, M.; Owen, P.; Myers, R.M.; Raff, J.; et al. Design, synthesis, and biological evaluation of an allosteric inhibitor of HSET that targets cancer cells with supernumerary centrosomes. Chem. Biol. 2013, 20, 1399–1410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Zhai, L.; Wang, Y.; Boohaker, R.J.; Lu, W.; Gupta, V.V.; Padmalayam, I.; Bostwick, R.J.; White, E.L.; Ross, L.J.; et al. Discovery of a novel inhibitor of kinesin-like protein KIFC1. Biochem. J. 2016, 473, 1027–1035. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Chandrashekar, D.; Varambally, S.; Creighton, C. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat. Commun. 2019, 10, 5679. [Google Scholar] [CrossRef] [Green Version]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.; Levine, D.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Farina, F.; Pierobon, P.; Delevoye, C.; Monnet, J.; Dingli, F.; Loew, D.; Quanz, M.; Dutreix, M.; Cappello, G. Kinesin KIFC1 actively transports bare double-stranded DNA. Nucleic Acids Res. 2013, 41, 4926–4937. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Zheng, Y.; Yang, C.; Huang, S.; He, X.; Tong, S. miR-635 targets KIFC1 to inhibit the progression of gastric cancer. J. Investig. Med. Off. Publ. Am. Fed. Clin. Res. 2020, 68, 1357–1363. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lu, W.; Chen, D.; Boohaker, R.; Zhai, L.; Padmalayam, I.; Wennerberg, K.; Xu, B.; Zhang, W. KIFC1 is a novel potential therapeutic target for breast cancer. Cancer Biol. Ther. 2015, 16, 1316–1322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Wang, F.; Lan, Y.; Wang, J.; Nie, C.; Liang, Y.; Song, R.; Zheng, T.; Pan, S.; Pei, T.; et al. KIFC1 regulated by miR-532-3p promotes epithelial-to-mesenchymal transition and metastasis of hepatocellular carcinoma via gankyrin/AKT signaling. Oncogene 2019, 38, 406–420. [Google Scholar] [CrossRef] [Green Version]
- Sampath, S.; Ohi, R.; Leismann, O.; Salic, A.; Pozniakovski, A.; Funabiki, H. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell 2004, 118, 187–202. [Google Scholar] [CrossRef] [Green Version]
- Wei, S.; Dai, M.; Zhang, C.; Teng, K.; Wang, F.; Li, H.; Sun, W.; Feng, Z.; Kang, T.; Guan, X.; et al. KIF2C: A novel link between Wnt/β-catenin and mTORC1 signaling in the pathogenesis of hepatocellular carcinoma. Protein Cell 2020, 12, 788–809. [Google Scholar] [CrossRef]
- Ono, T.; Losada, A.; Hirano, M.; Myers, M.; Neuwald, A.; Hirano, T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 2003, 115, 109–121. [Google Scholar] [CrossRef] [Green Version]
- Neumayer, G.; Belzil, C.; Gruss, O.; Nguyen, M. TPX2: Of spindle assembly, DNA damage response, and cancer. Cell. Mol. Life Sci. CMLS 2014, 71, 3027–3047. [Google Scholar] [CrossRef]
- Ladurner, R.; Kreidl, E.; Ivanov, M.; Ekker, H.; Idarraga-Amado, M.; Busslinger, G.; Wutz, G.; Cisneros, D.; Peters, J. Sororin actively maintains sister chromatid cohesion. EMBO J. 2016, 35, 635–653. [Google Scholar] [CrossRef]
- Koedoot, E.; van Steijn, E.; Vermeer, M.; González-Prieto, R.; Vertegaal, A.; Martens, J.; Le Dévédec, S.; van de Water, B. Splicing factors control triple-negative breast cancer cell mitosis through SUN2 interaction and sororin intron retention. J. Exp. Clin. Cancer Res. CR 2021, 40, 82. [Google Scholar] [CrossRef]
- Sisken, J.; Bonner, S.; Grasch, S.; Powell, D.; Donaldson, E. Alterations in metaphase durations in cells derived from human tumours. Cell Tissue Kinet. 1985, 18, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Rezabkova, L.; Hua, S.; Liu, Q.; Capitani, G.; Altelaar, A.; Heck, A.; Kammerer, R.; Steinmetz, M.; Akhmanova, A. Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex. Nat. Cell Biol. 2017, 19, 480–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capecchi, M.; Pozner, A. ASPM regulates symmetric stem cell division by tuning Cyclin E ubiquitination. Nat. Commun. 2015, 6, 8763. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hsu, C.; Wang, T.; Li, C.; Hou, Y.; Chu, J.; Lee, C.; Liu, M.; Su, J.; Jian, K.; et al. A gene expression signature of epithelial tubulogenesis and a role for ASPM in pancreatic tumor progression. Gastroenterology 2013, 145, 1110–1120. [Google Scholar] [CrossRef]
- Wenta, T.; Jarzab, M.; Rychlowski, M.; Borysiak, M.; Latala, A.; Zurawa-Janicka, D.; Filipek, A.; Lipinska, B. Cellular substrates and pro-apoptotic function of the human HtrA4 protease. J. Proteom. 2019, 209, 103505. [Google Scholar] [CrossRef]
- Janke, C.; Magiera, M. The tubulin code and its role in controlling microtubule properties and functions. Nat. Reviews. Mol. Cell Biol. 2020, 21, 307–326. [Google Scholar] [CrossRef]
- Kavallaris, M. Microtubules and resistance to tubulin-binding agents. Nat. Rev. Cancer 2010, 10, 194–204. [Google Scholar] [CrossRef]
- Kent, L.N.; Leone, G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer 2019, 19, 326–338. [Google Scholar] [CrossRef]
- Gabay, M.; Li, Y.; Felsher, D.W. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 2014, 4, a014241. [Google Scholar] [CrossRef] [Green Version]
- Bieging, K.T.; Mello, S.S.; Attardi, L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 2014, 14, 359–370. [Google Scholar] [CrossRef] [Green Version]
- Sekino, Y.; Pham, Q.T.; Kobatake, K.; Kitano, H.; Ikeda, K.; Goto, K.; Hayashi, T.; Nakahara, H.; Sentani, K.; Oue, N.; et al. KIFC1 Is Associated with Basal Type, Cisplatin Resistance, PD-L1 Expression and Poor Prognosis in Bladder Cancer. J. Clin. Med. 2021, 10, 4837. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, X.; Yuan, X.; Shan, Q.; Wang, Z.; Wu, Y.; Xie, J. Kinesin Family Member C1 Increases Temozolomide Resistance of Glioblastoma Through Promoting DNA Damage Repair. Cell Transplant. 2021, 30, 963689721991466. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Erin, N.; Grahovac, J.; Brozovic, A.; Efferth, T. Tumor microenvironment and epithelial mesenchymal transition as targets to overcome tumor multidrug resistance. Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer. Chemother. 2020, 53, 100715. [Google Scholar] [CrossRef]
- Lin, S.; Zhang, X.; Huang, G.; Cheng, L.; Lv, J.; Zheng, D.; Lin, S.; Wang, S.; Wu, Q.; Long, Y.; et al. Myeloid-derived suppressor cells promote lung cancer metastasis by CCL11 to activate ERK and AKT signaling and induce epithelial-mesenchymal transition in tumor cells. Oncogene 2021, 40, 1476–1489. [Google Scholar] [CrossRef]
- Wang, L.; Chang, E.W.; Wong, S.C.; Ong, S.M.; Chong, D.Q.; Ling, K.L. Increased myeloid-derived suppressor cells in gastric cancer correlate with cancer stage and plasma S100A8/A9 proinflammatory proteins. J. Immunol. 2013, 190, 794–804. [Google Scholar] [CrossRef]
- Alban, T.J.; Alvarado, A.G.; Sorensen, M.D.; Bayik, D.; Volovetz, J.; Serbinowski, E.; Mulkearns-Hubert, E.E.; Sinyuk, M.; Hale, J.S.; Onzi, G.R.; et al. Global immune fingerprinting in glioblastoma patient peripheral blood reveals immune-suppression signatures associated with prognosis. JCI Insight 2018, 3, e122264. [Google Scholar] [CrossRef] [Green Version]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [Green Version]
- Picarda, E.; Ohaegbulam, K.C.; Zang, X. Molecular Pathways: Targeting B7-H3 (CD276) for Human Cancer Immunotherapy. Clin. Cancer Res. 2016, 22, 3425–3431. [Google Scholar] [CrossRef] [Green Version]
- Qin, S.; Xu, L.; Yi, M.; Yu, S.; Wu, K.; Luo, S. Novel immune checkpoint targets: Moving beyond PD-1 and CTLA-4. Mol. Cancer 2019, 18, 155. [Google Scholar] [CrossRef]
- Sharpe, A.H.; Pauken, K.E. The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol. 2018, 18, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Schoenfeld, A.J.; Hellmann, M.D. Acquired Resistance to Immune Checkpoint Inhibitors. Cancer Cell 2020, 37, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Weber, R.; Fleming, V.; Hu, X.; Nagibin, V.; Groth, C.; Altevogt, P.; Utikal, J.; Umansky, V. Myeloid-Derived Suppressor Cells Hinder the Anti-Cancer Activity of Immune Checkpoint Inhibitors. Front. Immunol. 2018, 9, 1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, B.; Lamb, M.L.; Zhang, T.; Hennessy, E.J.; Grewal, G.; Sha, L.; Zambrowski, M.; Block, M.H.; Dowling, J.E.; Su, N.; et al. Discovery of potent KIFC1 inhibitors using a method of integrated high-throughput synthesis and screening. J. Med. Chem. 2014, 57, 9958–9970. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, H.; Duan, Y.; Gong, S.; Zhu, Q.; Liu, X.; Liu, Z. An Integrative Pan-Cancer Analysis of Kinesin Family Member C1 (KIFC1) in Human Tumors. Biomedicines 2022, 10, 637. https://doi.org/10.3390/biomedicines10030637
Wu H, Duan Y, Gong S, Zhu Q, Liu X, Liu Z. An Integrative Pan-Cancer Analysis of Kinesin Family Member C1 (KIFC1) in Human Tumors. Biomedicines. 2022; 10(3):637. https://doi.org/10.3390/biomedicines10030637
Chicago/Turabian StyleWu, Hao, Yingjuan Duan, Siming Gong, Qiang Zhu, Xuanyou Liu, and Zhenguo Liu. 2022. "An Integrative Pan-Cancer Analysis of Kinesin Family Member C1 (KIFC1) in Human Tumors" Biomedicines 10, no. 3: 637. https://doi.org/10.3390/biomedicines10030637
APA StyleWu, H., Duan, Y., Gong, S., Zhu, Q., Liu, X., & Liu, Z. (2022). An Integrative Pan-Cancer Analysis of Kinesin Family Member C1 (KIFC1) in Human Tumors. Biomedicines, 10(3), 637. https://doi.org/10.3390/biomedicines10030637





