Fibrin, Bone Marrow Cells and Macrophages Interactively Modulate Cardiomyoblast Fate
Abstract
:1. Introduction
2. Methods
2.1. In Vivo Study
2.1.1. Animals
2.1.2. Bone-Marrow Derived Cells Isolation BMCs and Characterization
2.1.3. Myocardial Infarction Model
2.1.4. Epicardial Treatment
2.1.5. High-Resolution Echocardiography
2.1.6. Histological Analysis
2.2. In Vitro Studies
2.2.1. Conditioned Medium Preparation
2.2.2. MS-Based Proteomics
2.2.3. Macrophage Isolation, Differentiation and Priming with Condition Media
2.2.4. Enzyme-Linked Immunosorbent Assay (ELISA)
2.2.5. Real-Time Polymerase Chain Reaction
2.2.6. H9C2 Rat Cardiomyoblasts
2.2.7. Real-Time Cell Analyzer System (RTCA) and EdU Cell Proliferation Assays
2.3. Statistical Analysis
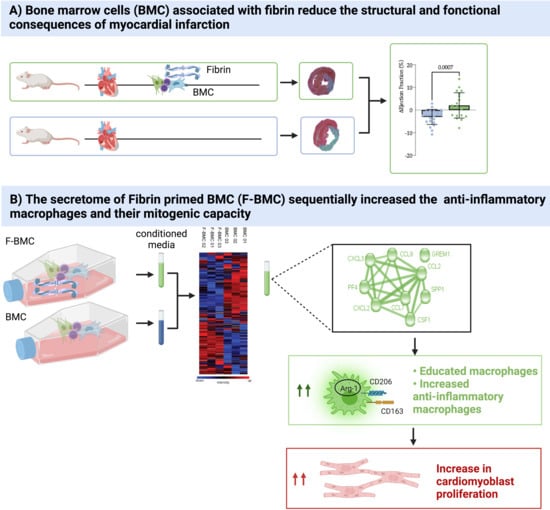
3. Results
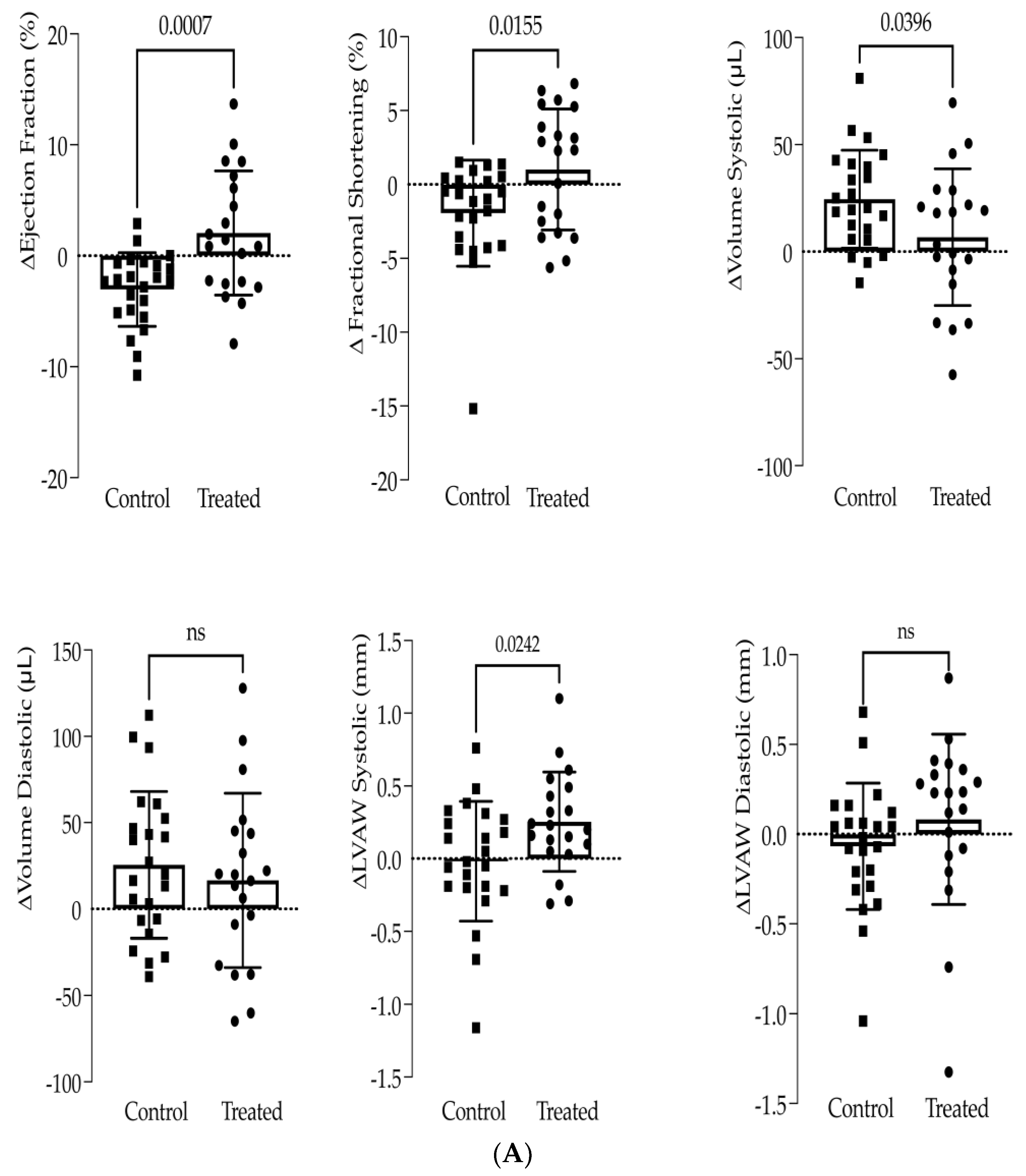
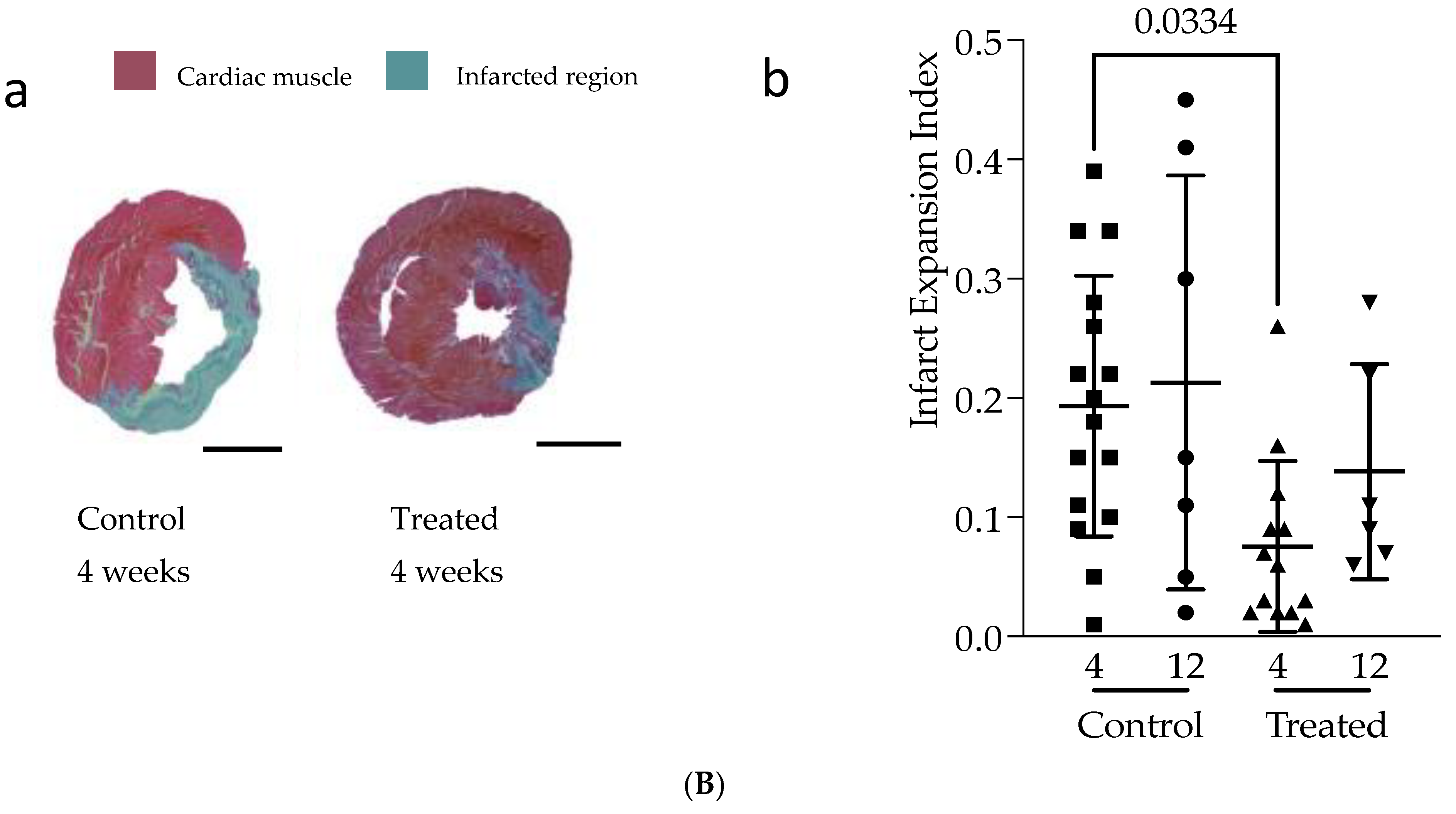
3.1. In Vivo Study
3.2. In Vitro Study
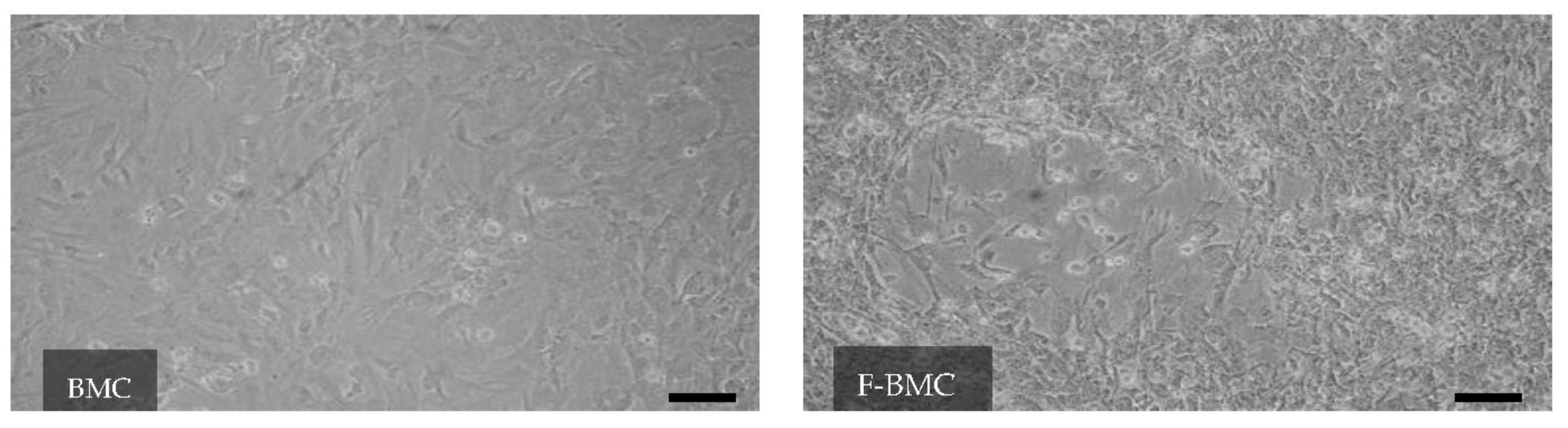
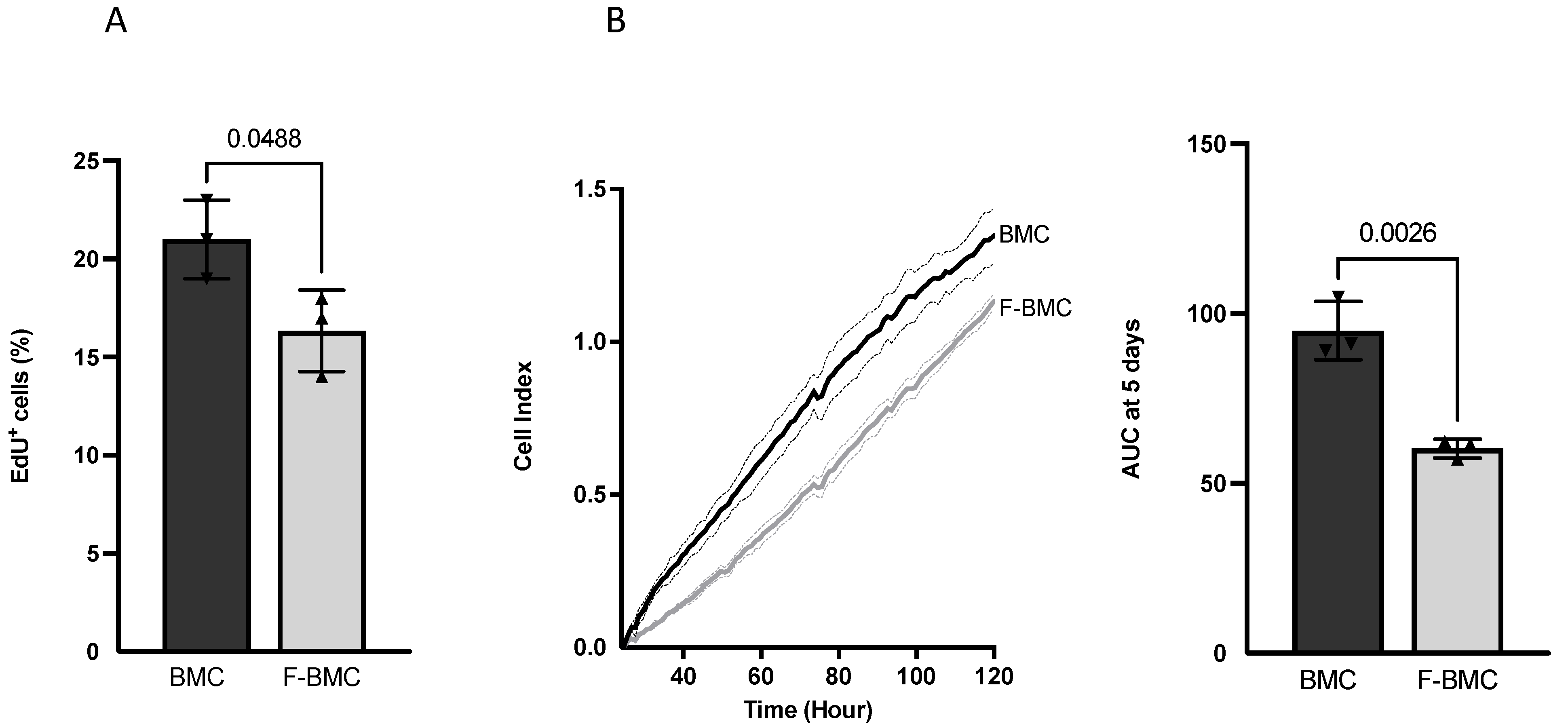
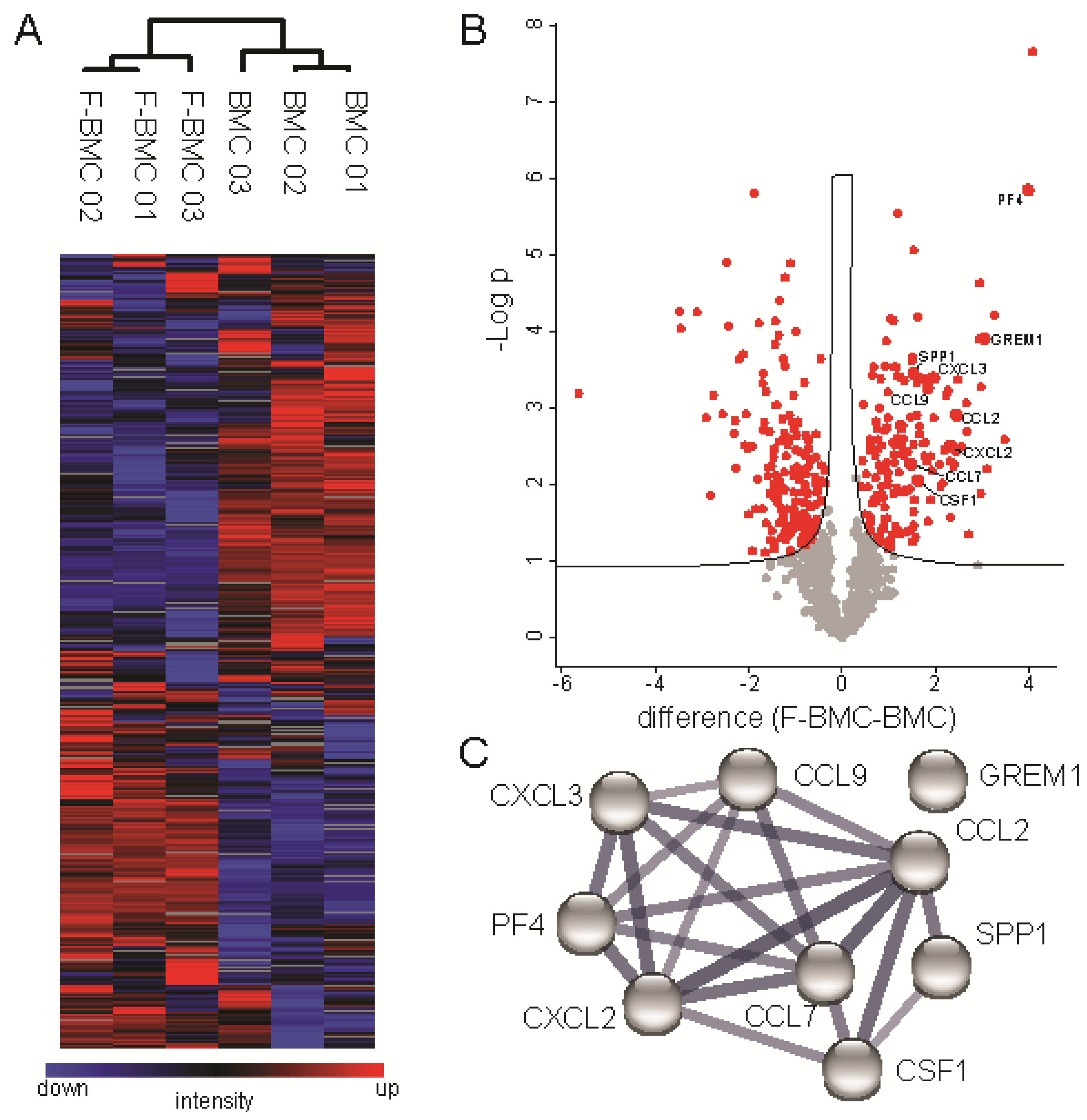
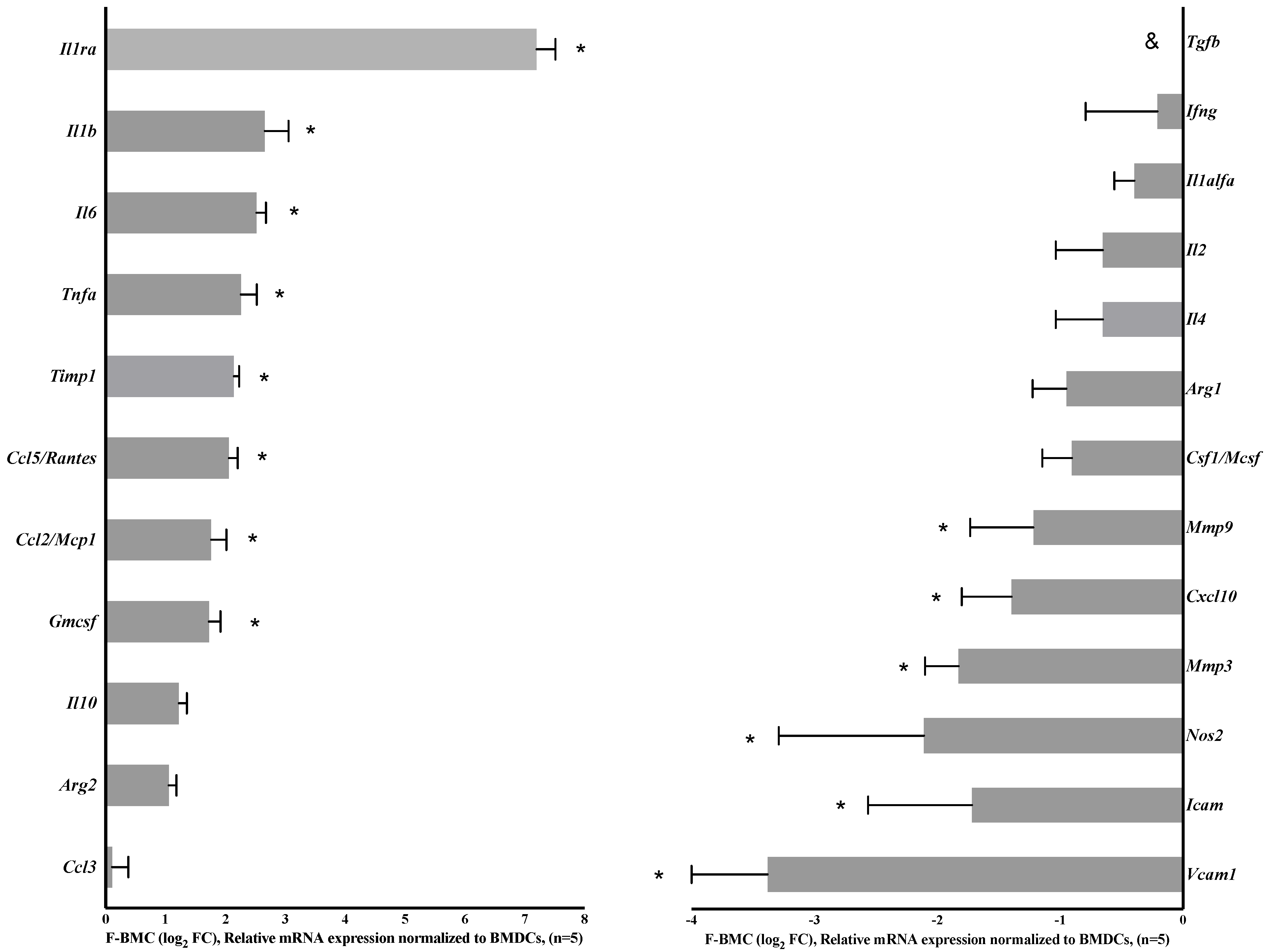
3.2.1. Unique Characteristics of F-BMC, Including Growth, Gene Expression and Secretion Profile Distinguish Them from BMC
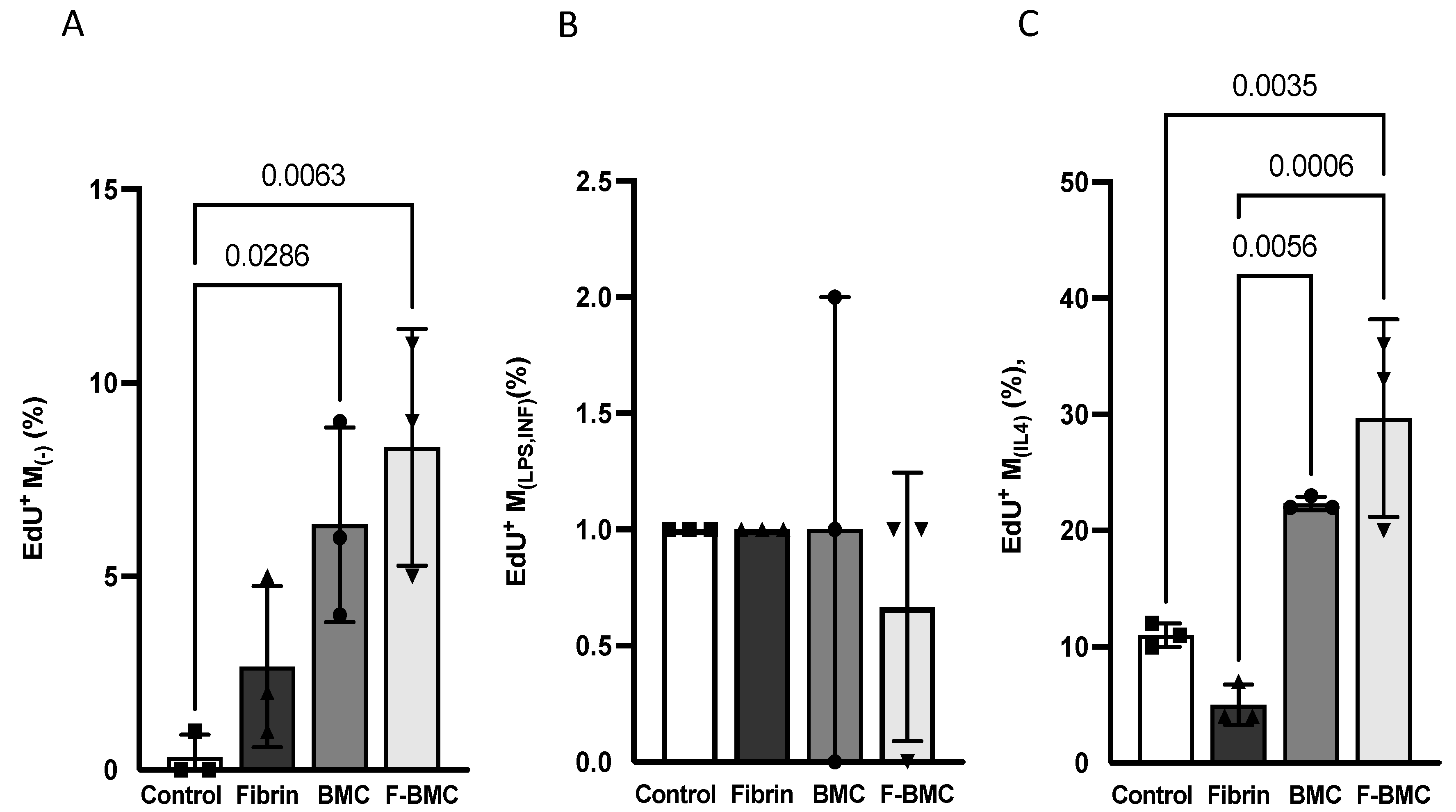
3.2.2. F-BMC Secretome Promotes the Proliferation of Undifferentiated and Anti-Inflammatory Macrophages
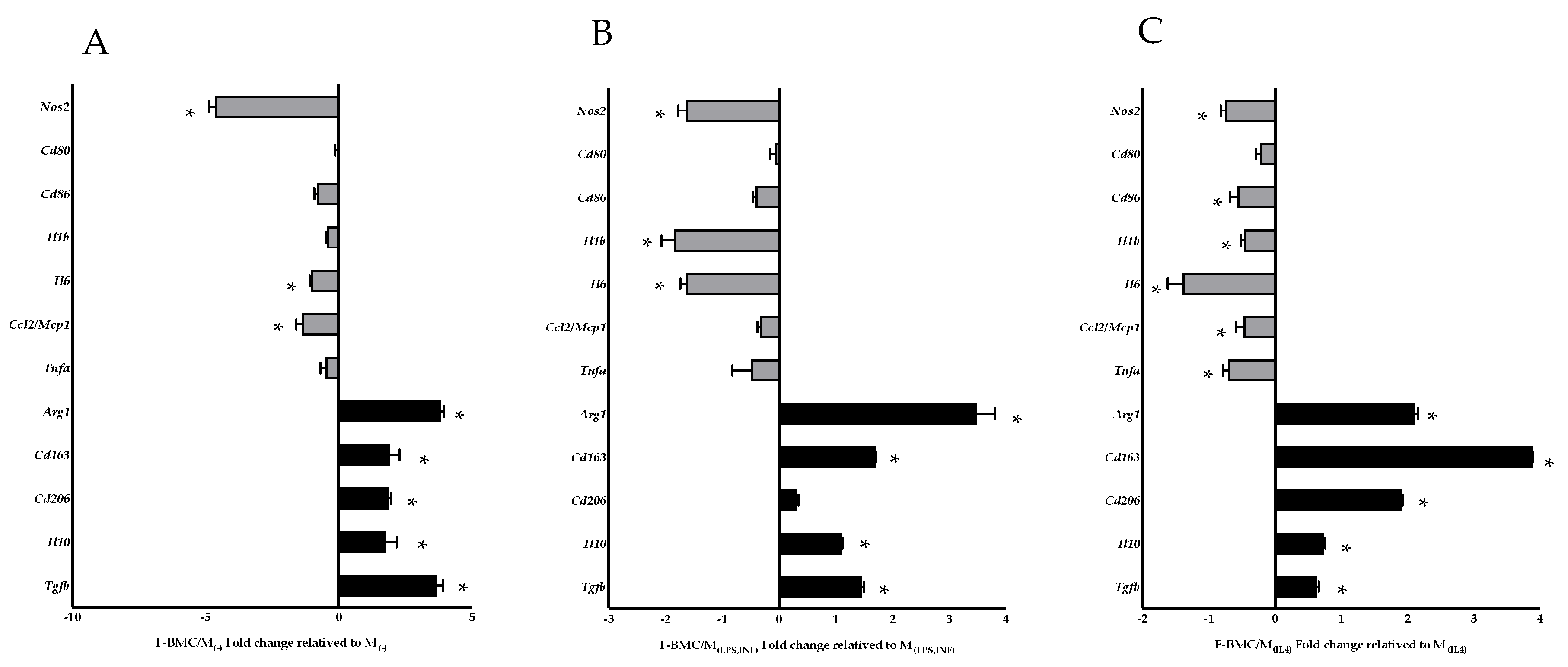
3.2.3. F-BMC Secretome Induces a Macrophage Phenotype Switch
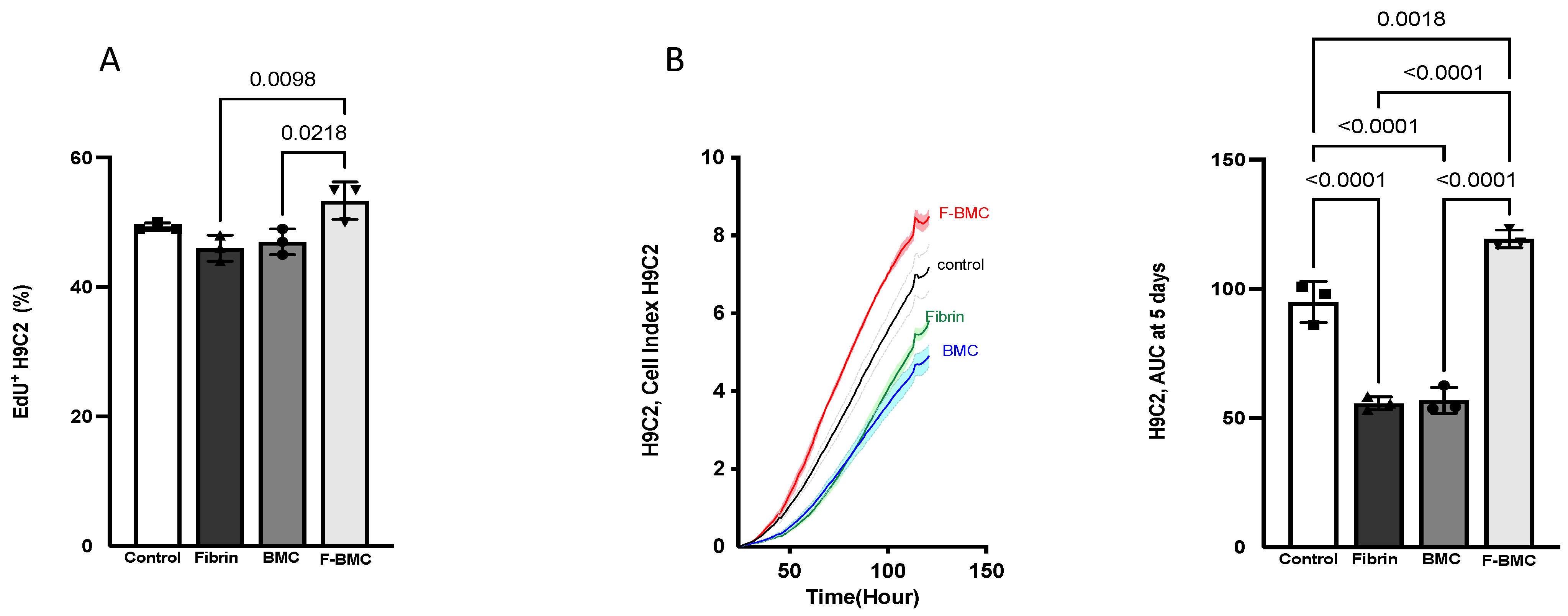
3.2.4. F-BMC Secretome Promotes Cardiomyoblast Spreading
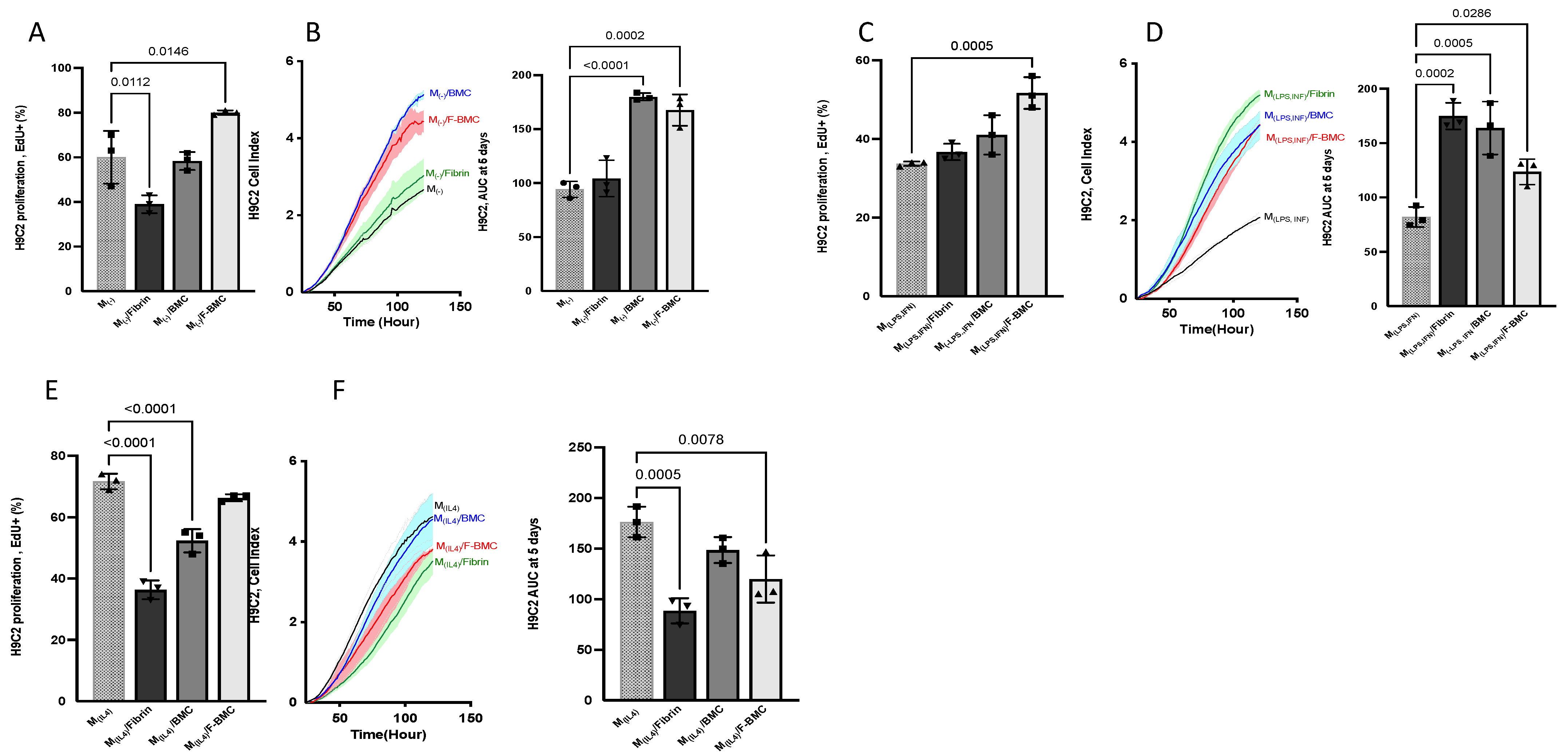
3.2.5. Alternatively-Activated Macrophages Promote Cardiomyoblast Proliferation
3.2.6. F-BMC-Educated-Macrophages Demonstrate Paracrine Mitogenic Properties on Cardiac Cells
4. Discussion
4.1. Impact of Fibrin on BMC and Their Properties
4.2. Cardiomyoblast Fate
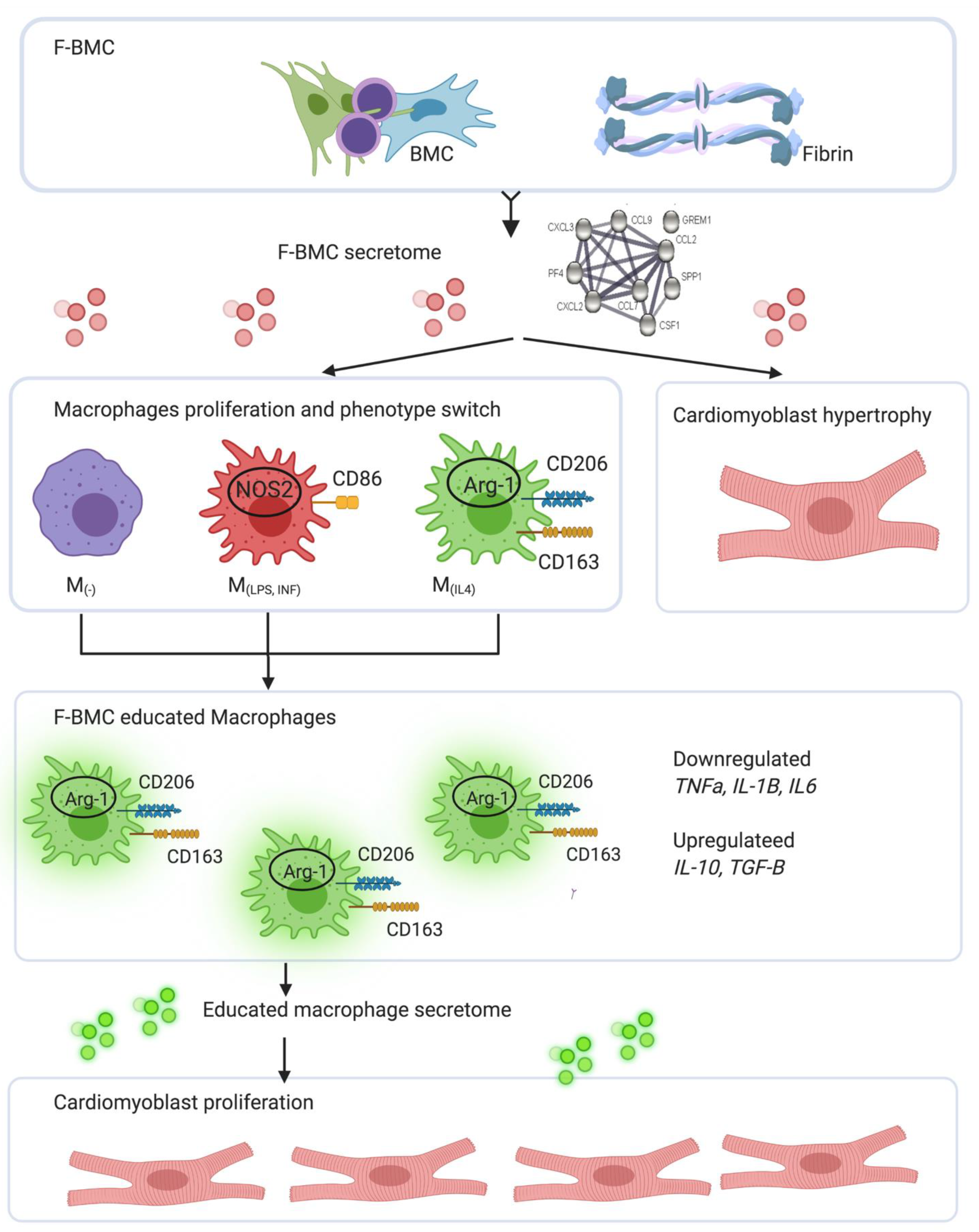
4.3. Integrated Concept
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lavine, K.J.; Pinto, A.R.; Epelman, S.; Kopecky, B.J.; Clemente-Casares, X.; Godwin, J.; Rosenthal, N.; Kovacic, J.C. The Macrophage in Cardiac Homeostasis and Disease: JACC Macrophage in CVD Series (Part 4). J. Am. Coll. Cardiol. 2018, 72, 2213–2230. [Google Scholar] [CrossRef] [PubMed]
- Frangogiannis, N.G. Regulation of the inflammatory response in cardiac repair. Circ. Res. 2012, 110, 159–173. [Google Scholar] [CrossRef]
- Aurora, A.B.; Porrello, E.R.; Tan, W.; Mahmoud, A.I.; Hill, J.A.; Bassel-Duby, R.; Sadek, H.A.; Olson, E.N. Macrophages are required for neonatal heart regeneration. J. Clin. Investig. 2014, 124, 1382–1392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef] [Green Version]
- Lafuse, W.P.; Wozniak, D.J.; Rajaram, M.V.S. Role of Cardiac Macrophages on Cardiac Inflammation, Fibrosis and Tissue Repair. Cells 2020, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Psarras, S.; Beis, D.; Nikouli, S.; Tsikitis, M.; Capetanaki, Y. Three in a Box: Understanding Cardiomyocyte, Fibroblast, and Innate Immune Cell Interactions to Orchestrate Cardiac Repair Processes. Front. Cardiovasc. Med. 2019, 6, 32. [Google Scholar] [CrossRef]
- Moskalik, A.; Niderla-Bielinska, J.; Ratajska, A. Multiple roles of cardiac macrophages in heart homeostasis and failure. Heart Fail. Rev. 2021. [Google Scholar] [CrossRef]
- Carotenuto, F.; Teodori, L.; Maccari, A.M.; Delbono, L.; Orlando, G.; Di Nardo, P. Turning regenerative technologies into treatment to repair myocardial injuries. J. Cell. Mol. Med. 2020, 24, 2704–2716. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Chen, R.; Chakrabarti, S.; Su, Z. Resident macrophages as potential therapeutic targets for cardiac ageing and injury. Clin. Transl. Immunol. 2020, 9, e1167. [Google Scholar] [CrossRef]
- Vagnozzi, R.J.; Maillet, M.; Sargent, M.A.; Khalil, H.; Johansen, A.K.Z.; Schwanekamp, J.A.; York, A.J.; Huang, V.; Nahrendorf, M.; Sadayappan, S.; et al. An acute immune response underlies the benefit of cardiac stem cell therapy. Nature 2020, 577, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.J.; Khan, M.; Mohsin, S. Healing the Broken Heart; The Immunomodulatory Effects of Stem Cell Therapy. Front. Immunol. 2020, 11, 639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Tian, H.; Yang, C.; Liu, J.; Zhang, H.; Wang, J.; Hu, S.; Sun, Z.; He, K.; Chen, G. Mesenchymal Stem Cells Promote the Resolution of Cardiac Inflammation After Ischemia Reperfusion Via Enhancing Efferocytosis of Neutrophils. J. Am. Heart Assoc. 2020, 9, e014397. [Google Scholar] [CrossRef]
- Podaru, M.N.; Fields, L.; Kainuma, S.; Ichihara, Y.; Hussain, M.; Ito, T.; Kobayashi, K.; Mathur, A.; D’Acquisto, F.; Lewis-McDougall, F.; et al. Reparative macrophage transplantation for myocardial repair: A refinement of bone marrow mononuclear cell-based therapy. Basic Res. Cardiol. 2019, 114, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carty, F.; Mahon, B.P.; English, K. The influence of macrophages on mesenchymal stromal cell therapy: Passive or aggressive agents? Clin. Exp. Immunol. 2017, 188, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khodayari, S.; Khodayari, H.; Amiri, A.Z.; Eslami, M.; Farhud, D.; Hescheler, J.; Nayernia, K. Inflammatory Microenvironment of Acute Myocardial Infarction Prevents Regeneration of Heart with Stem Cells Therapy. Cell Physiol. Biochem. 2019, 53, 887–909. [Google Scholar]
- Deng, S.; Zhou, X.; Ge, Z.; Song, Y.; Wang, H.; Liu, X.; Zhang, D. Exosomes from adipose-derived mesenchymal stem cells ameliorate cardiac damage after myocardial infarction by activating S1P/SK1/S1PR1 signaling and promoting macrophage M2 polarization. Int. J. Biochem. Cell. Biol. 2019, 114, 105564. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Han, B.; Wang, Y.; Wang, C.; Zhang, H.; Xue, J.; Wang, X.; Niu, T.; Niu, Z.; Chen, Y. Mesenchymal stem cell-secreted extracellular vesicles carrying TGF-beta1 upregulate miR-132 and promote mouse M2 macrophage polarization. J. Cell. Mol. Med. 2020, 24, 12750–12764. [Google Scholar] [CrossRef]
- Madonna, R.; Van Laake, L.W.; Botker, H.E.; Davidson, S.M.; De Caterina, R.; Engel, F.B.; Eschenhagen, T.; Fernandez-Aviles, F.; Hausenloy, D.J.; Hulot, J.S.; et al. ESC Working Group on Cellular Biology of the Heart: Position paper for Cardiovascular Research: Tissue engineering strategies combined with cell therapies for cardiac repair in ischaemic heart disease and heart failure. Cardiovasc. Res. 2019, 115, 488–500. [Google Scholar] [CrossRef] [Green Version]
- Valles, G.; Bensiamar, F.; Crespo, L.; Arruebo, M.; Vilaboa, N.; Saldana, L. Topographical cues regulate the crosstalk between MSCs and macrophages. Biomaterials 2015, 37, 124–133. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Shu, Z.; Qian, K.; Wang, J.; Zhu, H. Harnessing the Properties of Biomaterial to Enhance the Immunomodulation of Mesenchymal Stem Cells. Tissue Eng. Part B Rev. 2019, 25, 492–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roura, S.; Galvez-Monton, C.; Bayes-Genis, A. Fibrin, the preferred scaffold for cell transplantation after myocardial infarction? An old molecule with a new life. J. Tissue Eng. Regen. Med. 2017, 11, 2304–2313. [Google Scholar] [CrossRef] [PubMed]
- Frobert, A.; Valentin, J.; Cook, S.; Lopes-Vicente, J.; Giraud, M.N. Cell-based therapy for heart failure in rat: Double thoracotomy for myocardial infarction and epicardial implantation of cells and biomatrix. J. Vis. Exp. 2014, 91, 51390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frobert, A.; Valentin, J.; Magnin, J.L.; Riedo, E.; Cook, S.; Giraud, M.N. Prognostic Value of Troponin I for Infarct Size to Improve Preclinical Myocardial Infarction Small Animal Models. Front. Physiol. 2015, 6, 353. [Google Scholar] [CrossRef] [Green Version]
- Valentin, J.; Frobert, A.; Ajalbert, G.; Cook, S.; Giraud, M.N. Histological Quantification of Chronic Myocardial Infarct in Rats. J. Vis. Exp. 2016, 118, e54914. [Google Scholar] [CrossRef]
- Vasandan, A.B.; Jahnavi, S.; Shashank, C.; Prasad, P.; Kumar, A.; Prasanna, S.J. Human Mesenchymal stem cells program macrophage plasticity by altering their metabolic status via a PGE2-dependent mechanism. Sci. Rep. 2016, 6, 38308. [Google Scholar] [CrossRef] [Green Version]
- Roshan Moniri, M.; Young, A.; Reinheimer, K.; Rayat, J.; Dai, L.J.; Warnock, G.L. Dynamic assessment of cell viability, proliferation and migration using real time cell analyzer system (RTCA). Cytotechnology 2015, 67, 379–386. [Google Scholar] [CrossRef] [Green Version]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic. Acids. Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Rafatian, G.; Davis, D.R. Concise Review: Heart-Derived Cell Therapy 2.0: Paracrine Strategies to Increase Therapeutic Repair of Injured Myocardium. Stem Cells 2018, 36, 1794–1803. [Google Scholar] [CrossRef]
- Guex, A.G.; Frobert, A.; Valentin, J.; Fortunato, G.; Hegemann, D.; Cook, S.; Carrel, T.P.; Tevaearai, H.T.; Giraud, M.N. Plasma-functionalized electrospun matrix for biograft development and cardiac function stabilization. Acta Biomater. 2014, 10, 2996–3006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaka, R.; Davis, D.R. State-of-play for cellular therapies in cardiac repair and regeneration. Stem Cells 2021, 39, 1579–1588. [Google Scholar] [CrossRef] [PubMed]
- Kanda, P.; Davis, D.R. Cellular mechanisms underlying cardiac engraftment of stem cells. Expert. Opin. Biol. Ther. 2017, 17, 1127–1143. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Hill, K.L.; Li, Q.; Suntharalingam, P.; Mansoor, A.; Wang, X.; Jameel, M.N.; Zhang, P.; Swingen, C.; Kaufman, D.S.; et al. A fibrin patch-based enhanced delivery of human embryonic stem cell-derived vascular cell transplantation in a porcine model of postinfarction left ventricular remodeling. Stem Cells 2011, 29, 367–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vegas, A.J.; Veiseh, O.; Doloff, J.C.; Ma, M.; Tam, H.H.; Bratlie, K.; Li, J.; Bader, A.R.; Langan, E.; Olejnik, K.; et al. Combinatorial hydrogel library enables identification of materials that mitigate the foreign body response in primates. Nat. Biotechnol. 2016, 34, 345–352. [Google Scholar] [CrossRef] [Green Version]
- Stevens, H.Y.; Bowles, A.C.; Yeago, C.; Roy, K. Molecular Crosstalk Between Macrophages and Mesenchymal Stromal Cells. Front. Cell. Dev. Biol. 2020, 8, 600160. [Google Scholar] [CrossRef]
- Haasper, C.; Breitbart, A.; Hankemeier, S.; Wehmeier, M.; Hesse, E.; Citak, M.; Krettek, C.; Zeichen, J.; Jagodzinski, M. Influence of fibrin glue on proliferation and differentiation of human bone marrow stromal cells seeded on a biologic 3-dimensional matrix. Technol. Health Care 2008, 16, 93–101. [Google Scholar] [CrossRef]
- Stolzing, A.; Colley, H.; Scutt, A. Effect of age and diabetes on the response of mesenchymal progenitor cells to fibrin matrices. Int. J. Biomater. 2011, 2011, 378034. [Google Scholar] [CrossRef]
- Luz-Crawford, P.; Djouad, F.; Toupet, K.; Bony, C.; Franquesa, M.; Hoogduijn, M.J.; Jorgensen, C.; Noel, D. Mesenchymal Stem Cell-Derived Interleukin 1 Receptor Antagonist Promotes Macrophage Polarization and Inhibits B Cell Differentiation. Stem Cells 2016, 34, 483–492. [Google Scholar] [CrossRef] [Green Version]
- Melief, S.M.; Geutskens, S.B.; Fibbe, W.E.; Roelofs, H. Multipotent stromal cells skew monocytes towards an anti-inflammatory interleukin-10-producing phenotype by production of interleukin-6. Haematologica 2013, 98, 888–895. [Google Scholar] [CrossRef] [Green Version]
- Chiossone, L.; Conte, R.; Spaggiari, G.M.; Serra, M.; Romei, C.; Bellora, F.; Becchetti, F.; Andaloro, A.; Moretta, L.; Bottino, C. Mesenchymal Stromal Cells Induce Peculiar Alternatively Activated Macrophages Capable of Dampening Both Innate and Adaptive Immune Responses. Stem Cells 2016, 34, 1909–1921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maggini, J.; Mirkin, G.; Bognanni, I.; Holmberg, J.; Piazzon, I.M.; Nepomnaschy, I.; Costa, H.; Canones, C.; Raiden, S.; Vermeulen, M.; et al. Mouse bone marrow-derived mesenchymal stromal cells turn activated macrophages into a regulatory-like profile. PLoS ONE 2010, 5, e9252. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Shapiro, L.; Flynn, A. The clinical application of mesenchymal stem cells and cardiac stem cells as a therapy for cardiovascular disease. Pharmacol. Ther. 2015, 151, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Shindo, K.; Donahue, R.R.; Abdel-Latif, A. Cardiac Cell Therapy: Insights into the Mechanisms of Tissue Repair. Int. J. Mol. Sci. 2021, 22, 1201. [Google Scholar] [CrossRef]
- Kim, Y.; Nurakhayev, S.; Nurkesh, A.; Zharkinbekov, Z.; Saparov, A. Macrophage Polarization in Cardiac Tissue Repair Following Myocardial Infarction. Int. J. Mol. Sci. 2021, 22, 2715. [Google Scholar] [CrossRef]
- Hobby, A.R.H.; Berretta, R.M.; Eaton, D.M.; Kubo, H.; Feldsott, E.; Yang, Y.; Headrick, A.L.; Koch, K.A.; Rubino, M.; Kurian, J.; et al. Cortical bone stem cells modify cardiac inflammation after myocardial infarction by inducing a novel macrophage phenotype. Am. J. Physiol. Heart Circ. Physiol. 2021, 321, H684–H701. [Google Scholar] [CrossRef]
- Braga, T.T.; Agudelo, J.S.; Camara, N.O. Macrophages During the Fibrotic Process: M2 as Friend and Foe. Front. Immunol. 2015, 6, 602. [Google Scholar] [CrossRef] [Green Version]
- Ortiz, L.A.; Dutreil, M.; Fattman, C.; Pandey, A.C.; Torres, G.; Go, K.; Phinney, D.G. Interleukin 1 receptor antagonist mediates the antiinflammatory and antifibrotic effect of mesenchymal stem cells during lung injury. Proc. Natl. Acad. Sci. USA 2007, 104, 11002–11007. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.Q.; Wan, L.Y.; He, X.M.; Ni, Y.R.; Wang, C.; Liu, C.B.; Wu, J.F. Gremlin1 Accelerates Hepatic Stellate Cell Activation Through Upregulation of TGF-Beta Expression. DNA Cell. Biol. 2017, 36, 603–610. [Google Scholar] [CrossRef]
- Hsieh, J.Y.; Smith, T.D.; Meli, V.S.; Tran, T.N.; Botvinick, E.L.; Liu, W.F. Differential regulation of macrophage inflammatory activation by fibrin and fibrinogen. Acta Biomater. 2017, 47, 14–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, C.M.; de Azevedo Queiroz, I.O.; Ervolino, E.; Cintra, L.T.A.; Gomes-Filho, J.E. RUNX-2, OPN and OCN expression induced by grey and white mineral trioxide aggregate in normal and hypertensive rats. Int. Endod. J. 2018, 51, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Bollini, S.; Emanueli, C. To serve and protect: A new heart patrolling and recycling role for macrophages. Cardiovasc. Res. 2021, 117, e17–e20. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Spinali, K.; Schmuck, E.G.; Kink, J.A.; Hematti, P.; Raval, A.N. Cardiac fibroblast derived matrix-educated macrophages express VEGF and IL-6, and recruit mesenchymal stromal cells. J. Immunol. Regen. Med. 2020, 10, 100033. [Google Scholar] [CrossRef] [PubMed]
- Hitscherich, P.; Lee, E.J. Crosstalk Between Cardiac Cells and Macrophages Postmyocardial Infarction: Insights from In Vitro Studies. Tissue Eng. Part B Rev. 2021, 27, 475–485. [Google Scholar] [CrossRef]
- Alvarez-Argote, S.; O’Meara, C.C. The Evolving Roles of Cardiac Macrophages in Homeostasis, Regeneration, and Repair. Int. J. Mol. Sci. 2021, 22, 7923. [Google Scholar] [CrossRef]
- Vagnozzi, R.J.; Molkentin, J.D. Resident macrophages keep mitochondria running in the heart. Cell Res. 2020, 30, 1057–1058. [Google Scholar] [CrossRef]
- Bartelt, A.; Weber, C. Mitochondrial Ejection for Cardiac Protection: The Macrophage Connection. Cell Metab. 2020, 32, 512–513. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borrego, I.; Frobert, A.; Ajalbert, G.; Valentin, J.; Kaltenrieder, C.; Fellay, B.; Stumpe, M.; Cook, S.; Dengjel, J.; Giraud, M.-N. Fibrin, Bone Marrow Cells and Macrophages Interactively Modulate Cardiomyoblast Fate. Biomedicines 2022, 10, 527. https://doi.org/10.3390/biomedicines10030527
Borrego I, Frobert A, Ajalbert G, Valentin J, Kaltenrieder C, Fellay B, Stumpe M, Cook S, Dengjel J, Giraud M-N. Fibrin, Bone Marrow Cells and Macrophages Interactively Modulate Cardiomyoblast Fate. Biomedicines. 2022; 10(3):527. https://doi.org/10.3390/biomedicines10030527
Chicago/Turabian StyleBorrego, Inês, Aurélien Frobert, Guillaume Ajalbert, Jérémy Valentin, Cyrielle Kaltenrieder, Benoît Fellay, Michael Stumpe, Stéphane Cook, Joern Dengjel, and Marie-Noëlle Giraud. 2022. "Fibrin, Bone Marrow Cells and Macrophages Interactively Modulate Cardiomyoblast Fate" Biomedicines 10, no. 3: 527. https://doi.org/10.3390/biomedicines10030527
APA StyleBorrego, I., Frobert, A., Ajalbert, G., Valentin, J., Kaltenrieder, C., Fellay, B., Stumpe, M., Cook, S., Dengjel, J., & Giraud, M.-N. (2022). Fibrin, Bone Marrow Cells and Macrophages Interactively Modulate Cardiomyoblast Fate. Biomedicines, 10(3), 527. https://doi.org/10.3390/biomedicines10030527