Evaluation and Comparison of Multi-Omics Data Integration Methods for Subtyping of Cutaneous Melanoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Preprocessing and Analysis
2.2. RNA-Seq Data Preprocessing
2.3. Joint Singular Value Decomposition
2.4. Performance Evaluation
2.5. Feature Analysis
2.6. Skin Cutaneous Melanoma Molecular and Clinical Features
3. Results
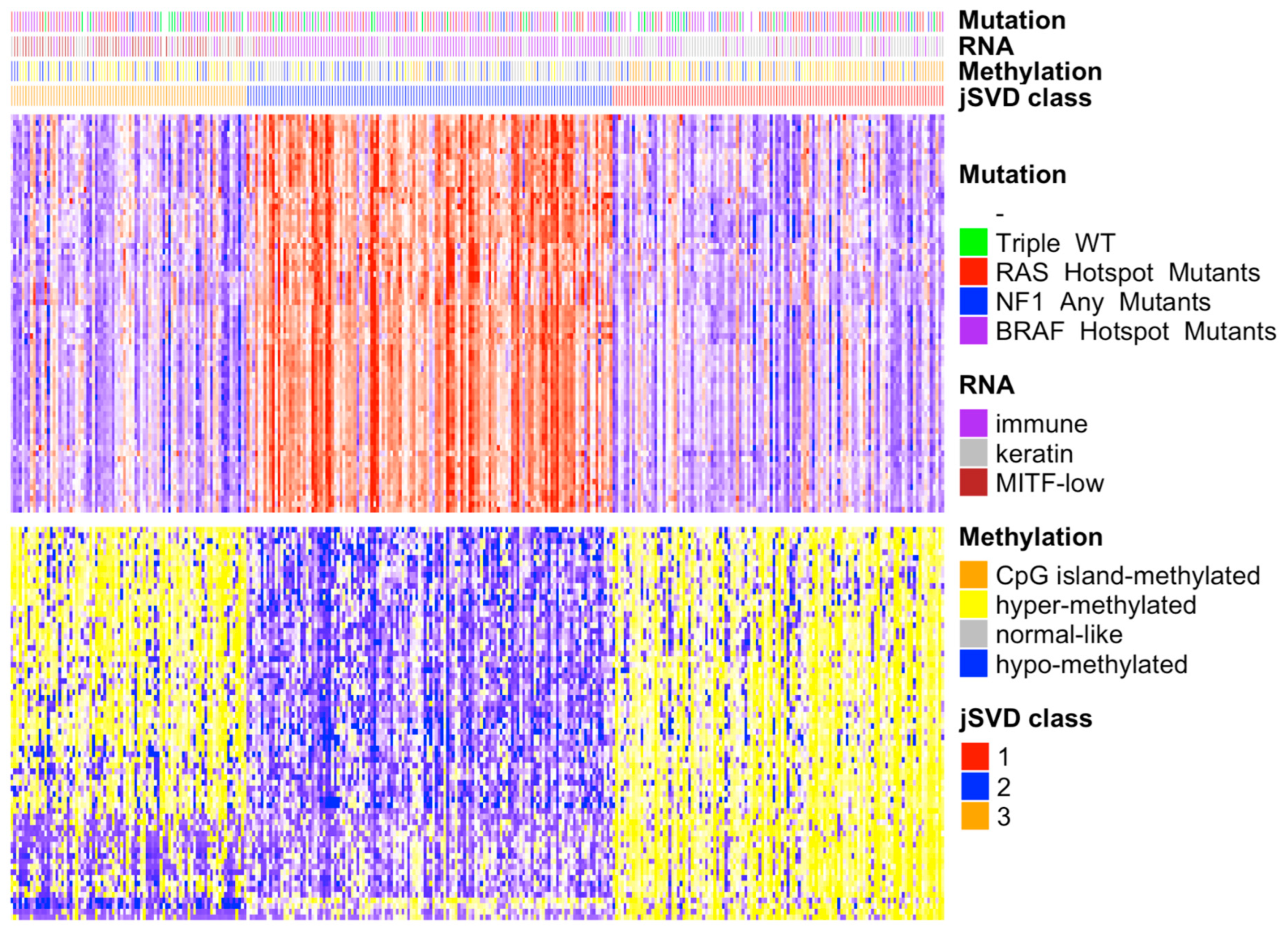
3.1. Data Fusion
3.2. Feature Extraction
3.3. Survival
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- MacArthur, J.; Bowler, E.; Cerezo, M.; Gil, L.; Hall, P.; Hastings, E.; Junkins, H.; McMahon, A.; Milano, A.; Morales, J.; et al. The New NHGRI-EBI Catalog of Published Genome-Wide Association Studies (GWAS Catalog). Nucleic Acids Res. 2017, 45, D896–D901. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Takaya, H.; Nakai, H.; Takamatsu, S.; Mandai, M.; Matsumura, N. Homologous Recombination Deficiency Status-Based Classification of High-Grade Serous Ovarian Carcinoma. Sci. Rep. 2020, 10, 2757. [Google Scholar] [CrossRef] [PubMed]
- Dotolo, S.; Esposito Abate, R.; Roma, C.; Guido, D.; Preziosi, A.; Tropea, B.; Palluzzi, F.; Giacò, L.; Normanno, N. Bioinformatics: From NGS Data to Biological Complexity in Variant Detection and Oncological Clinical Practice. Biomedicines 2022, 10, 2074. [Google Scholar] [CrossRef] [PubMed]
- Sahnane, N.; Carnevali, I.; Formenti, G.; Casarin, J.; Facchi, S.; Bombelli, R.; Di Lauro, E.; Memoli, D.; Salvati, A.; Rizzo, F.; et al. BRCA Methylation Testing Identifies a Subset of Ovarian Carcinomas without Germline Variants That Can Benefit from PARP Inhibitor. Int. J. Mol. Sci. 2020, 21, 9708. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, M.; Uschmajew, A.; Amaro, A.; Pfeffer, U. Data Fusion Techniques for the Integration of Multi-Domain Genomic Data from Uveal Melanoma. Cancers 2019, 11, 1434. [Google Scholar] [CrossRef]
- Gliozzo, J.; Mesiti, M.; Notaro, M.; Petrini, A.; Patak, A.; Puertas-Gallardo, A.; Paccanaro, A.; Valentini, G.; Casiraghi, E. Heterogeneous Data Integration Methods for Patient Similarity Networks. Brief. Bioinform. 2022, 23, bbac207. [Google Scholar] [CrossRef]
- Wang, B.; Mezlini, A.M.; Demir, F.; Fiume, M.; Tu, Z.; Brudno, M.; Haibe-Kains, B.; Goldenberg, A. Similarity Network Fusion for Aggregating Data Types on a Genomic Scale. Nat. Methods 2014, 11, 333–337. [Google Scholar] [CrossRef]
- John, C.R.; Watson, D.; Barnes, M.R.; Pitzalis, C.; Lewis, M.J. Spectrum: Fast Density-Aware Spectral Clustering for Single and Multi-Omic Data. Bioinform. Oxf. Engl. 2020, 36, 1159–1166. [Google Scholar] [CrossRef]
- Yu, T. AIME: Autoencoder-Based Integrative Multi-Omics Data Embedding That Allows for Confounder Adjustments. PLoS Comput. Biol. 2022, 18, e1009826. [Google Scholar] [CrossRef]
- Duan, R.; Gao, L.; Gao, Y.; Hu, Y.; Xu, H.; Huang, M.; Song, K.; Wang, H.; Dong, Y.; Jiang, C.; et al. Evaluation and Comparison of Multi-Omics Data Integration Methods for Cancer Subtyping. PLoS Comput. Biol. 2021, 17, e1009224. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, B.; Schimek, M.G. A Hierarchical Clustering and Data Fusion Approach for Disease Subtype Discovery. J. Biomed. Inform. 2021, 113, 103636. [Google Scholar] [CrossRef] [PubMed]
- Rappoport, N.; Shamir, R. NEMO: Cancer Subtyping by Integration of Partial Multi-Omic Data. Bioinform. Oxf. Engl. 2019, 35, 3348–3356. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [CrossRef]
- Robbins, S.L.; Kumar, V.; Cotran, R.S. Robbins and Cotran Pathologic Basis of Disease, 8th ed.; Saunders/Elsevier: Philadelphia, PA, USA, 2010; ISBN 978-1-4160-3121-5. [Google Scholar]
- Rossi, M.; Pellegrini, C.; Cardelli, L.; Ciciarelli, V.; Di Nardo, L.; Fargnoli, M.C. Familial Melanoma: Diagnostic and Management Implications. Dermatol. Pract. Concept. 2019, 9, 10–16. [Google Scholar] [CrossRef]
- Gu, F.; Chen, T.-H.; Pfeiffer, R.M.; Fargnoli, M.C.; Calista, D.; Ghiorzo, P.; Peris, K.; Puig, S.; Menin, C.; De Nicolo, A.; et al. Combining Common Genetic Variants and Non-Genetic Risk Factors to Predict Risk of Cutaneous Melanoma. Hum. Mol. Genet. 2018, 27, 4145–4156. [Google Scholar] [CrossRef]
- Nissan, M.H.; Pratilas, C.A.; Jones, A.M.; Ramirez, R.; Won, H.; Liu, C.; Tiwari, S.; Kong, L.; Hanrahan, A.J.; Yao, Z.; et al. Loss of NF1 in Cutaneous Melanoma Is Associated with RAS Activation and MEK Dependence. Cancer Res. 2014, 74, 2340–2350. [Google Scholar] [CrossRef]
- Conway, K.; Tsai, Y.S.; Edmiston, S.N.; Parker, J.S.; Parrish, E.A.; Hao, H.; Kuan, P.F.; Scott, G.A.; Frank, J.S.; Googe, P.; et al. Characterization of the CpG Island Hypermethylated Phenotype Subclass in Primary Melanomas. J. Investig. Dermatol. 2022, 142, 1869–1881.e10. [Google Scholar] [CrossRef]
- Koroknai, V.; Szász, I.; Hernandez-Vargas, H.; Fernandez-Jimenez, N.; Cuenin, C.; Herceg, Z.; Vízkeleti, L.; Ádány, R.; Ecsedi, S.; Balázs, M. DNA Hypermethylation Is Associated with Invasive Phenotype of Malignant Melanoma. Exp. Dermatol. 2020, 29, 39–50. [Google Scholar] [CrossRef]
- Samur, M.K. RTCGAToolbox: A New Tool for Exporting TCGA Firehose Data. PLoS ONE 2014, 9, e106397. [Google Scholar] [CrossRef]
- Townsend, J.; Koep, N.; Weinchwald, S. Pymanopt: A Python Toolbox for Optimization on Manifolds Using Automatic Differentiation. J. Mach. Learn. Res. 2016, 17, 1–5. [Google Scholar]
- Sato, H. Joint Singular Value Decomposition Algorithm Based on the Riemannian Trust-Region Method. JSIAM Lett. 2015, 7, 13–16. [Google Scholar] [CrossRef]
- Wilkerson, M.D.; Hayes, D.N. ConsensusClusterPlus: A Class Discovery Tool with Confidence Assessments and Item Tracking. Bioinformatics 2010, 26, 1572–1573. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Hubert, L.; Arabie, P. Comparing Partitions. J. Classif. 1985, 2, 193–218. [Google Scholar] [CrossRef]
- Azzalini, A.; Menardi, G. Clustering via Nonparametric Density Estimation: The R Package PdfCluster. arXiv 2013, arXiv:1301.6559. [Google Scholar]
- Mächer, M.; Rousseeuw, P.; Stryuf, A.; Hubert, M.; Hornik, K. Cluster: Cluster Analysis Basics and Extensions; ETH Zürich: Zürich, Switzerland, 2012. [Google Scholar]
- Tibishirani, R.; Seo, M.J.; Chu, G.; Balasubramanian, N.; Jun, L. SAM: Significance Analysis of Microarrays; R Package Version 3.0. 2018. Available online: https://cran.r-project.org/web/packages/samr/samr.pdf (accessed on 6 December 2022).
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Lauss, M.; Nsengimana, J.; Staaf, J.; Newton-Bishop, J.; Jönsson, G. Consensus of Melanoma Gene Expression Subtypes Converges on Biological Entities. J. Investig. Dermatol. 2016, 136, 2502–2505. [Google Scholar] [CrossRef]
- Therneau, T. A Package for Survival Analysis in R; R Package Version 3.2-11. 2022. Available online: https://github.com/therneau/survival (accessed on 6 December 2022).
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data: Extending the Cox Model (Statistics for Biology and Health); Springer: New York, NY, USA, 2000; ISBN 978-0-387-98784-2. [Google Scholar]
- Kassambara, A.; Kosinski, M.; Biecek, P. Survminer: Drawing Survival Curves Using “Ggplot2”, R Package Version 0.4.9. 2021. Available online: https://cran.r-project.org/web/packages/survminer/survminer.pdf (accessed on 6 December 2022).
- Rossi, E.; Croce, M.; Reggiani, F.; Schinzari, G.; Ambrosio, M.; Gangemi, R.; Tortora, G.; Pfeffer, U.; Amaro, A. Uveal Melanoma Metastasis. Cancers 2021, 13, 5684. [Google Scholar] [CrossRef]
- Robertson, A.G.; Shih, J.; Yau, C.; Gibb, E.A.; Oba, J.; Mungall, K.L.; Hess, J.M.; Uzunangelov, V.; Walter, V.; Danilova, L.; et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell 2017, 32, 204–220.e15. [Google Scholar] [CrossRef] [PubMed]
- Andreoletti, G.; Pal, L.R.; Moult, J.; Brenner, S.E. Reports from the Fifth Edition of CAGI: The Critical Assessment of Genome Interpretation. Hum. Mutat. 2019, 40, 1197–1201. [Google Scholar] [CrossRef] [PubMed]
- Walsh, I.; Fishman, D.; Garcia-Gasulla, D.; Titma, T.; Pollastri, G.; ELIXIR Machine Learning Focus Group; Harrow, J.; Psomopoulos, F.E.; Tosatto, S.C.E. DOME: Recommendations for Supervised Machine Learning Validation in Biology. Nat. Methods 2021, 18, 1122–1127. [Google Scholar] [CrossRef] [PubMed]
- Monzon, A.M.; Carraro, M.; Chiricosta, L.; Reggiani, F.; Han, J.; Ozturk, K.; Wang, Y.; Miller, M.; Bromberg, Y.; Capriotti, E.; et al. Performance of Computational Methods for the Evaluation of Pericentriolar Material 1 Missense Variants in CAGI-5. Hum. Mutat. 2019, 40, 1474–1485. [Google Scholar] [CrossRef] [PubMed]
- Carraro, M.; Minervini, G.; Giollo, M.; Bromberg, Y.; Capriotti, E.; Casadio, R.; Dunbrack, R.; Elefanti, L.; Fariselli, P.; Ferrari, C.; et al. Performance of in Silico Tools for the Evaluation of P16INK4a (CDKN2A) Variants in CAGI. Hum. Mutat. 2017, 38, 1042–1050. [Google Scholar] [CrossRef]
- Reggiani, F.; Carraro, M.; Belligoli, A.; Sanna, M.; Dal Prà, C.; Favaretto, F.; Ferrari, C.; Vettor, R.; Tosatto, S.C.E. In Silico Prediction of Blood Cholesterol Levels from Genotype Data. PLoS ONE 2020, 15, e0227191. [Google Scholar] [CrossRef]
- Noushmehr, H.; Weisenberger, D.J.; Diefes, K.; Phillips, H.S.; Pujara, K.; Berman, B.P.; Pan, F.; Pelloski, C.E.; Sulman, E.P.; Bhat, K.P.; et al. Identification of a CpG Island Methylator Phenotype That Defines a Distinct Subgroup of Glioma. Cancer Cell 2010, 17, 510–522. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor Package for Integrative Analysis of TCGA Data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- da Silva, V.; Ramos, M.; Groenen, M.; Crooijmans, R.; Johansson, A.; Regitano, L.; Coutinho, L.; Zimmer, R.; Waldron, L.; Geistlinger, L. CNVRanger: Association Analysis of CNVs with Gene Expression and Quantitative Phenotypes. Bioinform. Oxf. Engl. 2020, 36, 972–973. [Google Scholar] [CrossRef]
- Huang, A.C.; Zappasodi, R. A Decade of Checkpoint Blockade Immunotherapy in Melanoma: Understanding the Molecular Basis for Immune Sensitivity and Resistance. Nat. Immunol. 2022, 23, 660–670. [Google Scholar] [CrossRef]
- Ortega, M.; Fraile-Martínez, O.; García-Honduvilla, N.; Coca, S.; Álvarez-Mon, M.; Buján, J.; Teus, M. Update on Uveal Melanoma: Translational Research from Biology to Clinical Practice (Review). Int. J. Oncol. 2020, 57, 1262–1279. [Google Scholar] [CrossRef] [PubMed]
- Castellanos, E.; Gel, B.; Rosas, I.; Tornero, E.; Santín, S.; Pluvinet, R.; Velasco, J.; Sumoy, L.; del Valle, J.; Perucho, M.; et al. A Comprehensive Custom Panel Design for Routine Hereditary Cancer Testing: Preserving Control, Improving Diagnostics and Revealing a Complex Variation Landscape. Sci. Rep. 2017, 7, 39348. [Google Scholar] [CrossRef] [PubMed]
- Dowle, M.; Srinivasan, A. Data.Table: Extension of “Data.Frame” R Package Version 1.14.2. 2022. Available online: https://cran.r-project.org/web/packages/data.table/data.table.pdf (accessed on 6 December 2022).
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A Flexible and Comprehensive Bioconductor Package for the Analysis of Infinium DNA Methylation Microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Mayakonda, A.; Lin, D.-C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and Comprehensive Analysis of Somatic Variants in Cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K. IlluminaHumanMethylation450kanno.Ilmn12.Hg19: Annotation for Illumina’s 450k Methylation Arrays. R Package Version 0.6.0. 2016. Available online: https://bioconductor.org/packages/release/data/annotation/html/IlluminaHumanMethylation450kanno.ilmn12.hg19.html (accessed on 6 December 2022).
- Carlson, M. Org.Hs.Eg.Db: Genome Wide Annotation for Human. R Package Version 3.15.0. 2019. Available online: https://bioconductor.org/packages/release/data/annotation/html/org.Hs.eg.db.html (accessed on 6 December 2022).



| Genomic Domain/DF Method | Mutation Subtypes (TCGA) | RNA-seq Cluster Consenhier (TCGA) | Methylation Types 2014 08 (TCGA) | Silhouette Score |
|---|---|---|---|---|
| RNA-seq | −0.014 | 0.287 | 0.117 | 0.07, 0.135, 0.018, 0.037 |
| Methylation | 0.004 | 0.061 | 0.514 | 0.025, 0.026, 0.058 |
| Spectrum | 0.039 | 0.128 | 0.38 | 0.024, 0.03, 0.026 |
| SNF | 0.027 | 0.152 | 0.275 | 0.008, 0.004, 0.004 |
| NEMO | 0.029 | 0.108 | 0.195 | 0.05, −0.017, −0.015, −0.03, −0.008, 0.005, 0.031, −0.014, −0.027, −0.019, −0.023, −0.009 |
| jSVD | 0 | 0.272 | 0.147 | 0.212, 0.339, 0.281 |
| Mutation subtypes (TCGA) | 1 | 0.007 | 0.021 | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amaro, A.; Pfeffer, M.; Pfeffer, U.; Reggiani, F. Evaluation and Comparison of Multi-Omics Data Integration Methods for Subtyping of Cutaneous Melanoma. Biomedicines 2022, 10, 3240. https://doi.org/10.3390/biomedicines10123240
Amaro A, Pfeffer M, Pfeffer U, Reggiani F. Evaluation and Comparison of Multi-Omics Data Integration Methods for Subtyping of Cutaneous Melanoma. Biomedicines. 2022; 10(12):3240. https://doi.org/10.3390/biomedicines10123240
Chicago/Turabian StyleAmaro, Adriana, Max Pfeffer, Ulrich Pfeffer, and Francesco Reggiani. 2022. "Evaluation and Comparison of Multi-Omics Data Integration Methods for Subtyping of Cutaneous Melanoma" Biomedicines 10, no. 12: 3240. https://doi.org/10.3390/biomedicines10123240
APA StyleAmaro, A., Pfeffer, M., Pfeffer, U., & Reggiani, F. (2022). Evaluation and Comparison of Multi-Omics Data Integration Methods for Subtyping of Cutaneous Melanoma. Biomedicines, 10(12), 3240. https://doi.org/10.3390/biomedicines10123240






