CircDOCK1 Regulates miR-186/DNMT3A to Promote Osteosarcoma Progression
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. Bioinformatics Analysis
2.3. Cell Culture and Transfection
2.4. RNA Extraction and qRT-PCR
2.5. Ribonuclease R (RNase R) Digestion
2.6. Sanger Sequencing
2.7. Fluorescence In Situ Hybridization (FISH)
2.8. Cell Proliferation
2.9. Colony Formation Assay
2.10. Transwell Analysis
2.11. Wound-Healing Assay
2.12. Dual-Luciferase Reporter Analysis
2.13. RNA Immunoprecipitation (RIP) Assay
2.14. Western Blotting
2.15. Xenograft Model
2.16. Statistical Analysis
3. Results
3.1. CircDOCK1 Expression Is Highly Expressed in OS Tissues and Cells
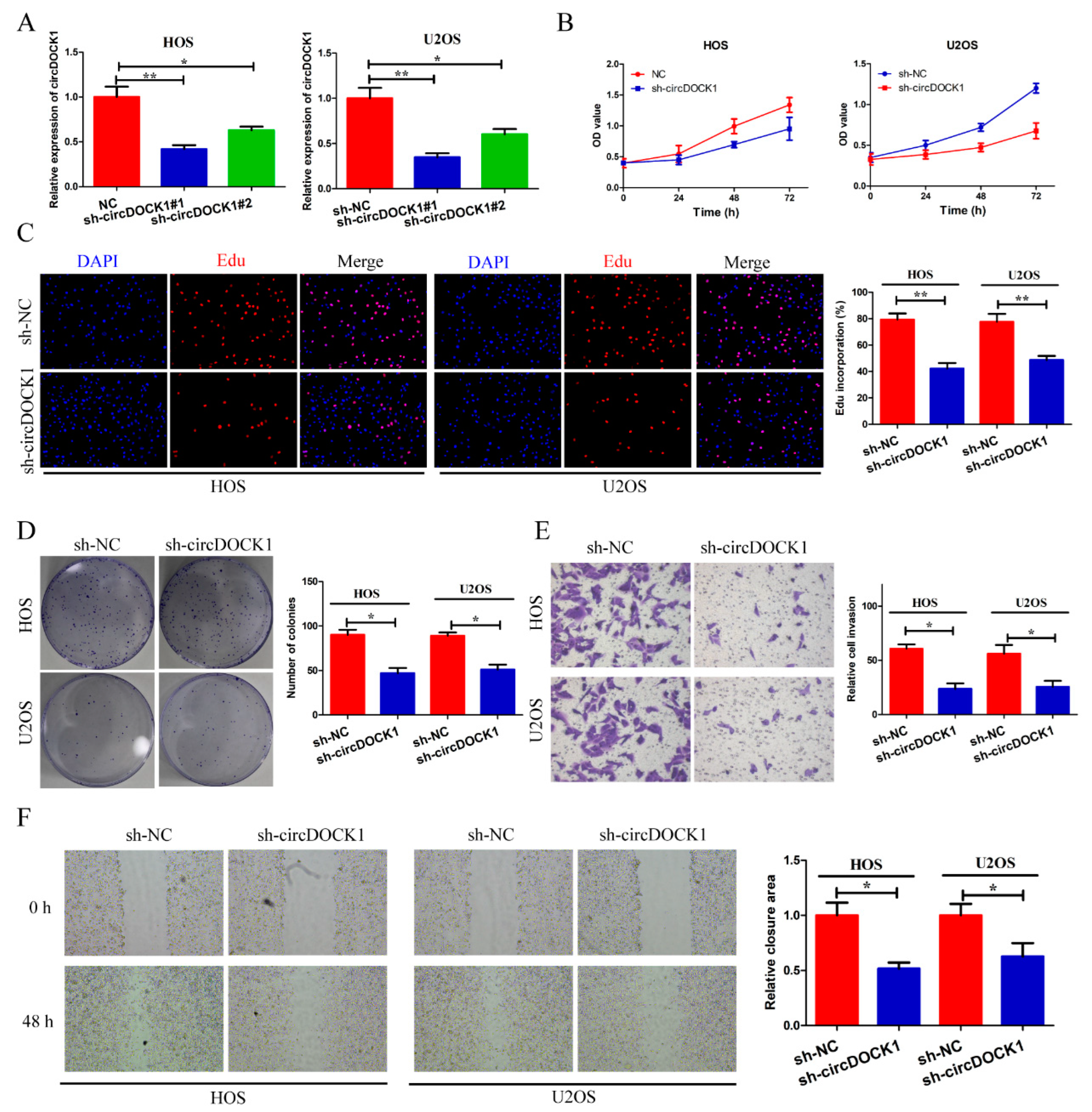
3.2. CircDOCK1 Knockdown Inhibits OS Progression In Vitro
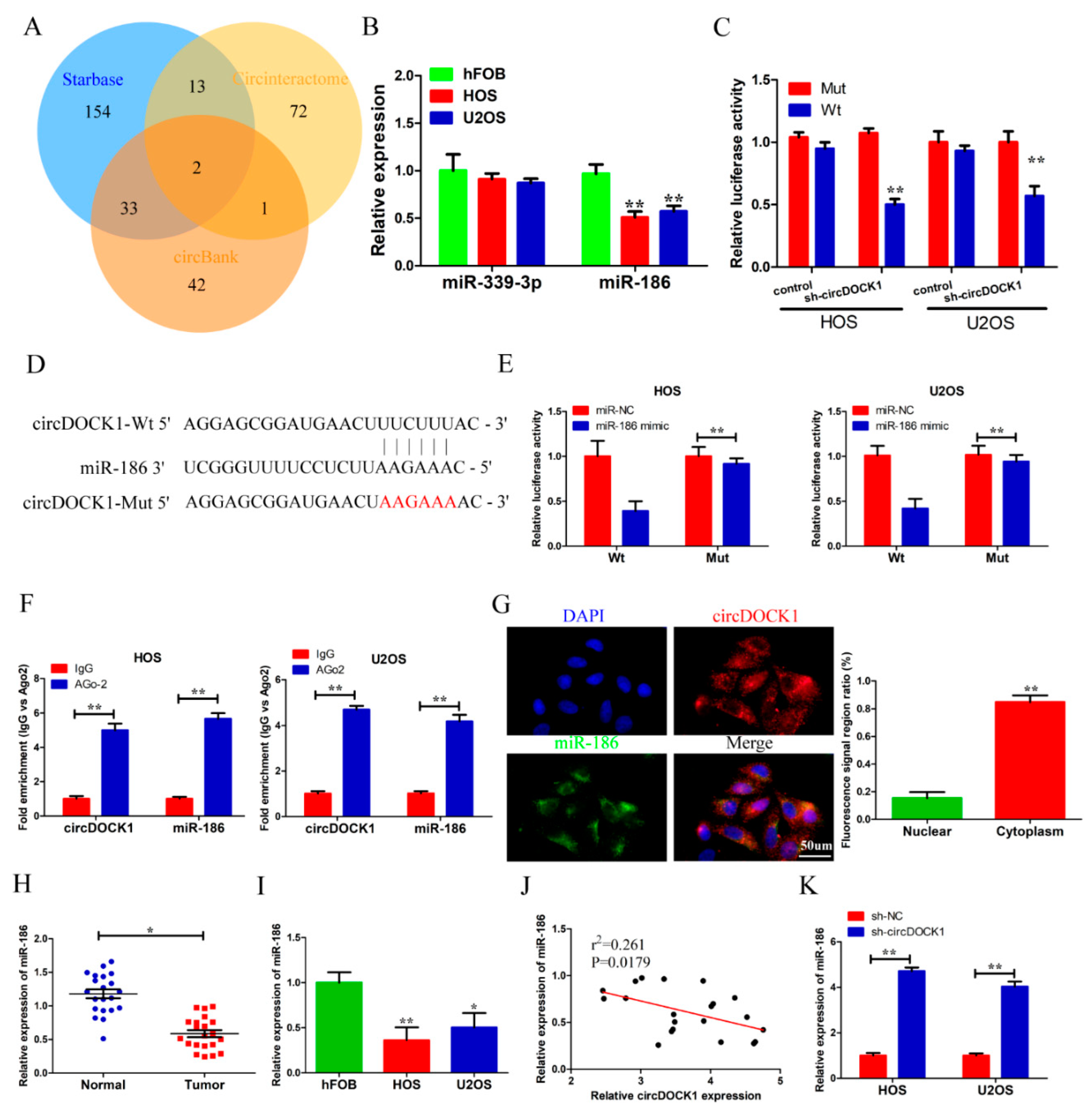
3.3. CircDOCK1 Acts as a Sponge for miR-186
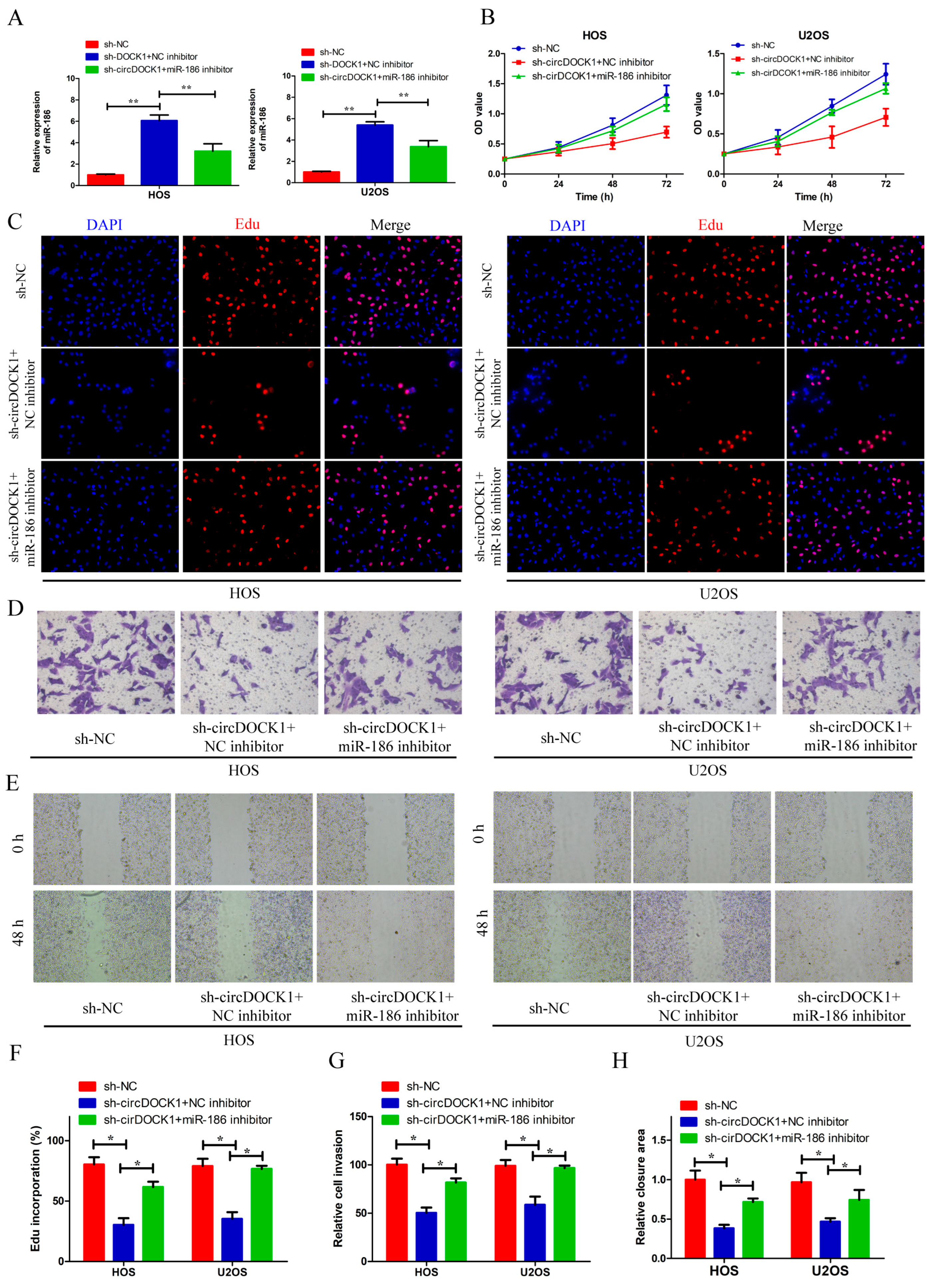
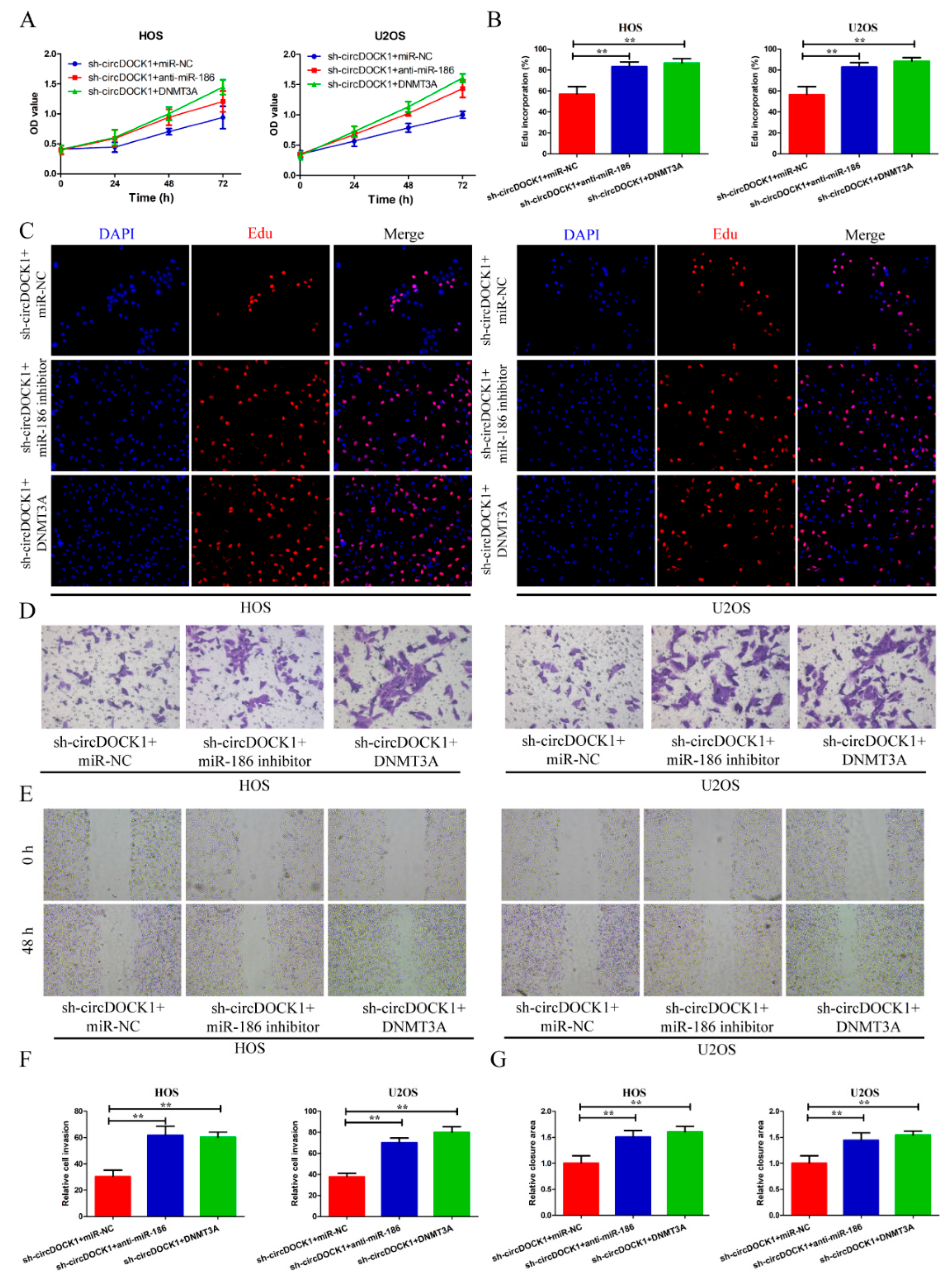
3.4. Knockdown of miR-186 Partly Reverses the Effect of sh-circDOCK1 on OS Progression
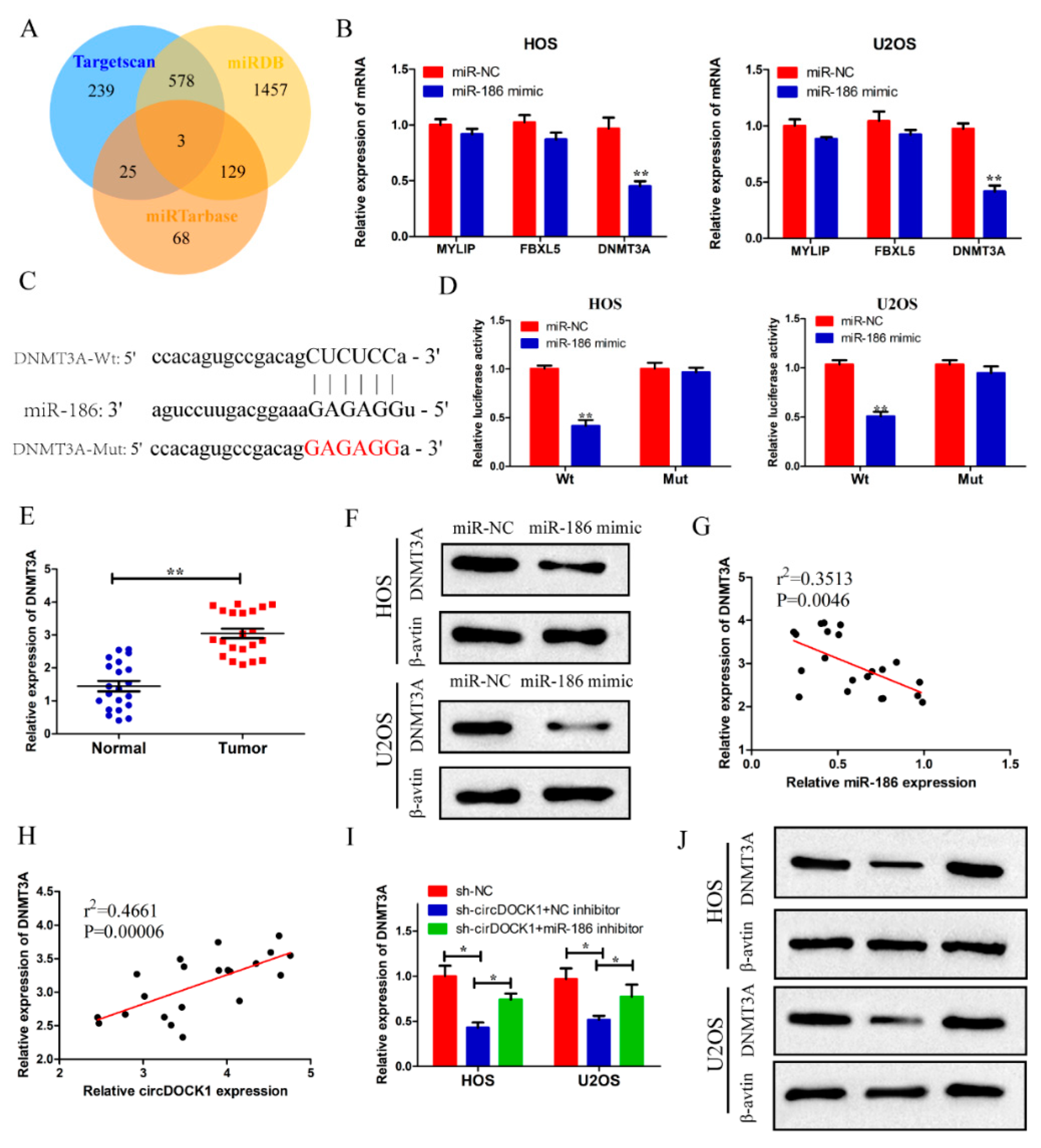
3.5. DNMT3A Was a Target of miR-186
3.6. CircDOCK1 Regulates OS Progression through the miR-186/DNMT3A Axis
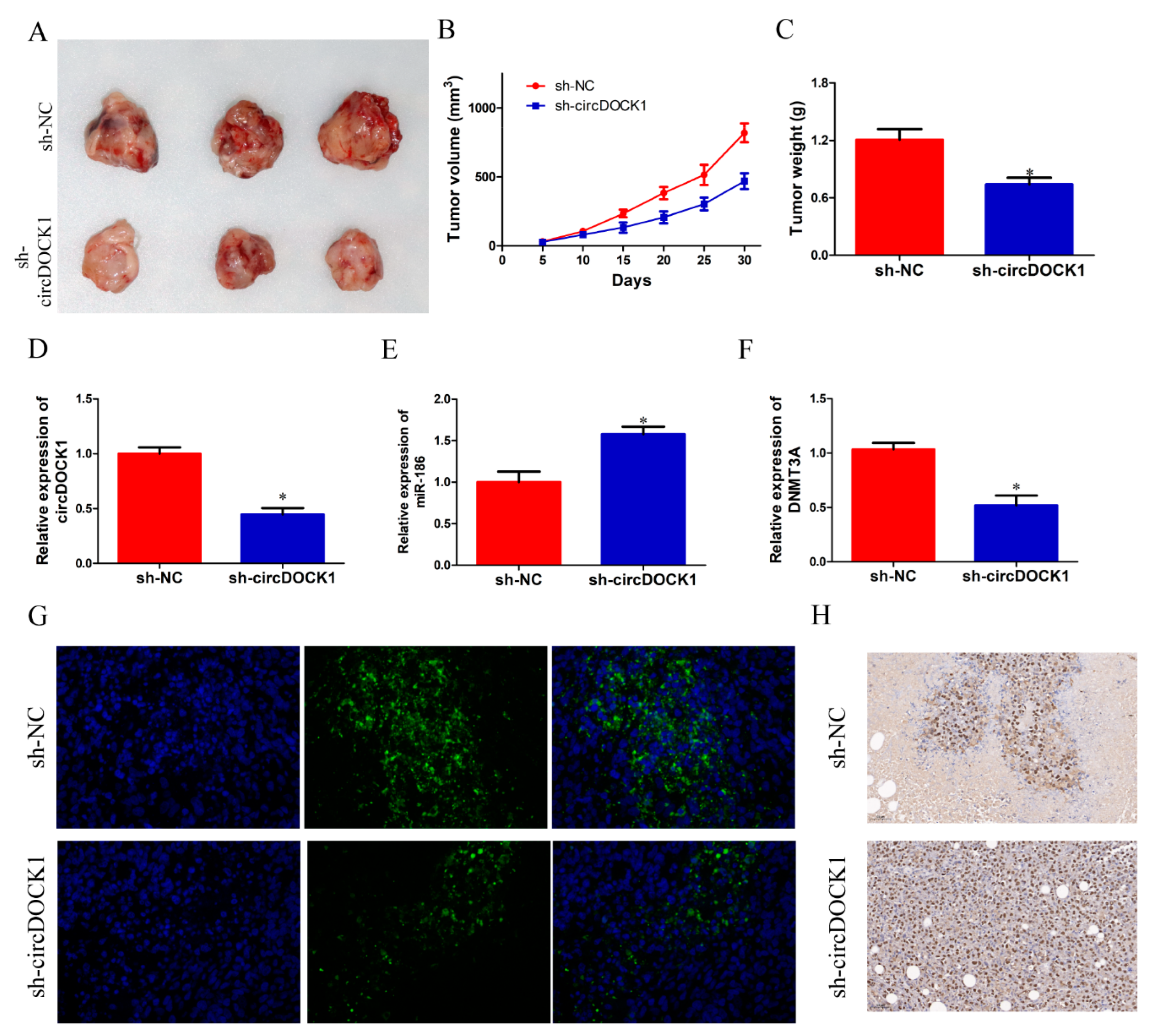
3.7. CircDOCK1 Knockdown Suppressed OS Growth In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Meazza, C.; Scanagatta, P. Metastatic osteosarcoma: A challenging multidisciplinary treatment. Expert Rev. Anticancer Ther. 2016, 16, 543–556. [Google Scholar] [CrossRef]
- Isakoff, M.S.; Bielack, S.S.; Meltzer, P.; Gorlick, R. Osteosarcoma: Current Treatment and a Collaborative Pathway to Success. J. Clin. Oncol. 2015, 33, 3029–3035. [Google Scholar] [CrossRef]
- Ferguson, J.L.; Turner, S.P. Bone Cancer: Diagnosis and Treatment Principles. Am. Fam. Physician 2018, 98, 205–213. [Google Scholar] [PubMed]
- Whelan, J.S.; Davis, L.E. Osteosarcoma, Chondrosarcoma, and Chordoma. J. Clin. Oncol. 2018, 36, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Simpson, E.; Brown, H.L. Understanding osteosarcomas. JAAPA 2018, 31, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Corre, I.; Verrecchia, F.; Crenn, V.; Redini, F.; Trichet, V. The Osteosarcoma Microenvironment: A Complex But Targetable Ecosystem. Cells 2020, 9, 976. [Google Scholar] [CrossRef]
- Shoaib, Z.; Fan, T.M.; Irudayaraj, J.M.K. Osteosarcoma mechanobiology and therapeutic targets. Br. J. Pharmacol. 2022, 179, 201–217. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nature reviews. Genetics 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Mo, Y.; Peng, M.; Tang, T.; Zhong, Y.; Deng, X.; Xiong, F.; Guo, C.; Wu, X.; Li, Y.; et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol. Cancer 2020, 19, 22. [Google Scholar] [CrossRef]
- Li, J.; Sun, D.; Pu, W.; Wang, J.; Peng, Y. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends Cancer 2020, 6, 319–336. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. TIG 2016, 32, 309–316. [Google Scholar] [CrossRef]
- Shi, Y.; Tian, Y.; Sun, X.; Qiu, Y.; Zhao, Y. Silencing circOMA1 Inhibits Osteosarcoma Progression by Sponging miR-1294 to Regulate c-Myc Expression. Front. Oncol. 2022, 12, 889583. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, F.; Zheng, K.; Wang, W.; Qiu, E.; Pei, Y.; Wang, S.; Zhang, J.; Zhang, X. CircDOCK1 promotes the tumorigenesis and cisplatin resistance of osteogenic sarcoma via the miR-339-3p/IGF1R axis. Mol. Cancer 2021, 20, 161. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Wu, X. Circular RNA hsa_circ_0032463 Acts as the Tumor Promoter in Osteosarcoma by Regulating the MicroRNA 498/LEF1 Axis. Mol. Cell. Biol. 2021, 41, e0010021. [Google Scholar] [CrossRef]
- Mao, X.; Guo, S.; Gao, L.; Li, G. Circ-XPR1 promotes osteosarcoma proliferation through regulating the miR-214-5p/DDX5 axis. Hum. Cell 2021, 34, 122–131. [Google Scholar] [CrossRef]
- Wang, L.; Li, B.; Yi, X.; Xiao, X.; Zheng, Q.; Ma, L. Circ_SMAD4 promotes gastric carcinogenesis by activating wnt/β-catenin pathway. Cell Prolif. 2021, 54, e12981. [Google Scholar] [CrossRef]
- Martins-Neves, S.R.; Sampaio-Ribeiro, G.; Gomes, C.M.F. Chemoresistance-Related Stem Cell Signaling in Osteosarcoma and Its Plausible Contribution to Poor Therapeutic Response: A Discussion That Still Matters. Int. J. Mol. Sci. 2022, 23, 1416. [Google Scholar] [CrossRef]
- Hattinger, C.M.; Patrizio, M.P.; Tavanti, E.; Luppi, S.; Magagnoli, F.; Picci, P.; Serra, M. Genetic testing for high-grade osteosarcoma: A guide for future tailored treatments? Expert Rev. Mol. Diagn. 2018, 18, 947–961. [Google Scholar] [CrossRef]
- Khadembaschi, D.; Jafri, M.; Praveen, P.; Parmar, S.; Breik, O. Does neoadjuvant chemotherapy provide a survival benefit in maxillofacial osteosarcoma: A systematic review and pooled analysis. Oral Oncol. 2022, 135, 106133. [Google Scholar] [CrossRef]
- Gao, X.; Gao, B.; Li, S. Extracellular vesicles: A new diagnostic biomarker and targeted drug in osteosarcoma. Front. Immunol. 2022, 13, 1002742. [Google Scholar] [CrossRef]
- Wang, H.; Zhou, X.; Li, C.; Yan, S.; Feng, C.; He, J.; Li, Z.; Tu, C. The emerging role of pyroptosis in pediatric cancers: From mechanism to therapy. J. Hematol. Oncol. 2022, 15, 140. [Google Scholar] [CrossRef]
- Han, J.; Kong, H.; Wang, X.; Zhang, X.A. Novel insights into the interaction between N6-methyladenosine methylation and noncoding RNAs in musculoskeletal disorders. Cell Prolif. 2022, 55, e13294. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Khoshbakht, T.; Hussen, B.M.; Sarfaraz, S.; Taheri, M.; Ayatollahi, S.A. Circ_CDR1as: A circular RNA with roles in the carcinogenesis. Pathol. Res. Pract. 2022, 236, 153968. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhao, X.; Wang, Y.; Ren, F.; Sun, D.; Yan, Y.; Kong, X.; Bu, J.; Liu, M.; Xu, S. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Na, N.; Liu, L.; Qiu, Y. CircRNA hsa_circ_0005909 Promotes Cell Proliferation of Osteosarcoma Cells by Targeting miR-338-3p/HMGA1 Axis. Cancer Manag. Res. 2021, 13, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Qian, Z.; Jiang, F.; Ge, D.; Tang, J.; Chen, H.; Yang, J.; Yao, Y.; Yan, J.; Zhao, L.; et al. CircRNA LRP6 promotes the development of osteosarcoma via negatively regulating KLF2 and APC levels. Am. J. Transl. Res. 2019, 11, 4126–4138. [Google Scholar]
- Shen, S.; Yao, T.; Xu, Y.; Zhang, D.; Fan, S.; Ma, J. CircECE1 activates energy metabolism in osteosarcoma by stabilizing c-Myc. Mol. Cancer 2020, 19, 151. [Google Scholar] [CrossRef]
- Lou, J.; Zhang, H.; Xu, J.; Ren, T.; Huang, Y.; Tang, X.; Guo, W. circUSP34 accelerates osteosarcoma malignant progression by sponging miR-16-5p. Cancer Sci. 2022, 113, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Nie, W.B.; Zhao, L.M.; Guo, R.; Wang, M.X.; Ye, F.G. Circular RNA circ-NT5C2 acts as a potential novel biomarker for prognosis of osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 6239–6244. [Google Scholar] [CrossRef]
- Liu, J.; Yang, L.; Fu, Q.; Liu, S. Emerging Roles and Potential Biological Value of CircRNA in Osteosarcoma. Front. Oncol. 2020, 10, 552236. [Google Scholar] [CrossRef]
- Zhu, K.P.; Zhang, C.L.; Ma, X.L.; Hu, J.P.; Cai, T.; Zhang, L. Analyzing the Interactions of mRNAs and ncRNAs to Predict Competing Endogenous RNA Networks in Osteosarcoma Chemo-Resistance. Mol. Ther. 2019, 27, 518–530. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Vishnoi, A.; Rani, S. MiRNA Biogenesis and Regulation of Diseases: An Overview. Methods Mol. Biol. 2017, 1509, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liao, M. Long noncoding RNA SNHG6 promotes papillary thyroid cancer cells proliferation via regulating miR-186/CDK6 axis. Gland Surg. 2021, 10, 2935–2944. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xie, R.; Wei, Q. Network analysis of miRNA targeting m6A-related genes in patients with esophageal cancer. PeerJ 2021, 9, e11893. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Y.; Bai, R.; Yang, K.; Tian, Z. MiR-186 inhibited aerobic glycolysis in gastric cancer via HIF-1α regulation. Oncogenesis 2016, 5, e224. [Google Scholar] [CrossRef]
- Tan, X.; Zeng, C.; Li, H.; Tan, Y.; Zhu, H. Circ0038632 modulates MiR-186/DNMT3A axis to promote proliferation and metastasis in osteosarcoma. Front. Oncol. 2022, 12, 939994. [Google Scholar] [CrossRef]
- Xiao, Q.; Wei, Z.; Li, Y.; Zhou, X.; Chen, J.; Wang, T.; Shao, G.; Zhang, M.; Zhang, Z. miR-186 functions as a tumor suppressor in osteosarcoma cells by suppressing the malignant phenotype and aerobic glycolysis. Oncol. Rep. 2018, 39, 2703–2710. [Google Scholar] [CrossRef]
- Tan, H.; Zhao, L. lncRNA nuclear-enriched abundant transcript 1 promotes cell proliferation and invasion by targeting miR-186-5p/HIF-1α in osteosarcoma. J. Cell. Biochem. 2019, 120, 6502–6514. [Google Scholar] [CrossRef]
- Cao, Q.; Wang, Z.; Wang, Y.; Liu, F.; Dong, Y.; Zhang, W.; Wang, L.; Ke, Z. TBL1XR1 promotes migration and invasion in osteosarcoma cells and is negatively regulated by miR-186-5p. Am. J. Cancer Res. 2018, 8, 2481–2493. [Google Scholar]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Zhang, Z.M.; Lu, R.; Wang, P.; Yu, Y.; Chen, D.; Gao, L.; Liu, S.; Ji, D.; Rothbart, S.B.; Wang, Y.; et al. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature 2018, 554, 387–391. [Google Scholar] [CrossRef]
- Cheng, S.; Wang, W. DNMT3A Regulates miR-149 DNA Methylation to Activate NOTCH1/Hedgehog Pathway to Promote the Development of Junctional Osteosarcoma. BioMed Res. Int. 2022, 2022, 3261213. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, Z.; Ye, J.; Chen, S.; Ren, Y.; Guo, W. CircDOCK1 Regulates miR-186/DNMT3A to Promote Osteosarcoma Progression. Biomedicines 2022, 10, 3013. https://doi.org/10.3390/biomedicines10123013
Jin Z, Ye J, Chen S, Ren Y, Guo W. CircDOCK1 Regulates miR-186/DNMT3A to Promote Osteosarcoma Progression. Biomedicines. 2022; 10(12):3013. https://doi.org/10.3390/biomedicines10123013
Chicago/Turabian StyleJin, Zhihui, Jia Ye, Sen Chen, Yijun Ren, and Weichun Guo. 2022. "CircDOCK1 Regulates miR-186/DNMT3A to Promote Osteosarcoma Progression" Biomedicines 10, no. 12: 3013. https://doi.org/10.3390/biomedicines10123013
APA StyleJin, Z., Ye, J., Chen, S., Ren, Y., & Guo, W. (2022). CircDOCK1 Regulates miR-186/DNMT3A to Promote Osteosarcoma Progression. Biomedicines, 10(12), 3013. https://doi.org/10.3390/biomedicines10123013






