A Few-Shot Learning-Based EEG and Stage Transition Sequence Generator for Improving Sleep Staging Performance
Abstract
1. Introduction
- (1)
- We propose a set of EEG-oriented progressive Wasserstein divergence GANs (WGAN-div) [19] that can adapt to sleep data and generate EEG epochs with few real data. The model can generate realistic 1D EEG epochs corresponding to different sleep stages and push the accuracy of the sleep staging model from 0.775 to 0.804.
- (2)
- We generated stage transition sequences based on a relational memory (RM) generator [20], which was used to generate a long text. This scenario is similar to stage transition sequence generation, and thus, we propose a few-shot learning-based model to generate plausible sequences such that the generated samples can be used in the training of the models based on RNNs [21], which have been proven to be capable of extracting sequential features from EEG data, thereby further pushing the accuracy of classification model from 0.804 to 0.831.
- (3)
- We evaluated our GANs by feeding both real data and EEG epochs and sleep stage transition sequences generated by us into a sleep staging model. In addition, we adopted the 1-NN method to ensure the efficiency of our GANs. The results showed that our GANs are capable of generating representative EEG epochs and plausible sleep stage transition sequences. With the help of the augmented data, the accuracy of the sleep staging model improved significantly after training with only a few samples.
2. Materials and Methods
2.1. Datasets
2.2. EEG Epoch Generation
2.3. Stage Transition Sequence Generation
3. Results
3.1. Choice of Hyperparameters and Metrics
3.2. Data Augmentation
3.3. Sequence Augmentation
3.4. Data Distribution Evaluation Via 1-NN Classifier
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zoubek, L.; Charbonnier, S.; Lecocq, S.; Buguet, A.; Chapotot, F. Feature selection for sleep/wake stages classification using data-driven methods. Biomed. Signal Process. Control 2007, 2, 171–179. [Google Scholar] [CrossRef]
- Supratak, A.; Dong, H.; Wu, C.; Guo, Y. DeepSleepNet: A Model for Automatic Sleep Stage Scoring Based on Raw Single-Channel EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 25, 1998–2008. [Google Scholar] [CrossRef] [PubMed]
- Hobson, J.A. A manual of standardized terminology, techniques and scoring system for sleep stages of human subjects. Electroencephalogr. Clin. Neurophysiol. 1969, 26, 644. [Google Scholar] [CrossRef]
- Iber, C.; Ancoli-Israel, S.; Chesson, A.L., Jr.; Quan, S.F. The AASM Manual for the Scoring of Sleep and Associated Events; American Academy of Sleep Medicine: Westchester, IL, USA, 2007. [Google Scholar]
- Taran, S.; Sharma, P.C.; Bajaj, V. Automatic sleep stages classification using optimise flexible analytic wavelet transform. Knowl.-Based Syst. 2020, 192, 10536. [Google Scholar] [CrossRef]
- Hassan, A.R.; Bhuiyan, M.I.H. Computer-aided sleep staging using complete ensemble empirical mode decomposition with adaptive noise and bootstrap aggregating. Biomed. Signal Process. Control 2016, 24, 1–10. [Google Scholar] [CrossRef]
- Da Silveira, T.L.T.; Kozakevicius, A.J.; Rodrigues, C.R. Single-channel EEG sleep stage classification based on a streamlined set of statistical features in the wavelet domain. Med. Biol. Eng. Comput. 2017, 55, 343–352. [Google Scholar] [CrossRef]
- Alickovic, E.; Subasi, A. Ensemble SVM Method for Automatic Sleep Stage Classification. IEEE Trans. Instrum. Meas. 2018, 67, 1258–1265. [Google Scholar] [CrossRef]
- Phan, H.; Andreotti, F.; Cooray, N.; Chén, O.Y.; de Vos, M. Joint Classification and Prediction CNN Framework for Automatic Sleep Stage Classification. IEEE Trans. Biomed. Eng. 2019, 66, 1285–1296. [Google Scholar] [CrossRef]
- Kanwal, S.; Uzair, M.; Ullah, H.; Khan, S.D.; Ullah, M.; Cheikh, F.A. An Image Based Prediction Model for Sleep Stage Identification. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; pp. 1366–1370. [Google Scholar]
- Phan, H.; Andreotti, F.; Cooray, N.; Chén, O.Y.; Vos, M.D. Automatic Sleep Stage Classification Using Single-Channel EEG: Learning Sequential Features with Attention-Based Recurrent Neural Networks. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 1452–1455. [Google Scholar]
- Zhu, T.; Luo, W.; Yu, F. Convolution- and Attention-Based Neural Network for Automated Sleep Stage Classification. Int. J. Environ. Res. Public Health 2020, 17, 4152. [Google Scholar] [CrossRef]
- Cai, Q.; Gao, Z.; An, J.; Gao, S.; Grebogi, C. A Graph-Temporal Fused Dual-Input Convolutional Neural Network for Detecting Sleep Stages from EEG Signals. IEEE Trans. Circuits Syst. II Express Briefs 2021, 68, 777–781. [Google Scholar] [CrossRef]
- Wang, Y.; Yao, Q.; Kwok, J.T.; Ni, L.M. Generalizing from a few examples: A survey on few-shot learning. ACM Comput. Surv. (CSUR) 2020, 53, 1–34. [Google Scholar] [CrossRef]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2014; Volume 27. [Google Scholar]
- Yang, J.; Yu, H.; Shen, T.; Song, Y.; Chen, Z. 4-Class Mi-EEG Signal Generation and Recognition With CVAE-GAN. Appl. Sci. 2021, 11, 1798. [Google Scholar] [CrossRef]
- Zhang, A.; Su, L.; Zhang, Y.; Fu, Y.; Wu, L.; Liang, S. EEG data augmentation for emotion recognition with a multiple generator conditional Wasserstein GAN. Complex Intell. Syst. 2021, 8, 3059–3071. [Google Scholar] [CrossRef]
- Hartmann, K.G.; Schirrmeister, R.T.; Ball, T. EEG-GAN: Generative adversarial networks for electroencephalographic (EEG) brain signals. arXiv 2018, arXiv:1806.01875. [Google Scholar]
- Wu, J.; Huang, Z.; Thoma, J.; Acharya, D.; Van Gool, L. Wasserstein divergence for gans. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018. [Google Scholar]
- Weili, N.; Narodytska, N.; Patel, A. Relgan: Relational generative adversarial networks for text generation. In Proceedings of the International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Hopfield, J.J. Neural networks and physical systems with emergent collective computational abilities. Proc. Natl. Acad. Sci. USA 1982, 79, 2554–2558. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Li, Q.; Xie, H.; Lau, R.Y.; Wang, Z.; Paul Smolley, S. Least squares generative adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017. [Google Scholar]
- Arjovsky, M.; Chintala, S.; Bottou, L. Wasserstein generative adversarial networks. In Proceedings of the 34th International Conference on Machine Learning, PMLR, Sydney, Australia, 6–11 August 2017. [Google Scholar]
- Gulrajani, I.; Ahmed, F.; Arjovsky, M.; Dumoulin, V.; Courville, A. Improved training of wasserstein gans. arXiv 2017, arXiv:1704.00028. [Google Scholar]
- Kodali, N.; Abernethy, J.; Hays, J.; Kira, Z. On convergence and stability of gans. arXiv 2017, arXiv:1705.07215. [Google Scholar]
- Hinz, T.; Fisher, M.; Wang, O.; Wermter, S. Improved techniques for training single-image gans. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Virtual, 5–9 January 2021. [Google Scholar]
- Shaham, T.R.; Dekel, T.; Michaeli, T. Singan: Learning a generative model from a single natural image. In Proceedings of the IEEE International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 4570–4580. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Isola, P.; Zhu, J.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]
- Jun-Yan, Z.; Richard, Z.; Pathak, D.; Darrell, T.; Efros, A.A.; Oliver, W.; Shechtman, E. Toward multimodal image-to-image translation. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2017; pp. 465–476. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 6–11 July 2015. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Březinová, V. Sleep cycle content and sleep cycle duration. Electroencephalogr. Clin. Neurophysiol. 1974, 36, 275–282. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Ba, J.L.; Kiros, J.R.; Hinton, G.E. Layer normalization. arXiv 2016, arXiv:1607.06450. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Abeywickrama, T.; Cheema, M.A.; Taniar, D. k-Nearest Neighbors on Road Networks: A Journey in Experimentation and In-Memory Implementation. arXiv 2016, arXiv:1601.01549. [Google Scholar] [CrossRef]
- Xu, Q.; Huang, G.; Yuan, Y.; Guo, C.; Sun, Y.; Wu, F.; Weinberger, K. An empirical study on evaluation metrics of generative adversarial networks. arXiv 2018, arXiv:1806.07755. [Google Scholar]
- Lopez-Paz, D.; Oquab, M. Revisiting Classifier Two-Sample Tests. arXiv 2016, arXiv:1610.06545. [Google Scholar]
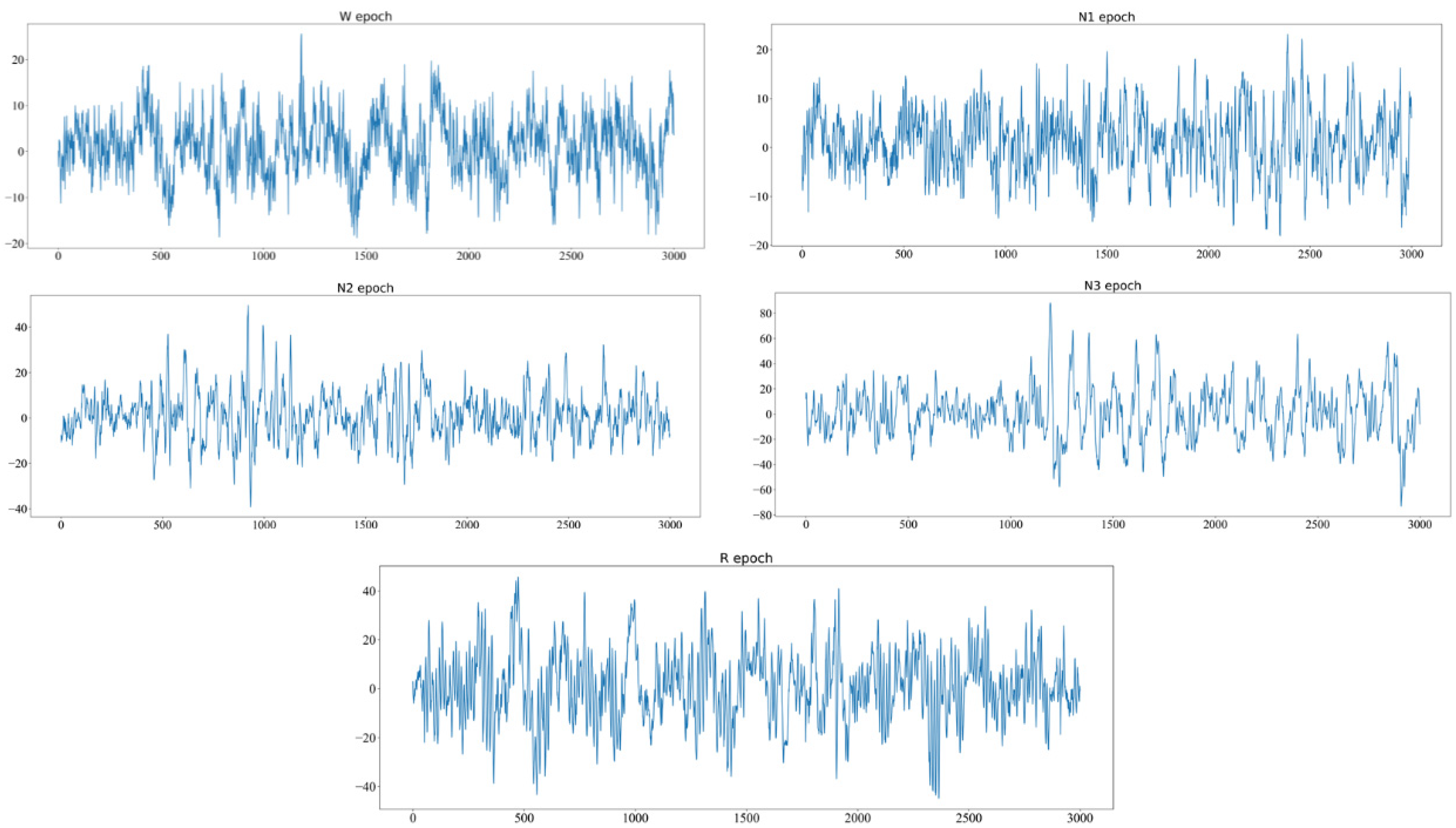

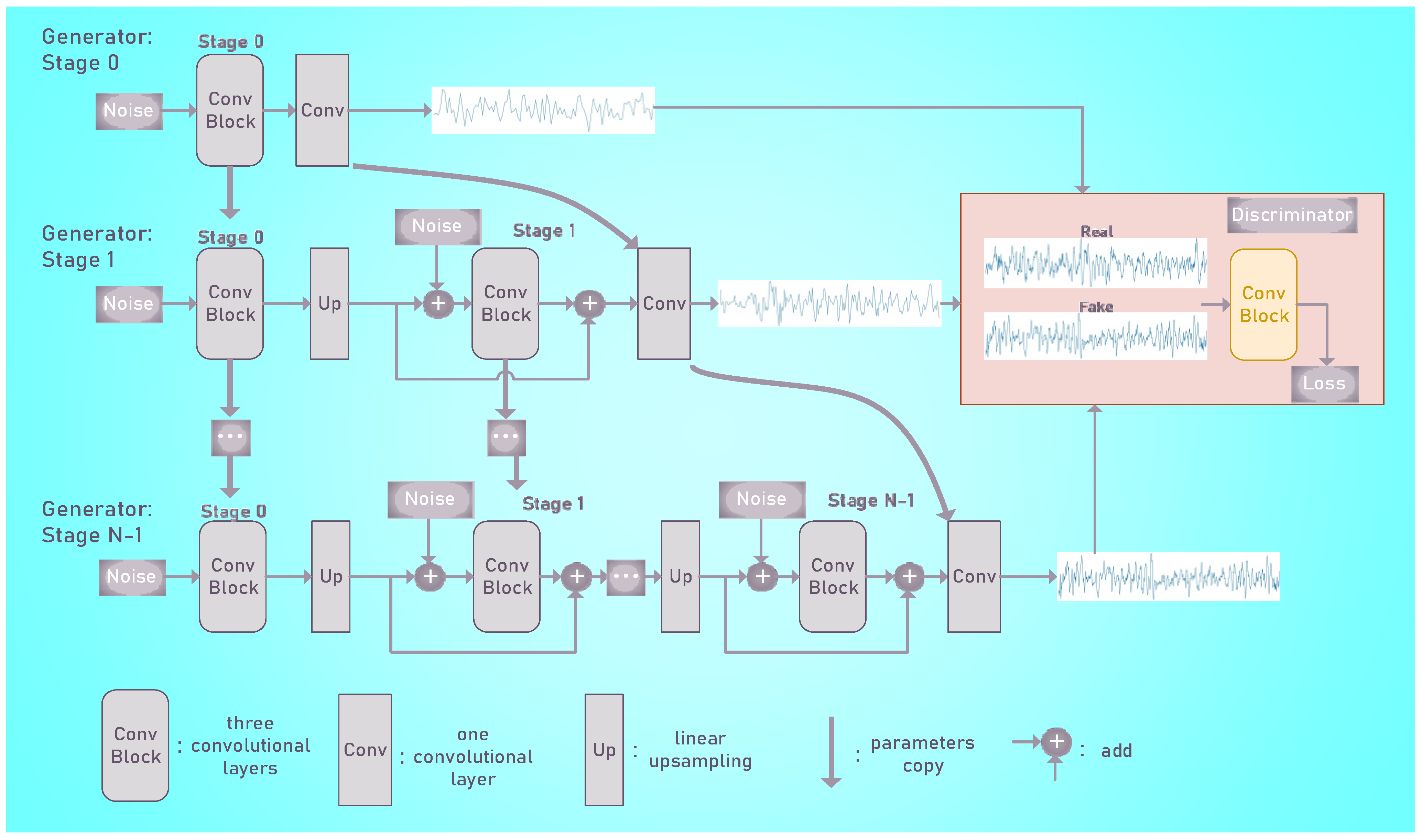
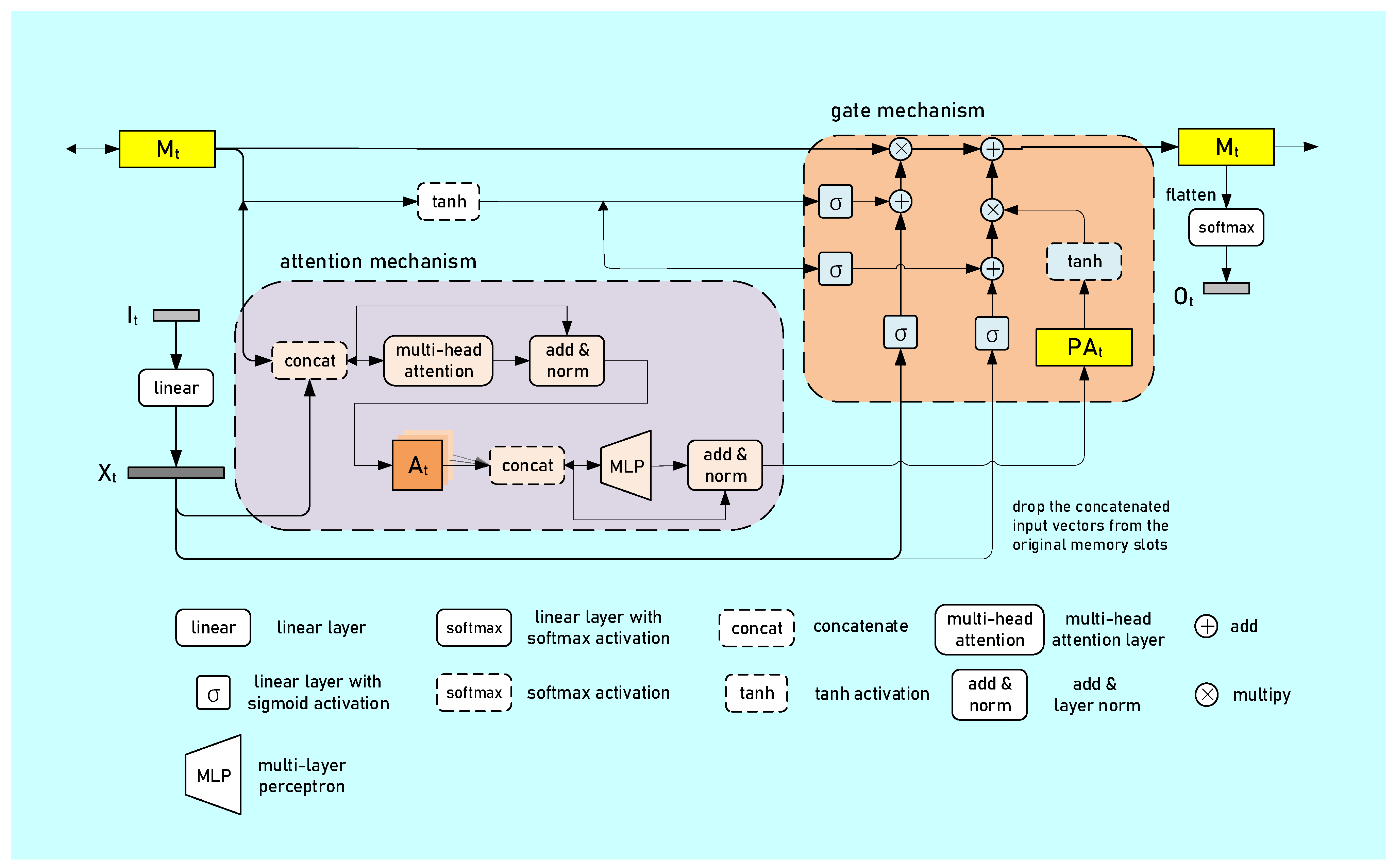
| Generator | ||
|---|---|---|
| Layer | Norm./Act. | Output Size |
| Latent noise vector | - | 1 × 100 |
| Stage 0: 3 × Conv 9 | Batch Norm./LreLU (0.05) | 32 × 100 |
| Stage 1: Upsampling 3 × Conv 9 | - Batch Norm./LreLU (0.05) | 32 × 230 32 × 230 |
| Stage 2: Upsampling 3 × Conv 9 | - Batch Norm./LreLU (0.05) | 32 × 965 32 × 965 |
| Stage 3: Upsampling 3 × Conv 9 | - Batch Norm./LreLU (0.05) | 32 × 3000 32 × 3000 |
| Conv 9 | -/Tanh | 1 × 3000 |
| Discriminator | ||
| Layer | Act. | Output size |
| Input signal | - | 1 × 3000 |
| 3×Conv 9 | LreLU (0.05) | 32 × 3000 |
| Conv 9 | - | 1 × 3000 |
| Training algorithm | ||
| General | ||
| Number of epochs to train per scale | 2000 | |
| Gamma and milestone of learning rate scheduler | 0.1, 1600 | |
| Batch size | 64 | |
| Generator: Stage n | ||
| Noise amplitude | 0.1 | |
| Number of stages N | 4 | |
| Concurrently trained stages | Last 3 stages | |
| Optimizer: Adam [37] | lr = 0.0005 × 0.1N-n-1 beta1 = 0.5 beta2 = 0.999 | |
| Generator inner steps | 3 | |
| Discriminator | ||
| Optimizer: Adam | lr = 0.0005 beta1 = 0.5 beta2 = 0.999 | |
| Generator inner steps | 3 | |
| Loss | ||
| WGAN-div loss | K = 2, p = 6 | |
| Model | |
|---|---|
| Number of memory slots | 1 |
| Number of heads | 2 |
| Head size | 64 |
| Memory size (number of heads × head size) | 2 × 64 = 128 |
| Number of layers of MLP in post attention | 2 |
| Hidden size of MLP | 128 |
| Activation of MLP | ReLU |
| Training algorithm | |
| Number of epochs | 200 |
| Sequence length | 180 |
| Batch size | 64 |
| Loss | MLE loss |
| Optimizer: Adam | lr = 1 × 10−3 beta1 = 0.9 beta2 = 0.999 |
| Generator inner steps | 3 |
| Test Name | Overall Metrics | Per-Class F1-Score(F1) | ||||||
|---|---|---|---|---|---|---|---|---|
| ACC | MF1 | k | W | N1 | N2 | N3 | REM | |
| 1-1 | 0.775 | 0.663 | 0.670 | 0.753 | 0.285 | 0.831 | 0.735 | 0.694 |
| 1-2 | 0.788 | 0.693 | 0.694 | 0.789 | 0.319 | 0.841 | 0.777 | 0.731 |
| 2-1 | 0.794 | 0.701 | 0.700 | 0.806 | 0.375 | 0.847 | 0.762 | 0.716 |
| 2-2 | 0.804 | 0.717 | 0.716 | 0.822 | 0.371 | 0.854 | 0.783 | 0.753 |
| Test Name | Overall Metrics | Per-Class F1-Score(F1) | ||||||
|---|---|---|---|---|---|---|---|---|
| ACC | MF1 | k | W | N1 | N2 | N3 | REM | |
| 2-1-1 | 0.811 | 0.708 | 0.719 | 0.721 | 0.418 | 0.850 | 0.756 | 0.792 |
| 2-1-2 | 0.814 | 0.714 | 0.723 | 0.741 | 0.423 | 0.850 | 0.741 | 0.797 |
| 2-2-1 | 0.829 | 0.735 | 0.745 | 0.745 | 0.438 | 0.864 | 0.769 | 0.844 |
| 2-2-2 | 0.831 | 0.742 | 0.747 | 0.767 | 0.450 | 0.864 | 0.771 | 0.844 |
| Results | Sleep Stages | ||||
|---|---|---|---|---|---|
| W | N1 | N2 | N3 | R | |
| Average | 52.37% | 54.84% | 53.74% | 57.75% | 50.63% |
| Variance | 0.003286 | 0.01009 | 0.008936 | 0.01277 | 0.000098 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
You, Y.; Guo, X.; Zhong, X.; Yang, Z. A Few-Shot Learning-Based EEG and Stage Transition Sequence Generator for Improving Sleep Staging Performance. Biomedicines 2022, 10, 3006. https://doi.org/10.3390/biomedicines10123006
You Y, Guo X, Zhong X, Yang Z. A Few-Shot Learning-Based EEG and Stage Transition Sequence Generator for Improving Sleep Staging Performance. Biomedicines. 2022; 10(12):3006. https://doi.org/10.3390/biomedicines10123006
Chicago/Turabian StyleYou, Yuyang, Xiaoyu Guo, Xuyang Zhong, and Zhihong Yang. 2022. "A Few-Shot Learning-Based EEG and Stage Transition Sequence Generator for Improving Sleep Staging Performance" Biomedicines 10, no. 12: 3006. https://doi.org/10.3390/biomedicines10123006
APA StyleYou, Y., Guo, X., Zhong, X., & Yang, Z. (2022). A Few-Shot Learning-Based EEG and Stage Transition Sequence Generator for Improving Sleep Staging Performance. Biomedicines, 10(12), 3006. https://doi.org/10.3390/biomedicines10123006





