Improving Pathological Assessment of Breast Cancer by Employing Array-Based Transcriptome Analysis
Abstract
:1. Introduction
2. The Discovery of Molecular Subtypes
3. Determination of Receptor Status
4. Prognostic Signatures
| Indication | IHC/FISH/RT-PCR-based tests | Ref. | Microarray-based tests | Ref. |
|---|---|---|---|---|
| Endrocine therapy | ESR1 * (I) (P) and PGR * (I) (P) | ESR1 | [29] | |
| H:I ratio (tamoxifen) | [43] | |||
| RecurrenceOnline | [40] | |||
| Targeted therapy | HER2 * (I) (F) (P) | HER2 | [29] | |
| RecurrenceOnline | [40] | |||
| Grade | FoxTop (P) | [60] | MapQuant DX | [41] |
| Chemotherapy response | PAM50 (P) | [44] | MapQuant DX | [41] |
| Oncotype DX (P) | [61] | |||
| Prognosis | Oncotype DX (P) CURIO (I) Celera Metastasis Score (P) BreastOncPx (P) | [10] [26] [56] [57] | 70 gene * RecurrenceOnline IGS HDP Rotterdam signature | [42] [40] [54] [55] [62] |
5. Solitary Genes of the Signatures
6. Validation Studies
| Gene | Probe | Cutoff | Basal | Luminal A | Luminal B | HER2 positive | |
|---|---|---|---|---|---|---|---|
| ESR1 | 205225_at | 500 | Low | High | High | High | Low |
| HER2 | 216836_s_at | 4800 | Low | Low | Low | High | High |
| MKI67 | 212021_s_at | 470 | N.R. | Low | High | N.R. | N.R. |
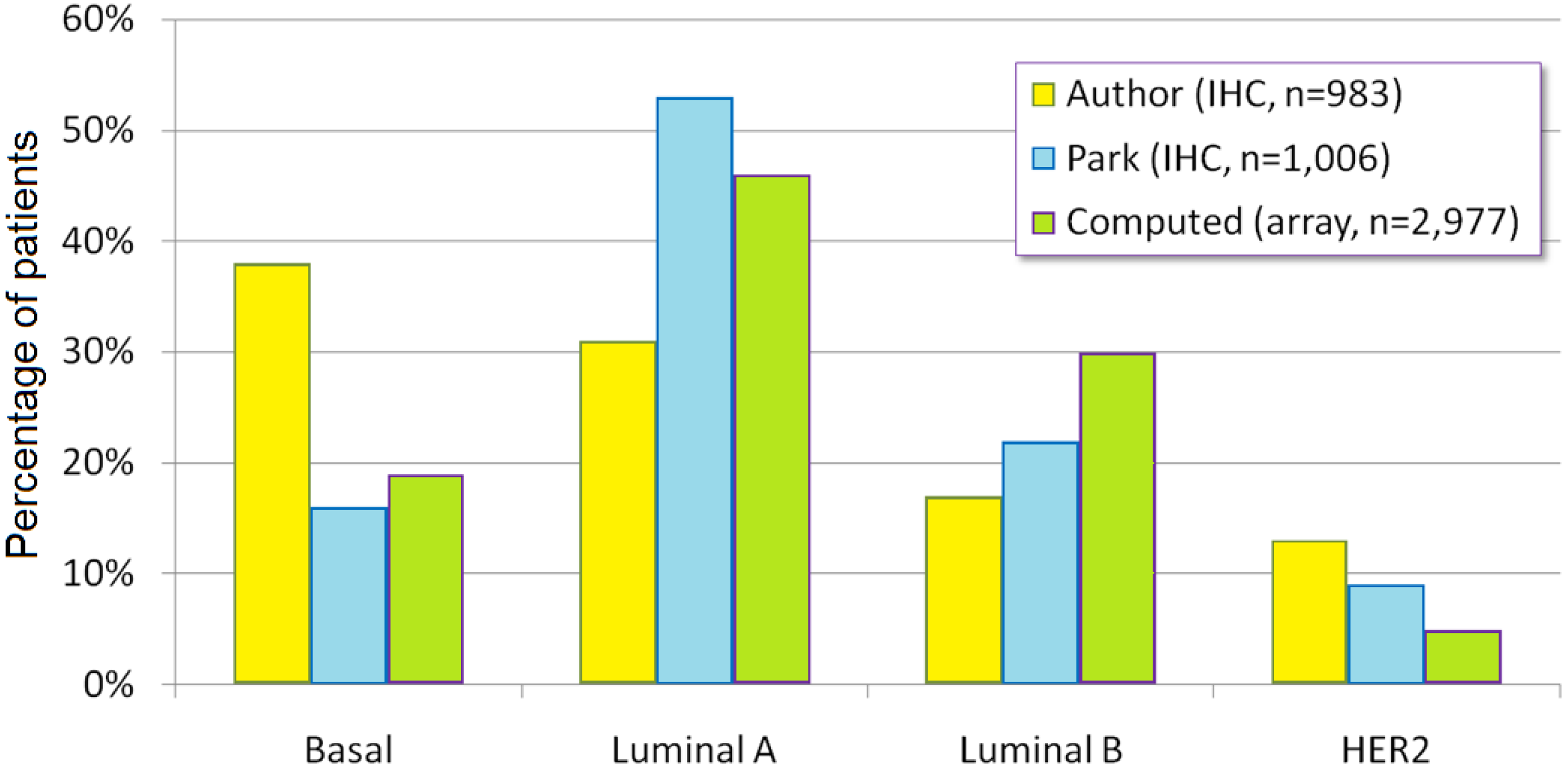
7. Conclusions
Acknowledgments
Conflicts of Interest
References
- Davies, C.; Godwin, J.; Gray, R.; Clarke, M.; Cutter, D.; Darby, S.; McGale, P.; Pan, H.C.; Taylor, C.; Wang, Y.C.; et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: Patient-level meta-analysis of randomised trials. Lancet 2011, 378, 771–784. [Google Scholar]
- Early Breast Cancer Trialists’ Collaborative Group. Tamoxifen for early breast cancer: An overview of the randomised trials. Lancet 1998, 351, 1451–1467. [CrossRef]
- Brufsky, A. Trastuzumab-based therapy for patients with HER2-positive breast cancer: From early scientific development to foundation of care. Am. J. Clin. Oncol. 2010, 33, 186–195. [Google Scholar]
- Dawood, S.; Broglio, K.; Buzdar, A.U.; Hortobagyi, G.N.; Giordano, S.H. Prognosis of women with metastatic breast cancer by HER2 status and trastuzumab treatment: An institutional-based review. J. Clin. Oncol. 2010, 28, 92–98. [Google Scholar] [CrossRef]
- Layfield, L.J.; Goldstein, N.; Perkinson, K.R.; Proia, A.D. Interlaboratory variation in results from immunohistochemical assessment of estrogen receptor status. Breast J. 2003, 9, 257–259. [Google Scholar] [CrossRef]
- Rhodes, A.; Jasani, B.; Balaton, A.J.; Barnes, D.M.; Miller, K.D. Frequency of oestrogen and progesterone receptor positivity by immunohistochemical analysis in 7,016 breast carcinomas: Correlation with patient age, assay sensitivity, threshold value, and mammographic screening. J. Clin. Pathol. 2000, 53, 688–696. [Google Scholar] [CrossRef]
- Rhodes, A.; Jasani, B.; Balaton, A.J.; Barnes, D.M.; Anderson, E.; Bobrow, L.G.; Miller, K.D. Study of interlaboratory reliability and reproducibility of estrogen and progesterone receptor assays in Europe. Documentation of poor reliability and identification of insufficient microwave antigen retrieval time as a major contributory element of unreliable assays. Am. J. Clin. Pathol. 2001, 115, 44–58. [Google Scholar] [CrossRef]
- Grabau, D.A.; Bendahl, P.O.; Ryden, L.; Stal, O.; Ferno, M. The prevalence of immunohistochemically determined oestrogen receptor positivity in primary breast cancer is dependent on the choice of antibody and method of heat-induced epitope retrieval—Prognostic implications? Acta Oncol. 2013. [Google Scholar] [CrossRef]
- Atkinson, R.; Mollerup, J.; Laenkholm, A.V.; Verardo, M.; Hawes, D.; Commins, D.; Engvad, B.; Correa, A.; Ehlers, C.C.; Nielsen, K.V. Effects of the change in cutoff values for human epidermal growth factor receptor 2 status by immunohistochemistry and fluorescence in situ hybridization: A study comparing conventional brightfield microscopy, image analysis-assisted microscopy, and interobserver variation. Arch. Pathol. Lab. Med. 2011, 135, 1010–1016. [Google Scholar] [CrossRef]
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Baehner, F.L.; Walker, M.G.; Watson, D.; Park, T.; et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004, 351, 2817–2826. [Google Scholar] [CrossRef]
- Kennecke, H.F.; Speers, C.H.; Ennis, C.A.; Gelmon, K.; Olivotto, I.A.; Hayes, M. Impact of routine pathology review on treatment for node-negative breast cancer. J. Clin. Oncol. 2012, 30, 2227–2231. [Google Scholar] [CrossRef]
- Gyorffy, B.; Molnar, B.; Lage, H.; Szallasi, Z.; Eklund, A.C. Evaluation of microarray preprocessing algorithms based on concordance with RT-PCR in clinical samples. PLoS One 2009, 4, e5645. [Google Scholar] [CrossRef]
- Consortium, M.; Shi, L.; Reid, L.H.; Jones, W.D.; Shippy, R.; Warrington, J.A.; Baker, S.C.; Collins, P.J.; de Longueville, F.; Kawasaki, E.S.; et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 2006, 24, 1151–1161. [Google Scholar] [CrossRef]
- Li, Q.; Birkbak, N.J.; Gyorffy, B.; Szallasi, Z.; Eklund, A.C. Jetset: Selecting the optimal microarray probe set to represent a gene. BMC Bioinformatics 2011, 12, 474. [Google Scholar] [CrossRef]
- Sotiriou, C.; Neo, S.Y.; McShane, L.M.; Korn, E.L.; Long, P.M.; Jazaeri, A.; Martiat, P.; Fox, S.B.; Harris, A.L.; Liu, E.T. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc. Natl. Acad. Sci. USA 2003, 100, 10393–10398. [Google Scholar] [CrossRef]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef]
- Sorlie, T.; Tibshirani, R.; Parker, J.; Hastie, T.; Marron, J.S.; Nobel, A.; Deng, S.; Johnsen, H.; Pesich, R.; Geisler, S.; et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. USA 2003, 100, 8418–8423. [Google Scholar] [CrossRef]
- Wang, Y.; Yin, Q.; Yu, Q.; Zhang, J.; Liu, Z.; Wang, S.; Lv, S.; Niu, Y. A retrospective study of breast cancer subtypes: The risk of relapse and the relations with treatments. Breast Cancer Res. Treat. 2011, 130, 489–498. [Google Scholar] [CrossRef]
- Gyorffy, B.; Serra, V.; Jurchott, K.; Abdul-Ghani, R.; Garber, M.; Stein, U.; Petersen, I.; Lage, H.; Dietel, M.; Schafer, R. Prediction of doxorubicin sensitivity in breast tumors based on gene expression profiles of drug-resistant cell lines correlates with patient survival. Oncogene 2005, 24, 7542–7551. [Google Scholar] [CrossRef]
- Colombo, P.E.; Milanezi, F.; Weigelt, B.; Reis-Filho, J.S. Microarrays in the 2010s: The contribution of microarray-based gene expression profiling to breast cancer classification, prognostication and prediction. Breast Cancer Res. 2011, 13, 212. [Google Scholar] [CrossRef]
- Valentin, M.D.; da Silva, S.D.; Privat, M.; Alaoui-Jamali, M.; Bignon, Y.J. Molecular insights on basal-like breast cancer. Breast Cancer Res. Treat. 2012, 134, 21–30. [Google Scholar] [CrossRef]
- Mackay, A.; Weigelt, B.; Grigoriadis, A.; Kreike, B.; Natrajan, R.; A’Hern, R.; Tan, D.S.; Dowsett, M.; Ashworth, A.; Reis-Filho, J.S. Microarray-based class discovery for molecular classification of breast cancer: Analysis of interobserver agreement. J. Natl. Cancer Inst. 2011, 103, 662–673. [Google Scholar] [CrossRef]
- Hu, Z.; Fan, C.; Oh, D.S.; Marron, J.S.; He, X.; Qaqish, B.F.; Livasy, C.; Carey, L.A.; Reynolds, E.; Dressler, L.; et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 2006, 7, 96. [Google Scholar] [CrossRef]
- Weigelt, B.; Horlings, H.M.; Kreike, B.; Hayes, M.M.; Hauptmann, M.; Wessels, L.F.; de Jong, D.; van de Vijver, M.J.; van’t Veer, L.J.; Peterse, J.L. Refinement of breast cancer classification by molecular characterization of histological special types. J. Pathol. 2008, 216, 141–150. [Google Scholar] [CrossRef]
- Park, S.; Park, B.W.; Kim, T.H.; Jeon, C.W.; Kang, H.S.; Choi, J.E.; Hwang, K.T.; Kim, I.C. Lack of either estrogen or progesterone receptor expression is associated with poor survival outcome among luminal A breast cancer subtype. Ann. Surg. Oncol. 2013, 20, 1505–1513. [Google Scholar] [CrossRef]
- Szasz, A.M.; Nemeth, Z.; Gyorffy, B.; Micsinai, M.; Krenacs, T.; Baranyai, Z.; Harsanyi, L.; Kiss, A.; Schaff, Z.; Tokes, A.M.; et al. Identification of a claudin-4 and E-cadherin score to predict prognosis in breast cancer. Cancer Sci. 2011, 102, 2248–2254. [Google Scholar] [CrossRef]
- Lu, S.; Singh, K.; Mangray, S.; Tavares, R.; Noble, L.; Resnick, M.B.; Yakirevich, E. Claudin expression in high-grade invasive ductal carcinoma of the breast: Correlation with the molecular subtype. Mod. Pathol. 2013, 26, 485–495. [Google Scholar] [CrossRef]
- Milde-Langosch, K.; Karn, T.; Muller, V.; Witzel, I.; Rody, A.; Schmidt, M.; Wirtz, R.M. Validity of the proliferation markers Ki67, TOP2A, and RacGAP1 in molecular subgroups of breast cancer. Breast Cancer Res. Treat. 2013, 137, 57–67. [Google Scholar] [CrossRef]
- Gong, Y.; Yan, K.; Lin, F.; Anderson, K.; Sotiriou, C.; Andre, F.; Holmes, F.A., Valero; Booser, D.; Pippen, J.E., Jr.; et al. Determination of oestrogen-receptor status and ERBB2 status of breast carcinoma: A gene-expression profiling study. Lancet Oncol. 2007, 8, 203–211. [Google Scholar] [CrossRef]
- Roepman, P.; Horlings, H.M.; Krijgsman, O.; Kok, M.; Bueno-de-Mesquita, J.M.; Bender, R.; Linn, S.C.; Glas, A.M.; van de Vijver, M.J. Microarray-based determination of estrogen receptor, progesterone receptor, and HER2 receptor status in breast cancer. Clin. Cancer Res. 2009, 15, 7003–7011. [Google Scholar] [CrossRef]
- Bastien, R.R.; Rodriguez-Lescure, A.; Ebbert, M.T.; Prat, A.; Munarriz, B.; Rowe, L.; Miller, P.; Ruiz-Borrego, M.; Anderson, D.; Lyons, B.; et al. PAM50 breast cancer subtyping by RT-qPCR and concordance with standard clinical molecular markers. BMC Med. Genomics 2012, 5, 44. [Google Scholar] [CrossRef]
- Zoubir, M.M.M.; Liedtke, C.; Bidard, F.; Delaloge, S.; Corley, L.; Spielmann, M.; Pusztai, L.; André, F.; Symmans, W.F. Predictive biomarkers for preoperative endocrine therapy of stage II-III breast cancer by tissue microarrays. J. Clin. Oncol. 2008, 26, 560. [Google Scholar]
- Bartlett, J.M.; Thomas, J.; Ross, D.T.; Seitz, R.S.; Ring, B.Z.; Beck, R.A.; Pedersen, H.C.; Munro, A.; Kunkler, I.H.; Campbell, F.M.; et al. Mammostrat as a tool to stratify breast cancer patients at risk of recurrence during endocrine therapy. Breast Cancer Res. 2010, 12, R47. [Google Scholar] [CrossRef]
- Hilborn, E.S.T.; Kot, A.; Fornander, T.; Skoog, L.; Nordenskjöld, B.; Stål, O.; Jansson, A. The importance of CXCL10 and CXCR3-A in breast cancer. Cancer Res. 2011, 71. [Google Scholar] [CrossRef]
- Surowiak, P.; Matkowski, R.; Materna, V.; Gyorffy, B.; Wojnar, A.; Pudelko, M.; Dziegiel, P.; Kornafel, J.; Zabel, M. Elevated metallothionein (MT) expression in invasive ductal breast cancers predicts tamoxifen resistance. Histol. Histopathol. 2005, 20, 1037–1044. [Google Scholar]
- Mihaly, Z.; Kormos, M.; Lanczky, A.; Dank, M.; Budczies, J.; Szasz, M.A.; Gyorffy, B. A meta-analysis of gene expression-based biomarkers predicting outcome after tamoxifen treatment in breast cancer. Breast Cancer Res. Treat. 2013, 140, 219–232. [Google Scholar] [CrossRef]
- Akhtar-Zaidi, B.; Cowper-Sal-lari, R.; Corradin, O.; Saiakhova, A.; Bartels, C.F.; Balasubramanian, D.; Myeroff, L.; Lutterbaugh, J.; Jarrar, A.; Kalady, M.F.; et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science 2012, 336, 736–739. [Google Scholar] [CrossRef]
- Magnani, L.; Stoeck, A.; Zhang, X.; Lanczky, A.; Mirabella, A.C.; Wang, T.L.; Gyorffy, B.; Lupien, M. Genome-wide reprogramming of the chromatin landscape underlies endocrine therapy resistance in breast cancer. Proc. Natl. Acad. Sci. USA 2013, 110, E1490–E1499. [Google Scholar] [CrossRef]
- Aguilar, H.; Sole, X.; Bonifaci, N.; Serra-Musach, J.; Islam, A.; Lopez-Bigas, N.; Mendez-Pertuz, M.; Beijersbergen, R.L.; Lazaro, C.; Urruticoechea, A.; et al. Biological reprogramming in acquired resistance to endocrine therapy of breast cancer. Oncogene 2010, 29, 6071–6083. [Google Scholar] [CrossRef]
- Gyorffy, B.; Benke, Z.; Lanczky, A.; Balazs, B.; Szallasi, Z.; Timar, J.; Schafer, R. RecurrenceOnline: An online analysis tool to determine breast cancer recurrence and hormone receptor status using microarray data. Breast Cancer Res. Treat. 2012, 132, 1025–1034. [Google Scholar] [CrossRef]
- Sotiriou, C.; Wirapati, P.; Loi, S.; Harris, A.; Fox, S.; Smeds, J.; Nordgren, H.; Farmer, P.; Praz, V.; Haibe-Kains, B.; et al. Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 2006, 98, 262–272. [Google Scholar] [CrossRef]
- Van de Vijver, M.J.; He, Y.D.; van’t Veer, L.J.; Dai, H.; Hart, A.A.; Voskuil, D.W.; Schreiber, G.J.; Peterse, J.L.; Roberts, C.; Marton, M.J.; et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002, 347, 1999–2009. [Google Scholar] [CrossRef]
- Ma, X.J.; Wang, Z.; Ryan, P.D.; Isakoff, S.J.; Barmettler, A.; Fuller, A.; Muir, B.; Mohapatra, G.; Salunga, R.; Tuggle, J.T.; et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell 2004, 5, 607–616. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Wittner, B.S.; Sgroi, D.C.; Ryan, P.D.; Bruinsma, T.J.; Glas, A.M.; Male, A.; Dahiya, S.; Habin, K.; Bernards, R.; Haber, D.A.; et al. Analysis of the MammaPrint breast cancer assay in a predominantly postmenopausal cohort. Clin. Cancer Res. 2008, 14, 2988–2993. [Google Scholar] [CrossRef]
- Habel, L.A.; Shak, S.; Jacobs, M.K.; Capra, A.; Alexander, C.; Pho, M.; Baker, J.; Walker, M.; Watson, D.; Hackett, J.; et al. A population-based study of tumor gene expression and risk of breast cancer death among lymph node-negative patients. Breast Cancer Res. 2006, 8, R25. [Google Scholar] [CrossRef]
- Esteva, F.J.; Sahin, A.A.; Cristofanilli, M.; Coombes, K.; Lee, S.J.; Baker, J.; Cronin, M.; Walker, M.; Watson, D.; Shak, S.; et al. Prognostic role of a multigene reverse transcriptase-PCR assay in patients with node-negative breast cancer not receiving adjuvant systemic therapy. Clin. Cancer Res. 2005, 11, 3315–3319. [Google Scholar] [CrossRef]
- Dowsett, C.W.; Forbes, J.; Mallon, L.; Salter, J.; Cuzick, J.; Wales, C.; Forbes, J.; Mallon, L.; Salter, J.; Quinn, E. Risk of distant recurrence using Oncotype DX in postmenopausal primary breast cancer patients treated with anastrozole or tamoxifen: A TransATAC study. Cancer Res. 2009, 69, 1059–1061. [Google Scholar]
- Gyorffy, B.; Schafer, R. Meta-analysis of gene expression profiles related to relapse-free survival in 1079 breast cancer patients. Breast Cancer Res. Treat. 2009, 118, 433–441. [Google Scholar] [CrossRef]
- Dabbs, D.J.; Klein, M.E.; Mohsin, S.K.; Tubbs, R.R.; Shuai, Y.; Bhargava, R. High false-negative rate of HER2 quantitative reverse transcription polymerase chain reaction of the Oncotype DX test: An independent quality assurance study. J. Clin. Oncol. 2011, 29, 4279–4285. [Google Scholar] [CrossRef]
- Ma, X.J.; Hilsenbeck, S.G.; Wang, W.; Ding, L.; Sgroi, D.C.; Bender, R.A.; Osborne, C.K.; Allred, D.C.; Erlander, M.G. The HOXB13:IL17BR expression index is a prognostic factor in early-stage breast cancer. J. Clin. Oncol. 2006, 24, 4611–4619. [Google Scholar] [CrossRef]
- Jerevall, P.L.; Brommesson, S.; Strand, C.; Gruvberger-Saal, S.; Malmstrom, P.; Nordenskjold, B.; Wingren, S.; Soderkvist, P.; Ferno, M.; Stal, O. Exploring the two-gene ratio in breast cancer—Independent roles for HOXB13 and IL17BR in prediction of clinical outcome. Breast Cancer Res. Treat. 2008, 107, 225–234. [Google Scholar] [CrossRef]
- Reid, J.F.; Lusa, L.; de Cecco, L.; Coradini, D.; Veneroni, S.; Daidone, M.G.; Gariboldi, M.; Pierotti, M.A. Limits of predictive models using microarray data for breast cancer clinical treatment outcome. J. Natl. Cancer Inst. 2005, 97, 927–930. [Google Scholar] [CrossRef]
- Liu, R.; Wang, X.; Chen, G.Y.; Dalerba, P.; Gurney, A.; Hoey, T.; Sherlock, G.; Lewicki, J.; Shedden, K.; Clarke, M.F. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N. Engl. J. Med. 2007, 356, 217–226. [Google Scholar] [CrossRef]
- Staaf, J.; Ringner, M.; Vallon-Christersson, J.; Jonsson, G.; Bendahl, P.O.; Holm, K.; Arason, A.; Gunnarsson, H.; Hegardt, C.; Agnarsson, B.A.; et al. Identification of subtypes in human epidermal growth factor receptor 2—Positive breast cancer reveals a gene signature prognostic of outcome. J. Clin. Oncol. 2010, 28, 1813–1820. [Google Scholar] [CrossRef]
- Cobleigh, M.A.; Tabesh, B.; Bitterman, P.; Baker, J.; Cronin, M.; Liu, M.L.; Borchik, R.; Mosquera, J.M.; Walker, M.G.; Shak, S. Tumor gene expression and prognosis in breast cancer patients with 10 or more positive lymph nodes. Clin. Cancer Res. 2005, 11, 8623–8631. [Google Scholar] [CrossRef]
- Tutt, A.; Wang, A.; Rowland, C.; Gillett, C.; Lau, K.; Chew, K.; Dai, H.; Kwok, S.; Ryder, K.; Shu, H.; et al. Risk estimation of distant metastasis in node-negative, estrogen receptor-positive breast cancer patients using an RT-PCR based prognostic expression signature. BMC Cancer 2008, 8, 339. [Google Scholar] [CrossRef]
- Chia, S.K.; Bramwell, V.H.; Tu, D.; Shepherd, L.E.; Jiang, S.; Vickery, T.; Mardis, E.; Leung, S.; Ung, K.; Pritchard, K.I.; et al. A 50-gene intrinsic subtype classifier for prognosis and prediction of benefit from adjuvant tamoxifen. Clin. Cancer Res. 2012, 18, 4465–4472. [Google Scholar] [CrossRef]
- Harvell, D.M.; Spoelstra, N.S.; Singh, M.; McManaman, J.L.; Finlayson, C.; Phang, T.; Trapp, S.; Hunter, L.; Dye, W.W.; Borges, V.F.; et al. Molecular signatures of neoadjuvant endocrine therapy for breast cancer: Characteristics of response or intrinsic resistance. Breast Cancer Res. Treat. 2008, 112, 475–488. [Google Scholar] [CrossRef]
- Szasz, A.M.; Li, Q.; Eklund, A.C.; Sztupinszki, Z.; Rowan, A.; Tokes, A.M.; Szekely, B.; Kiss, A.; Szendroi, M.; Gyorffy, B.; et al. The CIN4 chromosomal instability qPCR classifier defines tumor aneuploidy and stratifies outcome in grade 2 breast cancer. PLoS One 2013, 8, e56707. [Google Scholar] [CrossRef]
- Paik, S.; Tang, G.; Shak, S.; Kim, C.; Baker, J.; Kim, W.; Cronin, M.; Baehner, F.L.; Watson, D.; Bryant, J.; et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J. Clin. Oncol. 2006, 24, 3726–3734. [Google Scholar] [CrossRef]
- Wang, Y.; Klijn, J.G.; Zhang, Y.; Sieuwerts, A.M.; Look, M.P.; Yang, F.; Talantov, D.; Timmermans, M.; Meijer-van Gelder, M.E.; Yu, J.; et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 2005, 365, 671–679. [Google Scholar]
- Miller, T.W.; Balko, J.M.; Ghazoui, Z.; Dunbier, A.; Anderson, H.; Dowsett, M.; Gonzalez-Angulo, A.M.; Mills, G.B.; Miller, W.R.; Wu, H.; et al. A gene expression signature from human breast cancer cells with acquired hormone independence identifies MYC as a mediator of antiestrogen resistance. Clin. Cancer Res. 2011, 17, 2024–2034. [Google Scholar] [CrossRef]
- Roberts, C.G.; Millar, E.K.; O’Toole, S.A.; McNeil, C.M.; Lehrbach, G.M.; Pinese, M.; Tobelmann, P.; McCloy, R.A.; Musgrove, E.A.; Sutherland, R.L.; et al. Identification of PUMA as an estrogen target gene that mediates the apoptotic response to tamoxifen in human breast cancer cells and predicts patient outcome and tamoxifen responsiveness in breast cancer. Oncogene 2011, 30, 3186–3197. [Google Scholar] [CrossRef]
- Munkacsy, G.; Abdul-Ghani, R.; Mihaly, Z.; Tegze, B.; Tchernitsa, O.; Surowiak, P.; Schafer, R.; Gyorffy, B. PSMB7 is associated with anthracycline resistance and is a prognostic biomarker in breast cancer. Br. J. Cancer 2010, 102, 361–368. [Google Scholar] [CrossRef]
- Wend, P.; Runke, S.; Wend, K.; Anchondo, B.; Yesayan, M.; Jardon, M.; Hardie, N.; Loddenkemper, C.; Ulasov, I.; Lesniak, M.S.; et al. WNT10B/β-catenin signalling induces HMGA2 and proliferation in metastatic triple-negative breast cancer. EMBO Mol. Med. 2013, 5, 264–279. [Google Scholar] [CrossRef]
- Gyorffy, B.; Surowiak, P.; Kiesslich, O.; Denkert, C.; Schafer, R.; Dietel, M.; Lage, H. Gene expression profiling of 30 cancer cell lines predicts resistance towards 11 anticancer drugs at clinically achieved concentrations. Int. J. Cancer 2006, 118, 1699–1712. [Google Scholar] [CrossRef]
- Baxter, R.C. Insulin-like growth factor binding protein-3 (IGFBP-3): Novel ligands mediate unexpected functions. J. Cell Commun. Signal. 2013, 7, 179–189. [Google Scholar] [CrossRef]
- Porter, D.C.; Farmaki, E.; Altilia, S.; Schools, G.P.; West, D.K.; Chen, M.; Chang, B.D.; Puzyrev, A.T.; Lim, C.U.; Rokow-Kittell, R.; et al. Cyclin-dependent kinase 8 mediates chemotherapy-induced tumor-promoting paracrine activities. Proc. Natl. Acad. Sci. USA 2012, 109, 13799–13804. [Google Scholar] [CrossRef]
- Molina, A.; Velot, L.; Ghouinem, L.; Abdelkarim, M.; Bouchet, B.P.; Luissint, A.C.; Bouhlel, I.; Morel, M.; Sapharikas, E.; di Tommaso, A.; et al. ATIP3, a novel prognostic marker of breast cancer patient survival, limits cancer cell migration and slows metastatic progression by regulating microtubule dynamics. Cancer Res. 2013, 73, 2905–2915. [Google Scholar] [CrossRef]
- Gene Expression Omnibus. Available online: http://www.ncbi.nlm.nih.gov/geo/ (accessed on 26 August 2013).
- The European Genome-phenome Archive. Available online: https://www.ebi.ac.uk/ega/ (accessed on 26 August 2013).
- Gyorffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef]
- Gyorffy, B.; Lanczky, A.; Szallasi, Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1,287 patients. Endocr. Relat. Cancer 2012, 19, 197–208. [Google Scholar] [CrossRef]
- Park, S.; Koo, J.S.; Kim, M.S.; Park, H.S.; Lee, J.S.; Lee, J.S.; Kim, S.I.; Park, B.W. Characteristics and outcomes according to molecular subtypes of breast cancer as classified by a panel of four biomarkers using immunohistochemistry. Breast 2012, 21, 50–57. [Google Scholar] [CrossRef]
- Pawitan, Y.; Bjohle, J.; Amler, L.; Borg, A.L.; Egyhazi, S.; Hall, P.; Han, X.; Holmberg, L.; Huang, F.; Klaar, S.; et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: Derived and validated in two population-based cohorts. Breast Cancer Res. 2005, 7, R953–R964. [Google Scholar] [CrossRef]
- Sabatier, R.; Finetti, P.; Cervera, N.; Lambaudie, E.; Esterni, B.; Mamessier, E.; Tallet, A.; Chabannon, C.; Extra, J.M.; Jacquemier, J.; et al. A gene expression signature identifies two prognostic subgroups of basal breast cancer. Breast Cancer Res. Treat. 2011, 126, 407–420. [Google Scholar] [CrossRef]
- Hatzis, C.; Pusztai, L.; Valero, V.; Booser, D.J.; Esserman, L.; Lluch, A.; Vidaurre, T.; Holmes, F.; Souchon, E.; Wang, H.; et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA 2011, 305, 1873–1881. [Google Scholar] [CrossRef]
- Dedeurwaerder, S.; Desmedt, C.; Calonne, E.; Singhal, S.K.; Haibe-Kains, B.; Defrance, M.; Michiels, S.; Volkmar, M.; Deplus, R.; Luciani, J.; et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol. Med. 2011, 3, 726–741. [Google Scholar] [CrossRef]
- Karn, T.; Pusztai, L.; Holtrich, U.; Iwamoto, T.; Shiang, C.Y.; Schmidt, M.; Muller, V.; Solbach, C.; Gaetje, R.; Hanker, L.; et al. Homogeneous datasets of triple negative breast cancers enable the identification of novel prognostic and predictive signatures. PLoS One 2011, 6, e28403. [Google Scholar] [CrossRef]
- Sircoulomb, F.; Bekhouche, I.; Finetti, P.; Adelaide, J.; Ben Hamida, A.; Bonansea, J.; Raynaud, S.; Innocenti, C.; Charafe-Jauffret, E.; Tarpin, C.; et al. Genome profiling of ERBB2-amplified breast cancers. BMC Cancer 2010, 10, 539. [Google Scholar] [CrossRef]
- KM Plotter. Available online: http://www.kmplot.com (accessed on 26 August 2013).
- Budczies, J.; Klauschen, F.; Sinn, B.V.; Gyorffy, B.; Schmitt, W.D.; Darb-Esfahani, S.; Denkert, C. Cutoff finder: A comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One 2012, 7, e51862. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mihály, Z.; Győrffy, B. Improving Pathological Assessment of Breast Cancer by Employing Array-Based Transcriptome Analysis. Microarrays 2013, 2, 228-242. https://doi.org/10.3390/microarrays2030228
Mihály Z, Győrffy B. Improving Pathological Assessment of Breast Cancer by Employing Array-Based Transcriptome Analysis. Microarrays. 2013; 2(3):228-242. https://doi.org/10.3390/microarrays2030228
Chicago/Turabian StyleMihály, Zsuzsanna, and Balázs Győrffy. 2013. "Improving Pathological Assessment of Breast Cancer by Employing Array-Based Transcriptome Analysis" Microarrays 2, no. 3: 228-242. https://doi.org/10.3390/microarrays2030228
APA StyleMihály, Z., & Győrffy, B. (2013). Improving Pathological Assessment of Breast Cancer by Employing Array-Based Transcriptome Analysis. Microarrays, 2(3), 228-242. https://doi.org/10.3390/microarrays2030228




