Pathogens 2022, 11(7), 790; https://doi.org/10.3390/pathogens11070790 - 12 Jul 2022
Cited by 2 | Viewed by 2937
Abstract
Decades of research on vaccinia virus (VACV) have provided a wealth of insights and tools that have proven to be invaluable in a broad range of studies of molecular virology and pathogenesis. Among the challenges that viruses face are intrinsic host cellular defenses,
[...] Read more.
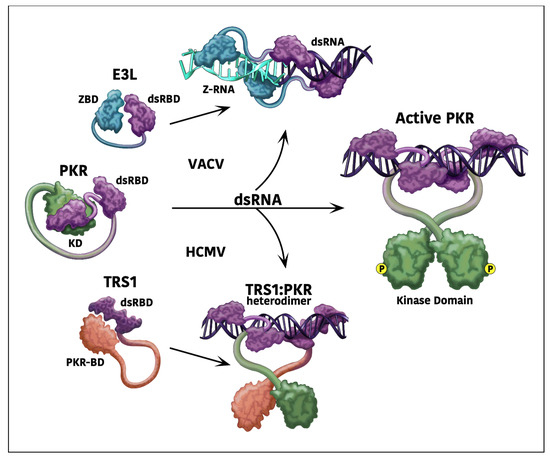
Decades of research on vaccinia virus (VACV) have provided a wealth of insights and tools that have proven to be invaluable in a broad range of studies of molecular virology and pathogenesis. Among the challenges that viruses face are intrinsic host cellular defenses, such as the protein kinase R pathway, which shuts off protein synthesis in response to the dsRNA that accumulates during replication of many viruses. Activation of PKR results in phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α), inhibition of protein synthesis, and limited viral replication. VACV encodes two well-characterized antagonists, E3L and K3L, that can block the PKR pathway and thus enable the virus to replicate efficiently. The use of VACV with a deletion of the dominant factor, E3L, enabled the initial identification of PKR antagonists encoded by human cytomegalovirus (HCMV), a prevalent and medically important virus. Understanding the molecular mechanisms of E3L and K3L function facilitated the dissection of the domains, species-specificity, and evolutionary potential of PKR antagonists encoded by human and nonhuman CMVs. While remaining cognizant of the substantial differences in the molecular virology and replication strategies of VACV and CMVs, this review illustrates how VACV can provide a valuable guide for the study of other experimentally less tractable viruses.
Full article
(This article belongs to the Special Issue Poxvirus-Driven Insights into Virus and Host Biology)
►
Show Figures