The Diversity of Ribonuclease P: Protein and RNA Catalysts with Analogous Biological Functions
Abstract
:1. Introduction
2. Diversity and Distribution of RNase Ps
2.1. RNA-Based RNase Ps Exist in Bacteria, Archaea, and Many Eukarya
2.2. PRORPs are Found Only in Eukarya
3. Structures of RNase Ps
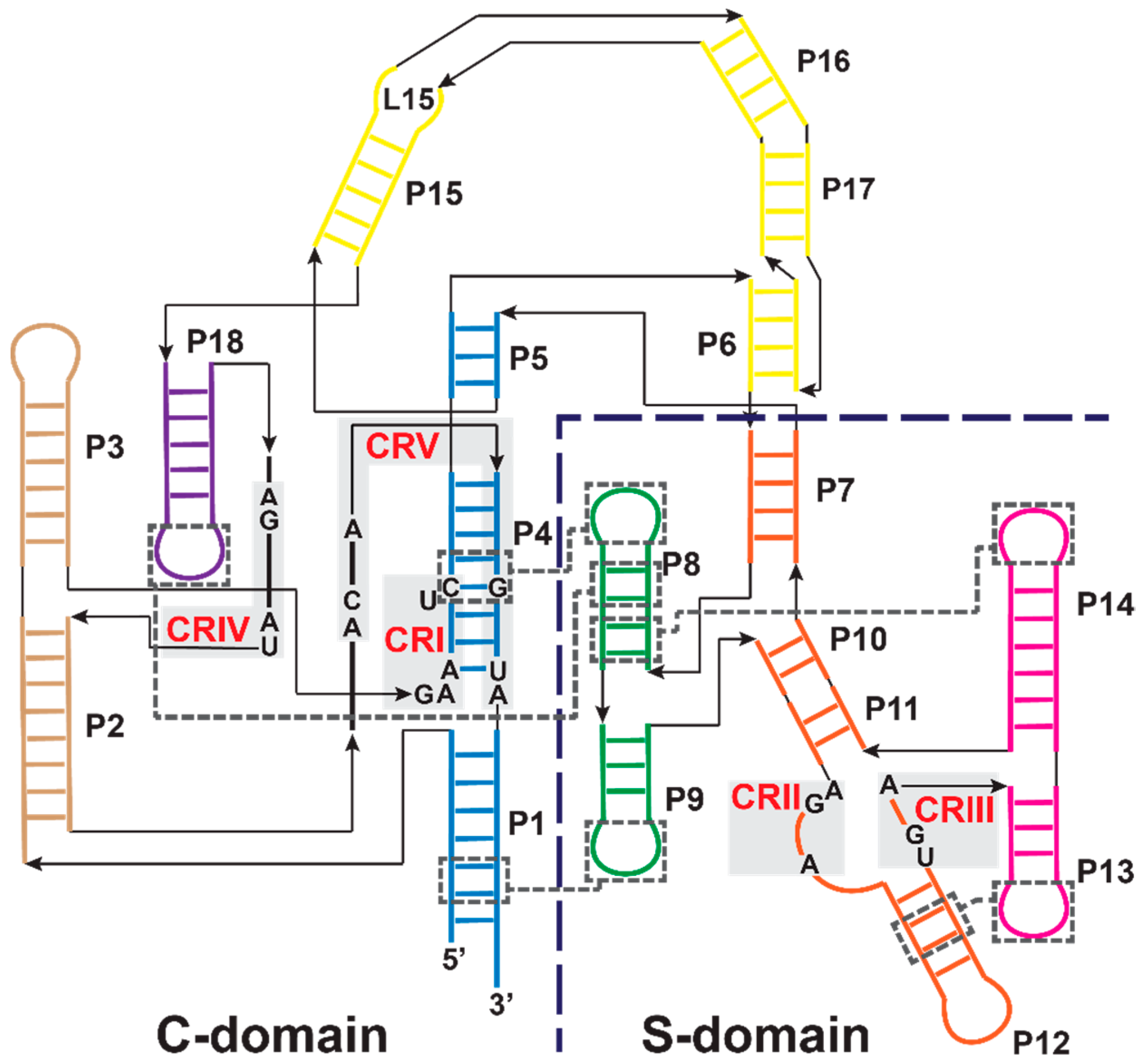
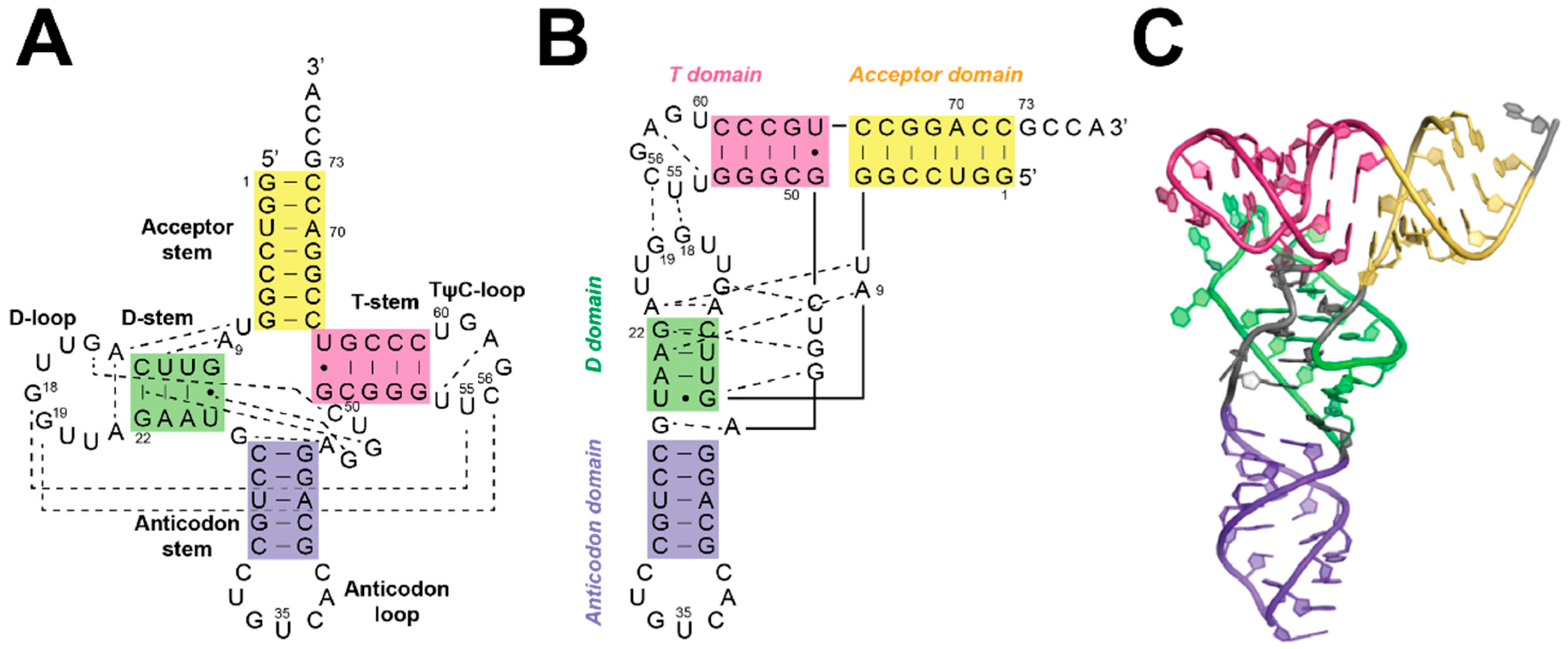
3.1. Bacterial RNase P Ribozyme
3.2. Archaeal and Eukaryotic RNA-Based RNase Ps
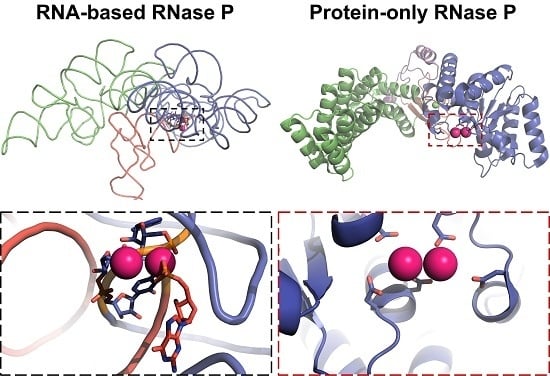
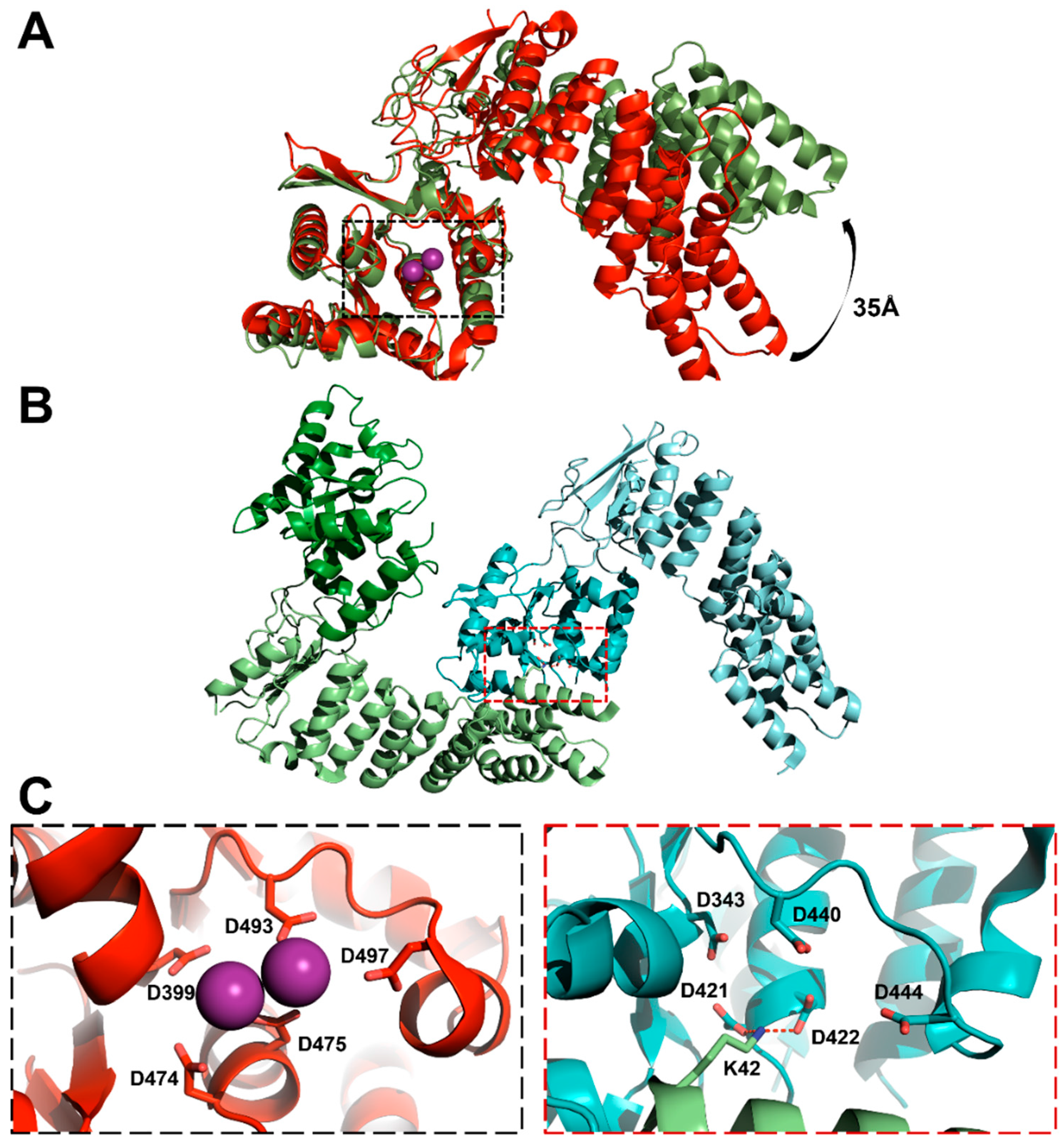
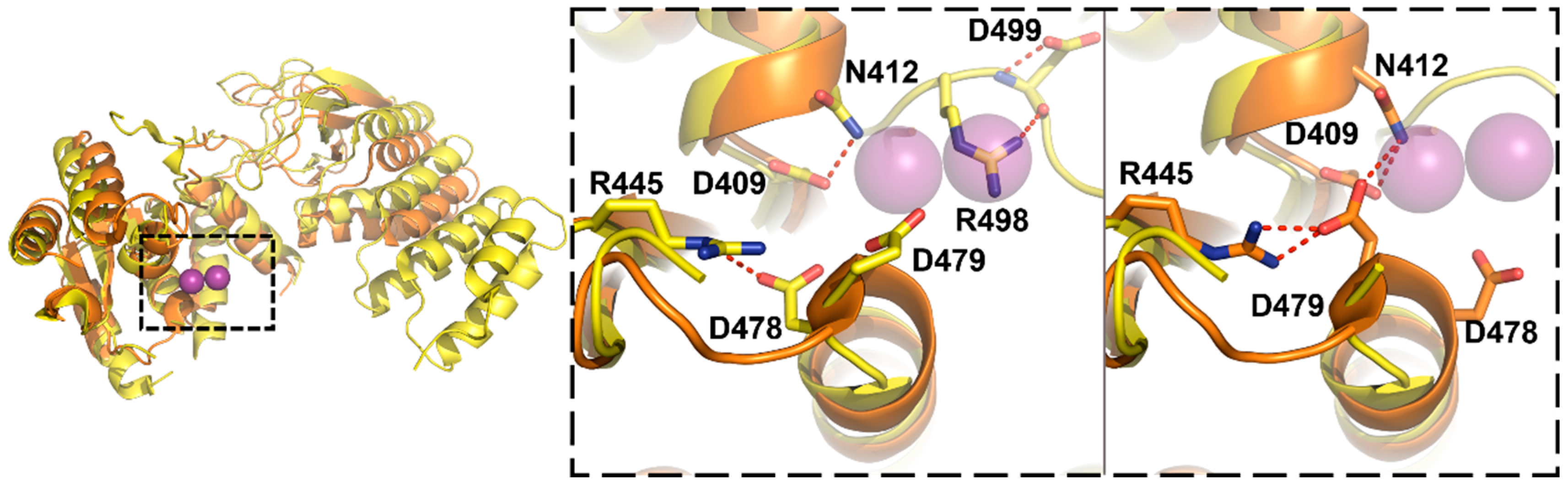
3.3. Protein-Only RNase Ps
4. Catalysis by RNase Ps
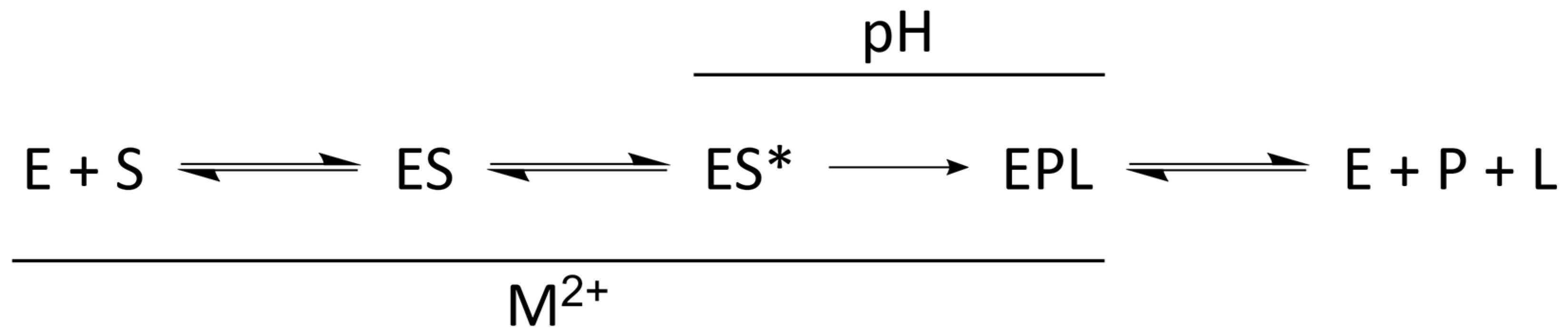
4.1. Kinetic Mechanism
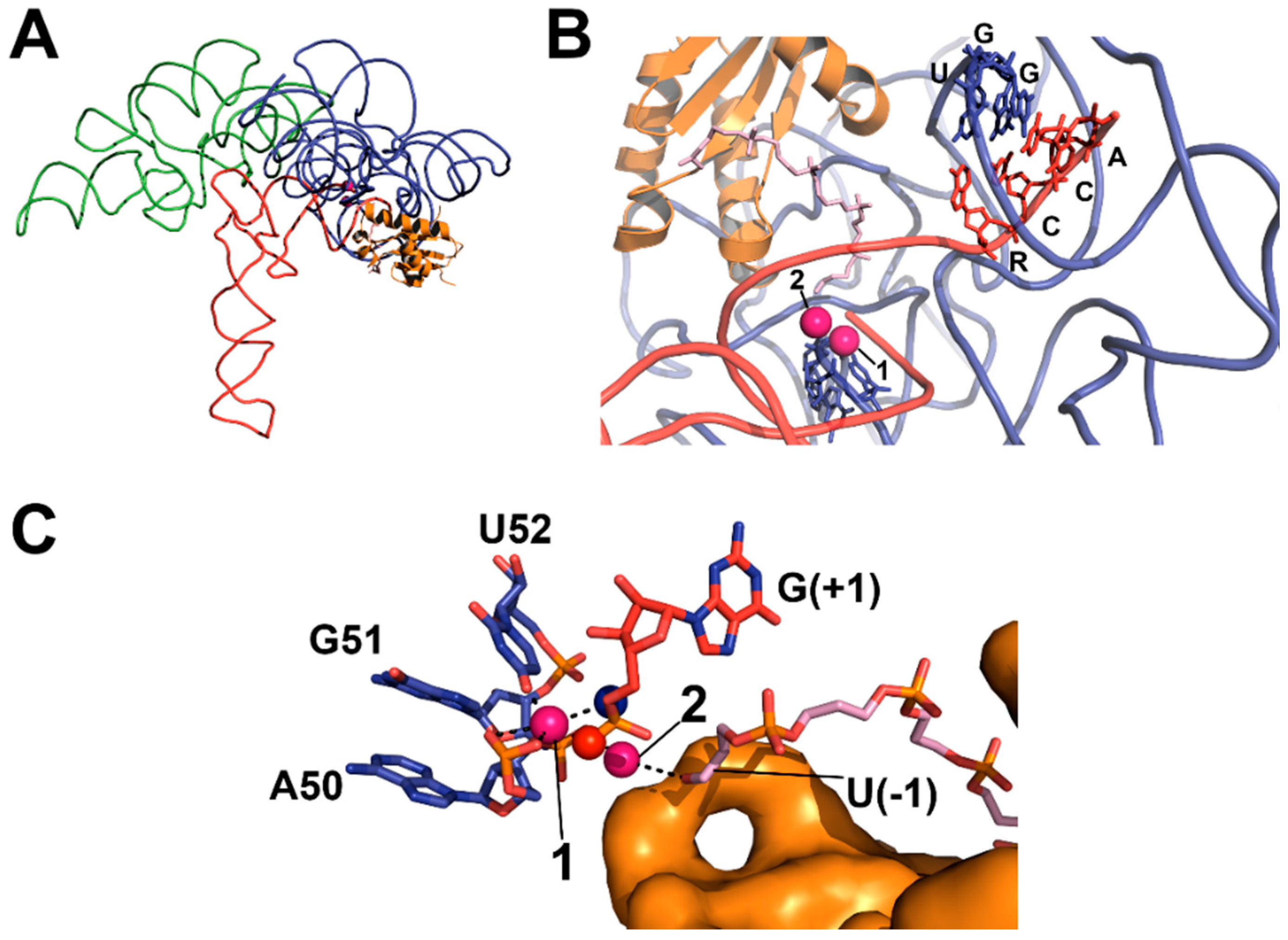
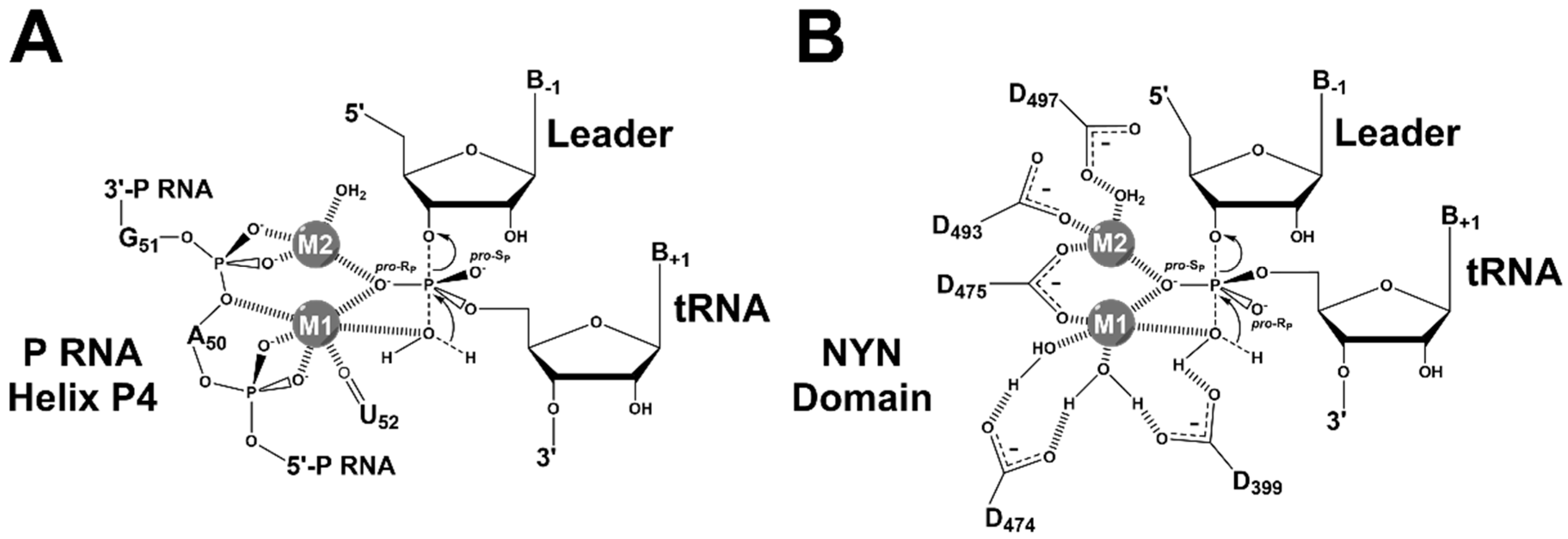
4.2. RNase P Ribozyme Metal-Binding Sites
4.3. Hydrolysis Requires Activation of a Metal-Bound Water Molecule
4.4. RNase Ps Utilize Similar Catalytic Strategies
5. Substrate Recognition by RNase Ps
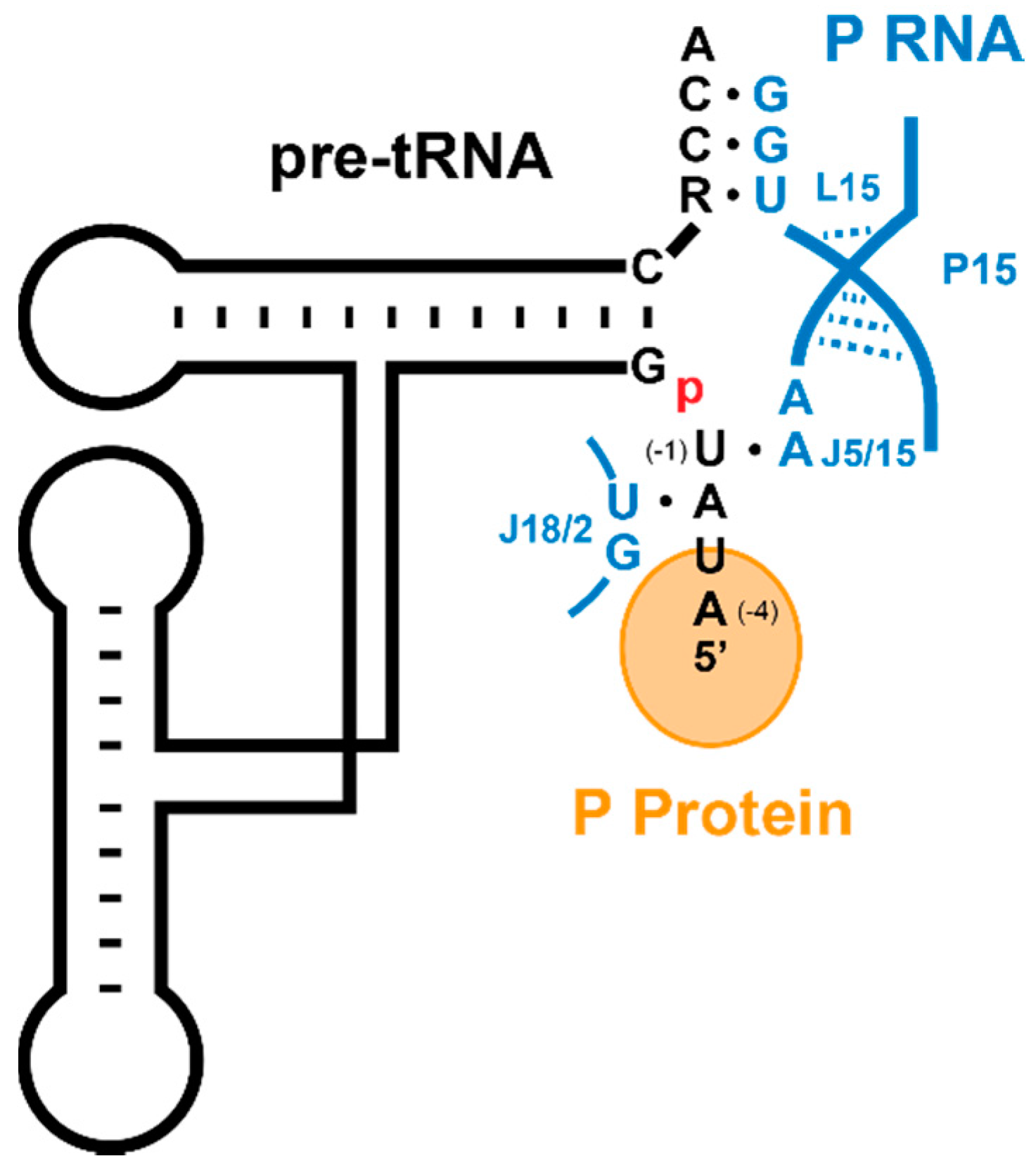
5.1. Recogntion by Bacterial RNase P
5.2. Substrate Recogntion by Archaeal and Eukaryotic RNase P
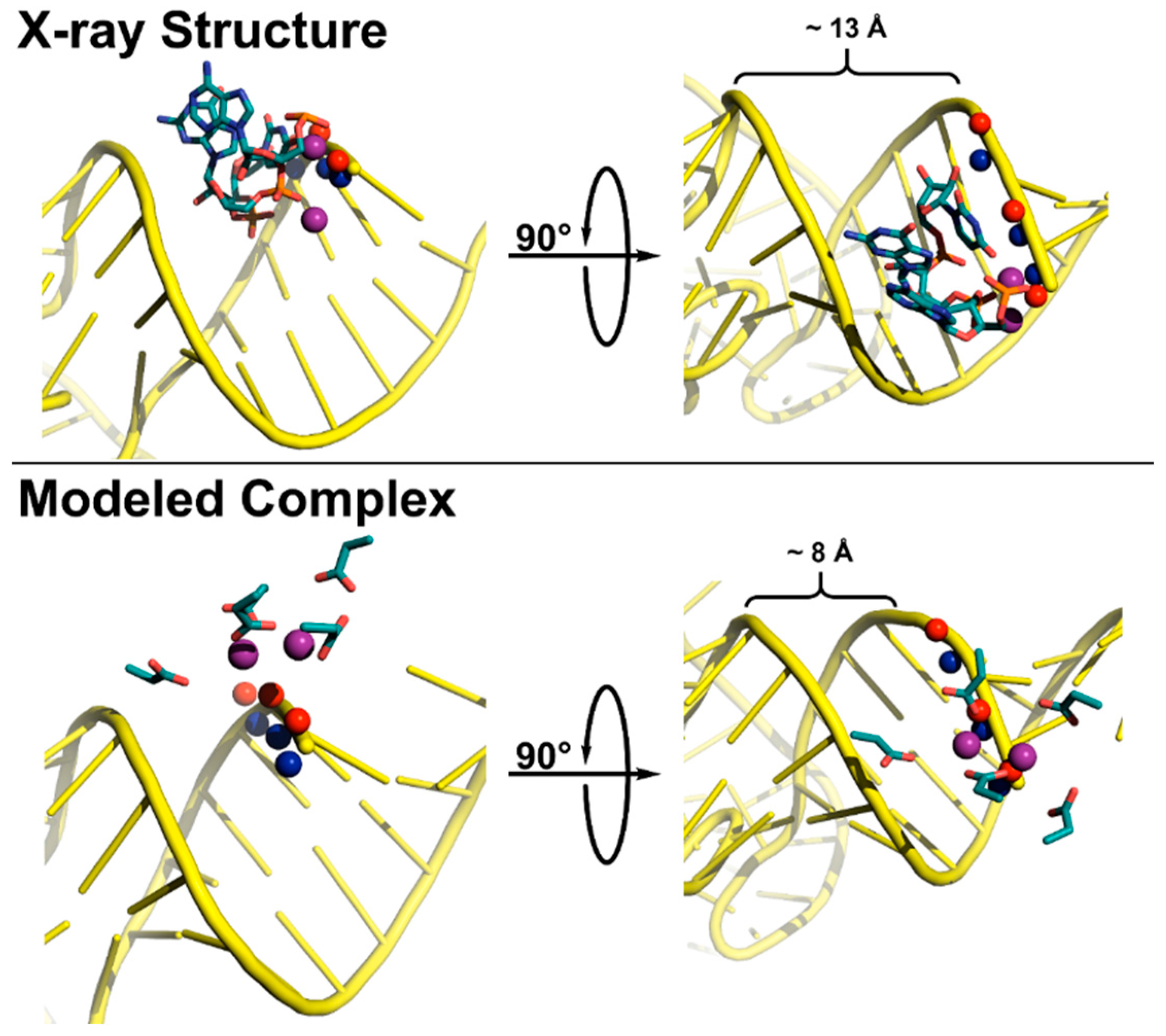
5.3. Substrate Recogntion by Protein-Only RNase Ps
5.4. Non-tRNA RNase P Substrates
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| RNase P | Ribonuclease P |
| RNA | ribonucleic acid |
| tRNA | transfer RNA |
| pre-tRNA | precursor tRNA |
| P RNA | RNase P RNA |
| PRORP | protein-only (alternatively: proteinaceous) Ribonuclease P |
| MRPP | mitochondrial RNase P protein |
| NYN | Nedd4-BP1, YacP nuclease |
| PPR | pentatricopeptide repeat |
References
- The Nobel Prize in Chemistry 1989. Available online: http://www.nobelprize.org/nobel_prizes/chemistry/laureates/1989/ (accessed on 15 January 2016).
- Robertson, H.D.; Altman, S.; Smith, J.D. Purification and properties of a specific Escherichia coli ribonuclease which cleaves a tyrosine transfer ribonucleic acid precursor. J. Biol. Chem. 1972, 247, 5243–5251. [Google Scholar] [PubMed]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.R.; Altman, S. The RNA moiety of Ribonuclease P is the catalytic subunit of the enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef]
- Frank, D.N.; Pace, N.R. Ribonuclease P: Unity and diversity in a trna processing ribozyme. Annu Rev Biochem 1998, 67, 153–180. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.C.; Engelke, D.R. Ribonuclease P: The evolution of an ancient RNA enzyme. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 77–102. [Google Scholar] [CrossRef] [PubMed]
- Hopper, A.K.; Huang, H.Y. Quality control pathways for nucleus-encoded eukaryotic tRNA biosynthesis and subcellular trafficking. Mol. Cell Biol. 2015, 35, 2052–2058. [Google Scholar] [CrossRef] [PubMed]
- Liu, X. Molecular recognition of inhibitors, metal ions and substrates by Ribonuclease P. Ph.D. Thesis, The University of Michigan, Ann Arbor, MI, USA, 2013. [Google Scholar]
- Randau, L.; Schröder, I.; Soll, D. Life without RNase P. Nature 2008, 453, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Randau, L. RNA processing in the minimal organism Nanoarchaeum equitans. Genome Biology 2012, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Howard, M.J.; Liu, X.; Lim, W.H.; Klemm, B.P.; Fierke, C.A.; Koutmos, M.; Engelke, D.R. RNase P enzymes: Divergent scaffolds for a conserved biological reaction. RNA Biol. 2013, 10, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.; Rossmanith, W.; Hartmann, R.K.; Thölken, C.; Gutmann, B.; Giegé, P.; Gobert, A. Distribution of ribonucleoprotein and protein–only RNase P in Eukarya. Mol. Biol. Evol. 2015, 32, 3186–3193. [Google Scholar] [CrossRef] [PubMed]
- Adl, S.M.; Simpson, A.G.; Lane, C.E.; Lukes, J.; Bass, D.; Bowser, S.S.; Brown, M.W.; Burki, F.; Dunthorn, M.; Hampl, V.; et al. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 2012, 59, 429–493. [Google Scholar] [CrossRef] [PubMed]
- Burki, F. The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb. Perspect. Biol. 2014, 6, a016147. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.L.; Pace, N.R. Identification of the universally conserved core of Ribonuclease P RNA. RNA 1997, 3, 557–560. [Google Scholar] [PubMed]
- Brown, J.W.; Haas, E.S.; James, B.D.; Hunt, D.A.; Liu, J.S.; Pace, N.R. Phylogenetic analysis and evolution of RNase P RNA in proteobacteria. J. Bacteriol. 1991, 173, 3855–3863. [Google Scholar] [PubMed]
- Haas, E.S.; Brown, J.W. Evolutionary variation in bacterial RNase P RNAs. Nucleic Acids Res. 1998, 26, 4093–4099. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.W. The Ribonuclease P database. Nucleic Acids Res. 1999, 27, 314. [Google Scholar] [CrossRef] [PubMed]
- Marquez, S.M.; Harris, J.K.; Kelley, S.T.; Brown, J.W.; Dawson, S.C.; Roberts, E.C.; Pace, N.R. Structural implications of novel diversity in eucaryal RNase P RNA. RNA 2005, 11, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.; Marquez, S.M.; Pace, N.R. RNase P: Interface of the RNA and protein worlds. Trends Biochem. Sci. 2006, 31, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.C.; Brown, J.W. The RNase P family. RNA Biol. 2009, 6, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y. Ribonuclease p catalysis: Metal ion interaction and conformational dynamics. Ph.D. Thesis, The University of Michigan, Ann Arbor, MI, USA, 2015. [Google Scholar]
- Reiter, N.J.; Osterman, A.; Torres-Larios, A.; Swinger, K.K.; Pan, T.; Mondragon, A. Structure of a bacterial Ribonuclease P holoenzyme in complex with tRNA. Nature 2010, 468, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Lim, W.H. Importance of substrate recognition and metal ions in the Ribonuclease P catalysis. Ph.D. Thesis, The University of Michigan, Ann Arbor, MI, USA, 2011. [Google Scholar]
- Kole, R.; Altman, S. Properties of purified Ribonuclease P from Escherichia coli. Biochemistry 1981, 20, 1902–1906. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.W.; Nolan, J.M.; Haas, E.S.; Rubio, M.A.; Major, F.; Pace, N.R. Comparative analysis of Ribonuclease P RNA using gene sequences from natural microbial populations reveals tertiary structural elements. Proc. Natl. Acad. Sci. USA 1996, 93, 3001–3006. [Google Scholar] [CrossRef] [PubMed]
- Massire, C.; Jaeger, L.; Westhof, E. Phylogenetic evidence for a new tertiary interaction in bacterial RNase P RNAs. RNA 1997, 3, 553–556. [Google Scholar] [PubMed]
- Massire, C.; Jaeger, L.; Westhof, E. Derivation of the three-dimensional architecture of bacterial Ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 1998, 279, 773–793. [Google Scholar] [CrossRef] [PubMed]
- Pomeranz Krummel, D.A.; Altman, S. Verification of phylogenetic predictions in vivo and the importance of the tetraloop motif in a catalytic RNA. Proc. Natl. Acad. Sci. USA 1999, 96, 11200–11205. [Google Scholar] [CrossRef] [PubMed]
- Haas, E.S.; Banta, A.B.; Harris, J.K.; Pace, N.R.; Brown, J.W. Structure and evolution of Ribonuclease P RNA in gram-positive bacteria. Nucleic Acids Res. 1996, 24, 4775–4782. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.W.; Banta, A.B.; Haas, E.S.; Brown, J.W.; Pace, N.R. Mycoplasma fermentans simplifies our view of the catalytic core of Ribonuclease P RNA. RNA 1996, 2, 452–462. [Google Scholar] [PubMed]
- Kurz, J.C.; Niranjanakumari, S.; Fierke, C.A. Protein component of Bacillus subtilis RNase P specifically enhances the affinity for precursor-tRNAAsp. Biochemistry 1998, 37, 2393–2400. [Google Scholar] [CrossRef] [PubMed]
- Kurz, J.C.; Fierke, C.A. The affinity of magnesium binding sites in the Bacillus subtilis RNase P pre-tRNA complex is enhanced by the protein subunit. Biochemistry 2002, 41, 9545–9558. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.; Andrews, A.J.; Fierke, C.A. Roles of protein subunits in RNA-protein complexes: Lessons from Ribonuclease P. Biopolymers 2004, 73, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.; Rueda, D.; Koutmou, K.S.; Koutmos, M.; Walter, N.G.; Fierke, C.A. A divalent cation stabilizes the active conformation of the B. subtilis RNase P pre-tRNA complex: A role for an inner-sphere metal ion in RNase P. J. Mol. Biol. 2010, 400, 28–51. [Google Scholar] [CrossRef] [PubMed]
- Koutmou, K.S.; Day-Storms, J.J.; Fierke, C.A. The RNR motif of B. subtilis RNase P protein interacts with both PRNA and pre-tRNA to stabilize an active conformer. RNA 2011, 17, 1225–1235. [Google Scholar] [PubMed]
- Biswas, R.; Ledman, D.W.; Fox, R.O.; Altman, S.; Gopalan, V. Mapping RNA-protein interactions in Ribonuclease P from Escherichia coli using disulfide-linked EDTA-Fe. J. Mol. Biol. 2000, 296, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Kirsebom, L.A.; Vioque, A. RNase P from Bacteria. Substrate recognition and function of the protein subunit. Mol. Biol. Rep. 1995, 22, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Jovanovic, M.; Sanchez, R.; Altman, S.; Gopalan, V. Elucidation of structure-function relationships in the protein subunit of bacterial RNase P using a genetic complementation approach. Nucleic Acids Res. 2002, 30, 5065–5073. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.Y.; Pulukkunat, D.K.; Woznick, W.K.; Gopalan, V. Functional reconstitution and characterization of Pyrococcus furiosus RNase P. Proc. Natl. Acad. Sci. USA 2006, 103, 16147–16152. [Google Scholar] [CrossRef] [PubMed]
- Waugh, D.S.; Pace, N.R. Complementation of an RNase P RNA (rnpb) gene deletion in Escherichia coli by homologous genes from distantly related Eubacteria. J. Bacteriol. 1990, 172, 6316–6322. [Google Scholar] [PubMed]
- Gößringer, M.; Hartmann, R.K. Function of heterologous and truncated RNase P proteins in Bacillus subtilis. Mol. Microbiol. 2007, 66, 801–813. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.K.; Haas, E.S.; Williams, D.; Frank, D.N.; Brown, J.W. New insight into RNase P RNA structure from comparative analysis of the archaeal RNA. RNA 2001, 7, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.B.; Chan, P.P.; Cozen, A.E.; Bernick, D.L.; Brown, J.W.; Gopalan, V.; Lowe, T.M. Discovery of a minimal form of RNase P in Pyrobaculum. Proc. Natl. Acad. Sci. USA 2010, 107, 22493–22498. [Google Scholar] [CrossRef] [PubMed]
- Pannucci, J.A.; Haas, E.S.; Hall, T.A.; Harris, J.K.; Brown, J.W. RNase P RNAs from some Archaea are catalytically active. Proc. Natl. Acad. Sci. USA 1999, 96, 7803–7808. [Google Scholar] [CrossRef] [PubMed]
- Andrews, A.J.; Hall, T.A.; Brown, J.W. Characterization of RNase P holoenzymes from Methanococcus jannaschii and Methanothermobacter thermoautotrophicus. Biol. Chem. 2001, 382, 1171–1177. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, E.; Hartmann, R.K. The enigma of Ribonuclease P evolution. Trends Genet. 2003, 19, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.M.; Lai, L.B.; Susanti, D.; Mukhopadhyay, B.; Gopalan, V. Ribosomal protein L7Ae is a subunit of archaeal RNase P. Proc. Natl. Acad. Sci. USA 2010, 107, 14573–14578. [Google Scholar] [CrossRef] [PubMed]
- Jarrous, N.; Gopalan, V. Archaeal/eukaryal RNase P: Subunits, functions and RNA diversification. Nucleic Acids Res. 2010, 38, 7885–7894. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, J.R.; Lee, Y.; Lane, W.S.; Engelke, D.R. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998, 12, 1678–1690. [Google Scholar] [CrossRef] [PubMed]
- Sinapah, S.; Wu, S.; Chen, Y.; Pettersson, B.M.; Gopalan, V.; Kirsebom, L.A. Cleavage of model substrates by archaeal RNase P: role of protein cofactors in cleavage-site selection. Nucleic Acids Res. 2011, 39, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Fukuhara, H.; Kifusa, M.; Watanabe, M.; Terada, A.; Honda, T.; Numata, T.; Kakuta, Y.; Kimura, M. A fifth protein subunit Ph1496p elevates the optimum temperature for the Ribonuclease P activity from Pyrococcus horikoshii OT3. Biochem. Biophys. Res. Commun. 2006, 343, 956–964. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Rohlman, C.E.; Molony, L.A.; Engelke, D.R. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol. Cell Biol. 1991, 11, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Lygerou, Z.; Mitchell, P.; Petfalski, E.; Seraphin, B.; Tollervey, D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 1994, 8, 1423–1433. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.; Zengel, J.M.; Lindahl, L. A novel protein shared by RNase MRP and RNase P. RNA 1997, 3, 382–391. [Google Scholar] [PubMed]
- Dichtl, B.; Tollervey, D. POP3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. EMBO J. 1997, 16, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Morales, M.J.; Dang, Y.L.; Lou, Y.C.; Sulo, P.; Martin, N.C. A 105-kDa protein is required for yeast mitochondrial RNase P activity. Proc. Natl. Acad. Sci. USA 1992, 89, 9875–9879. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.L.; Martin, N.C. Yeast mitochondrial RNase P. Sequence of the RPM2 gene and demonstration that its product is a protein subunit of the enzyme. J. Biol. Chem. 1993, 268, 19791–19796. [Google Scholar] [PubMed]
- Jarrous, N. Human Ribonuclease P: Subunits, function, and intranuclear localization. RNA 2002, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Marvin, M.C.; Engelke, D.R. Broadening the mission of an RNA enzyme. J. Cell Biochem. 2009, 108, 1244–1251. [Google Scholar] [CrossRef] [PubMed]
- Holzmann, J.; Frank, P.; Loffler, E.; Bennett, K.L.; Gerner, C.; Rossmanith, W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 2008, 135, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Kikovska, E.; Svard, S.G.; Kirsebom, L.A. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc. Natl. Acad. Sci. USA 2007, 104, 2062–2067. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.B.; Bernal-Bayard, P.; Mohannath, G.; Lai, S.M.; Gopalan, V.; Vioque, A. A functional RNase P protein subunit of bacterial origin in some eukaryotes. Mol. Genet. Genomics 2011, 286, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Gobert, A.; Butmann, B.; Taschner, A.; Gößringer, M.; Holzmann, J.; Hartmann, R.K.; Rossmanith, W.; Giegé, P. A single Arabidopsis organellar protein has RNase P activity. Nat. Struct. Mol. Biol. 2010, 17, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.; Hartig, A.; Hartmann, R.K.; Rossmanith, W. Playing RNase P evolution: Swapping the RNA catalyst for a protein reveals functional uniformity of highly divergent enzyme forms. PLoS Genet. 2014, 10, e1004506. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.I.; Mercer, T.R.; Davies, S.M.; Shearwood, A.M.; Nygard, K.K.; Richman, T.R.; Mattick, J.S.; Rackham, O.; Filipovska, A. RNA processing in human mitochondria. Cell Cycle 2011, 10, 2904–2916. [Google Scholar] [CrossRef] [PubMed]
- Howard, M.J.; Lim, W.H.; Fierke, C.A.; Koutmos, M. Mitochondrial Ribonuclease P structure provides insight into the evolution of catalytic strategies for precursor-tRNA 5′ processing. Proc. Natl. Acad. Sci. USA 2012, 109, 16149–16154. [Google Scholar] [CrossRef] [PubMed]
- Jackman, J.E.; Montange, R.K.; Malik, H.S.; Phizicky, E.M. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA 2003, 9, 574–585. [Google Scholar] [CrossRef] [PubMed]
- Vilardo, E.; Nachbagauer, C.; Buzet, A.; Taschner, A.; Holzmann, J.; Rossmanith, W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase-extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 2012, 40, 11583–11593. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, L.; Sridhara, S.; Hallberg, B.M. Structure of the nuclease subunit of human mitochondrial RNase P. Nucleic Acids Res. 2015, 43, 5664–5672. [Google Scholar] [CrossRef] [PubMed]
- Taschner, A.; Weber, C.; Buzet, A.; Hartmann, R.K.; Hartig, A.; Rossmanith, W. Nuclear RNase P of Trypanosoma brucei: A single protein in place of the multicomponent RNA-protein complex. Cell reports 2012, 2, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, B.; Gobert, A.; Giegé, P. PRORP proteins support RNase P activity in both organelles and the nucleus in Arabidopsis. Genes Dev. 2012, 26, 1022–1027. [Google Scholar] [CrossRef] [PubMed]
- Sugita, C.; Komura, Y.; Tanaka, K.; Kometani, K.; Satoh, H.; Sugita, M. Molecular characterization of three PRORP proteins in the moss Physcomitrella patens: Nuclear PRORP protein is not essential for moss viability. PLoS ONE 2014, 9, e108962. [Google Scholar] [CrossRef] [PubMed]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef] [PubMed]
- Salavati, R.; Panigrahi, A.K.; Stuart, K.D. Mitochondrial Ribonuclease P activity of Trypanosoma brucei. Mol. Biochem. Parasitol. 2001, 115, 109–117. [Google Scholar] [CrossRef]
- Green, C.J.; Rivera–Leon, R.; Vold, B.S. The catalytic core of RNase P. Nucleic Acids Res. 1996, 24, 1497–1503. [Google Scholar] [CrossRef] [PubMed]
- Loria, A.; Pan, T. Modular construction for function of a ribonucleoprotein enzyme: The catalytic domain of Bacillus subtilis RNase P complexed with B. subtilis RNase P protein. Nucleic Acids Res. 2001, 29, 1892–1897. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Kikovska, E.; Lindell, M.; Kirsebom, L.A. Cleavage mediated by the catalytic domain of bacterial RNase P RNA. J. Mol. Biol. 2012, 422, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.E.; Nolan, J.M.; Malhotra, A.; Brown, J.W.; Harvey, S.C.; Pace, N.R. Use of photoaffinity crosslinking and molecular modeling to analyze the global architecture of Ribonuclease P RNA. EMBO J. 1994, 13, 3953–3963. [Google Scholar] [PubMed]
- Westhof, E.; Altman, S. Three-dimensional working model of M1 RNA, the catalytic RNA subunit of Ribonuclease P from Escherichia coli. Proc. Natl. Acad. Sci. USA 1994, 91, 5133–5137. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Loria, A.; Zhong, K. Probing of tertiary interactions in RNA: 2′-hydroxyl-base contacts between the RNase P RNA and pre-tRNA. Proc. Natl. Acad. Sci USA 1995, 92, 12510–12514. [Google Scholar] [CrossRef] [PubMed]
- Loria, A.; Pan, T. Domain structure of the ribozyme from eubacterial Ribonuclease P. RNA 1996, 2, 551–563. [Google Scholar] [PubMed]
- Pan, T.; Jakacka, M. Multiple substrate binding sites in the ribozyme from Bacillus subtilis RNase P. EMBO J. 1996, 15, 2249–2255. [Google Scholar] [PubMed]
- Torres-Larios, A.; Swinger, K.K.; Krasilnikov, A.S.; Pan, T.; Mondragon, A. Crystal structure of the RNA component of bacterial Ribonuclease P. Nature 2005, 437, 584–587. [Google Scholar] [CrossRef] [PubMed]
- Kazantsev, A.V.; Krivenko, A.A.; Harrington, D.J.; Holbrook, S.R.; Adams, P.D.; Pace, N.R. Crystal structure of a bacterial Ribonuclease P RNA. Proc. Natl. Acad. Sci. USA 2005, 102, 13392–13397. [Google Scholar] [CrossRef] [PubMed]
- Stams, T.; Niranjanakumari, S.; Fierke, C.A.; Christianson, D.W. Ribonuclease P protein structure: evolutionary origins in the translational apparatus. Science 1998, 280, 752–755. [Google Scholar] [CrossRef] [PubMed]
- Kazantsev, A.V.; Krivenko, A.A.; Harrington, D.J.; Carter, R.J.; Holbrook, S.R.; Adams, P.D.; Pace, N.R. High-resolution structure of RNase P protein from Thermotoga maritima. Proc. Natl. Acad. Sci. USA 2003, 100, 7497–7502. [Google Scholar] [CrossRef] [PubMed]
- Spitzfaden, C.; Nicholson, N.; Jones, J.J.; Guth, S.; Lehr, R.; Prescott, C.D.; Hegg, L.A.; Eggleston, D.S. The structure of Ribonuclease P protein from Staphylococcus aureus reveals a unique binding site for single-stranded RNA. J. Mol. Biol. 2000, 295, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Shannon, R. Revised effective ionic radii and systematic studies of interatomic distances in halides and chalcogenides. Acta Crystallogr. Sect. A: Found Adv. 1976, 32, 751–767. [Google Scholar] [CrossRef]
- Kazantsev, A.V.; Krivenko, A.A.; Pace, N.R. Mapping metal-binding sites in the catalytic domain of bacterial RNase P RNA. RNA 2009, 15, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, M.; Tinoco, I. Solution structure and metal-ion binding of the P4 element from bacterial RNase P RNA. RNA 2000, 6, 1212–1225. [Google Scholar] [CrossRef] [PubMed]
- Getz, M.M.; Andrews, A.J.; Fierke, C.A.; Al-Hashimi, H.M. Structural plasticity and Mg2+ binding properties of rnase p p4 from combined analysis of nmr residual dipolar couplings and motionally decoupled spin relaxation. RNA 2007, 13, 251–266. [Google Scholar] [CrossRef] [PubMed]
- Koutmou, K.S.; Casiano-Negroni, A.; Getz, M.M.; Pazicni, S.; Andrews, A.J.; Penner-Hahn, J.E.; Al-Hashimi, H.M.; Fierke, C.A. NMR and XAS reveal an inner-sphere metal binding site in the P4 helix of the metallo-ribozyme Ribonuclease P. Proc. Natl. Acad. Sci. USA 2010, 107, 2479–2484. [Google Scholar] [CrossRef] [PubMed]
- Kufel, J.; Kirsebom, L.A. The P15-loop of Escherichia coli RNase P RNA is an autonomous divalent metal ion binding domain. RNA 1998, 4, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Kakuta, Y.; Ishimatsu, I.; Numata, T.; Kimura, K.; Yao, M.; Tanaka, I.; Kimura, M. Crystal structure of a Ribonuclease P protein Ph1601p from Pyrococcus horikoshii OT3: An archaeal homologue of human nuclear Ribonuclease P protein Rpp21. Biochemistry 2005, 44, 12086–12093. [Google Scholar] [CrossRef] [PubMed]
- Amero, C.D.; Boomershine, W.P.; Xu, Y.; Foster, M. Solution structure of Pyrococcus furiosus RPP21, a component of the archaeal RNase P holoenzyme, and interactions with its RPP29 protein partner. Biochemistry 2008, 47, 11704–11710. [Google Scholar] [CrossRef] [PubMed]
- Boomershine, W.P.; McElroy, C.A.; Tsai, H.Y.; Wilson, R.C.; Gopalan, V.; Foster, M.P. Structure of Mth11/Mth Rpp29, an essential protein subunit of archaeal and eukaryotic RNase P. Proc. Natl. Acad. Sci. USA 2003, 100, 15398–15403. [Google Scholar] [CrossRef] [PubMed]
- Sidote, D.J.; Heideker, J.; Hoffman, D.W. Crystal structure of archaeal Ribonuclease P protein aRpp29 from Archaeoglobus fulgidus. Biochemistry 2004, 43, 14128–14138. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Watanabe, M.; Kakuta, Y.; Kamachi, R.; Numata, T.; Tanaka, I.; Kimura, M. Crystal structure of the Ribonuclease P protein Ph1877p from hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biochem. Biophys. Res. Commun. 2004, 319, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Charron, C.; Manival, X.; Clery, A.; Senty-Segault, V.; Charpentier, B.; Marmier-Gourrier, N.; Branlant, C.; Aubry, A. The archaeal sRNA binding protein L7Ae has a 3D structure very similar to that of its eukaryal counterpart while having a broader RNA-binding specificity. J. Mol. Biol. 2004, 342, 757–773. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Lilley, D.M. Structure of a rare non-standard sequence k-turn bound by L7Ae protein. Nucleic Acids Res. 2014, 42, 4734–4740. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.C.; Bohlen, C.J.; Foster, M.P.; Bell, C.E. Structure of Pfu POP5, an archaeal RNase P protein. Proc. Natl. Acad. Sci. USA 2006, 103, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Kakuta, Y.; Kimura, K.; Saho, J.; Kimura, M. Structure of an archaeal homolog of the human protein complex Rpp21–Rpp29 that is a key core component for the assembly of active Ribonuclease P. J. Mol. Biol. 2008, 384, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Amero, C.D.; Pulukkunat, D.K.; Gopalan, V.; Foster, M.P. Solution structure of an archaeal RNase P binary protein complex: Formation of the 30-kDa complex between Pyrococcus furiosus RPP21 and RPP29 is accompanied by coupled protein folding and highlights critical features for protein-protein and protein-RNA interactions. J. Mol. Biol. 2009, 393, 1043–1055. [Google Scholar] [PubMed]
- Perederina, A.; Esakova, O.; Quan, C.; Khanova, E.; Krasilnikov, A.S. Eukaryotic Ribonucleases P/MRP: the crystal structure of the P3 domain. EMBO J. 2010, 29, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Hipp, K.; Galani, K.; Batisse, C.; Prinz, S.; Bottcher, B. Modular architecture of eukaryotic RNase P and RNase MRP revealed by electron microscopy. Nucleic Acids Res. 2012, 40, 3275–3288. [Google Scholar] [CrossRef] [PubMed]
- Karasik, A.; Shanmuganathan, A.; Howard, M.J.; Fierke, C.A.; Koutmos, M. Nuclear protein-only Ribonuclease P2 structure and biochemical characterization provide insight into the conserved properties of tRNA 5′ end processing enzymes. J. Mol. Biol. 2015, 428, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Liu, X.; Zhou, W.; Yang, X.; Shen, Y. Auto-inhibitory mechanism of the human mitochondrial RNase P protein complex. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Kissinger, C.R.; Rejto, P.A.; Pelletier, L.A.; Thomson, J.A.; Showalter, R.E.; Abreo, M.A.; Agree, C.S.; Margosiak, S.; Meng, J.J.; Aust, R.M.; et al. Crystal structure of human ABAD/HSD10 with a bound inhibitor: implications for design of Alzheimer’s disease therapeutics. J. Mol. Biol. 2004, 342, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Van Laer, B.; Roovers, M.; Wauters, L.; Kasprzak, J.M.; Dyzma, M.; Deyaert, E.; Kumar Singh, R.; Feller, A.; Bujnicki, J.M.; Droogmans, L.; et al. Structural and functional insights into tRNA binding and adenosine N1-methylation by an archaeal Trm10 homologue. Nucleic Acids Res. 2016, 44, 940–953. [Google Scholar] [CrossRef] [PubMed]
- Howard, M.H.; Klemm, B.P.; Fierke, C.A. Mechanistic studies reveal similar catalytic strategies for phosphodiester bond hydrolysis by protein-only and RNA-dependent Ribonuclease P. J. Biol. Chem. 2015, 290, 13454–13464. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Nakamura, T.; Maeda, T.; Nakayama, K.; Gao, X.; Nakashima, T.; Kakuta, Y.; Kimura, M. Pentatricopeptide repeat motifs in the processing enzyme PRORP1 in Arabidopsis thaliana play a crucial role in recognition of nucleotide bases at TψC loop in precursor tRNAs. Biochem. Biophys. Res. Commun. 2014, 450, 1541–1546. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.; Fierke, C.A. Conformational change in the Bacillus subtilis RNase P holoenzyme–pre-tRNA complex enhances substrate affinity and limits cleavage rate. RNA 2009, 15, 1565–1577. [Google Scholar] [CrossRef] [PubMed]
- Beebe, J.A.; Fierke, C.A. A kinetic mechanism for cleavage of precursor tRNAAsp catalyzed by the RNA component of Bacillus subtilis Ribonuclease P. Biochemistry 1994, 33, 10294–10304. [Google Scholar] [CrossRef] [PubMed]
- Stanford, N.P.; Halford, S.E.; Baldwin, G.S. DNA cleavage by the EcoRV restriction endonuclease: pH dependence and proton transfers in catalysis. J. Mol. Biol. 1999, 288, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Bastock, J.A.; Webb, M.; Grasby, J.A. The pH-dependence of the Escherichia coli RNase HII-catalysed reaction suggests that an active site carboxylate group participates directly in catalysis. J. Mol. Biol. 2007, 368, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Crary, S.M.; Niranjanakumari, S.; Fierke, C.A. The protein component of Bacillus subtilis Ribonuclease P increases catalytic efficiency by enhancing interactions with the 5′ leader sequence of pre-tRNAAsp. Biochemistry 1998, 37, 9409–9416. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Campbell, F.E.; Zahler, N.H.; Harris, M.E. Evidence that substrate-specific effects of C5 protein lead to uniformity in binding and catalysis by RNase P. EMBO J. 2006, 25, 3998–4007. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Cid, A.; Aguirre-Sampieri, S.; Diaz-Vilchis, A.; Torres-Larios, A. Ribonucleases P/MRP and the expanding ribonucleoprotein world. IUBMB Life 2012, 64, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Pomeranz Krummel, D.A.; Kent, O.; MacMillan, A.M.; Altman, S. Evidence for helical unwinding of an RNA substrate by the RNA enzyme RNase P: use of an interstrand disulfide crosslink in substrate. J. Mol. Biol. 2000, 295, 1113–1118. [Google Scholar] [CrossRef] [PubMed]
- Zahler, N.H.; Christian, E.L.; Harris, M.E. Recognition of the 5′ leader of pre-tRNA substrates by the active site of Ribonuclease P. RNA 2003, 9, 734–745. [Google Scholar] [CrossRef] [PubMed]
- Niranjanakumari, S.; Day-Storms, J.J.; Ahmed, M.; Hsieh, J.; Zahler, N.H.; Venters, R.A.; Fierke, C.A. Probing the architecture of the B. subtilis RNase P holoenzyme active site by crosslinking and affinity cleavage. RNA 2007, 13, 512–535. [Google Scholar]
- Buck, A.H.; Kazantsev, A.V.; Dalby, A.B.; Pace, N.R. Structural perspective on the activation of RNase P RNA by protein. Nat. Struct. Mol. Biol. 2005, 12, 958–964. [Google Scholar] [CrossRef] [PubMed]
- Niranjanakumari, S.; Stams, T.; Crary, S.M.; Christianson, D.W.; Fierke, C.A. Protein component of the ribozyme Ribonuclease P alters substrate recognition by directly contacting precursor tRNA. Proc. Natl. Acad. Sci. USA 1998, 95, 15212–15217. [Google Scholar] [CrossRef] [PubMed]
- Rueda, D.; Hsieh, J.; Day-Storms, J.J.; Fierke, C.A.; Walter, N.G. The 5′ leader of precursor tRNAAsp bound to the Bacillus subtilis RNase P holoenzyme has an extended conformation. Biochemistry 2005, 44, 16130–16139. [Google Scholar] [CrossRef] [PubMed]
- Loria, A.; Pan, T. Recognition of the 5′ leader and the acceptor stem of a pre-tRNA substrate by the ribozyme from Bacillus subtilis RNase P. Biochemistry 1998, 37, 10126–10133. [Google Scholar] [CrossRef] [PubMed]
- Loria, A.; Pan, T. The cleavage step of Ribonuclease P catalysis is determined by ribozyme-substrate interactions both distal and proximal to the cleavage site. Biochemistry 1999, 38, 8612–8620. [Google Scholar] [CrossRef] [PubMed]
- Brännvall, M.; Kikovska, E.; Wu, S.; Kirsebom, L.A. Evidence for induced fit in bacterial RNase P RNA-mediated cleavage. J. Mol. Biol. 2007, 372, 1149–1164. [Google Scholar] [CrossRef] [PubMed]
- Kazantsev, A.V.; Pace, N.R. Bacterial RNase P: a new view of an ancient enzyme. Nat. Rev. Microbiol. 2006, 4, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Kazantsev, A.V.; Rambo, R.P.; Karimpour, S.; Santalucia, J., Jr.; Tainer, J.A.; Pace, N.R. Solution structure of RNase P RNA. RNA 2011, 17, 1159–1171. [Google Scholar] [CrossRef] [PubMed]
- Misra, V.K.; Draper, D.E. On the role of magnesium ions in RNA stability. Biopolymers 1998, 48, 113–135. [Google Scholar] [CrossRef]
- Beebe, J.A.; Kurz, J.C.; Fierke, C.A. Magnesium ions are required by Bacillus subtilis Ribonuclease P RNA for both binding and cleaving precursor tRNAAsp. Biochemistry 1996, 35, 10493–10505. [Google Scholar] [CrossRef] [PubMed]
- Pan, T. Higher order folding and domain analysis of the ribozyme from Bacillus subtilis Ribonuclease P. Biochemistry 1995, 34, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.; Pace, N.R. Multiple magnesium ions in the Ribonuclease P reaction mechanism. Biochemistry 1993, 32, 5273–5281. [Google Scholar] [CrossRef] [PubMed]
- Christian, E.L.; Smith, K.M.; Perera, N.; Harris, M.E. The P4 metal binding site in RNase P RNA affects active site metal affinity through substrate positioning. RNA 2006, 12, 1463–1467. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.E.; Pace, N.R. Identification of phosphates involved in catalysis by the ribozyme RNase P RNA. RNA 1995, 1, 210–218. [Google Scholar] [PubMed]
- Warnecke, J.M.; Fürste, J.P.; Hardt, W.D.; Erdmann, V.A.; Hartmann, R.K. Ribonuclease P (RNase P) RNA is converted to a Cd2+-ribozyme by a single Rp-phosphorothioate modification in the precursor tRNA at the RNase P cleavage site. Proc. Natl. Acad. Sci. USA 1996, 93, 8924–8928. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, X.; Gegenheimer, P. Ribonuclease P catalysis requires Mg2+ coordinated to the pro-RP oxygen of the scissile bond. Biochemistry 1997, 36, 2425–2438. [Google Scholar] [PubMed]
- Kazantsev, A.V.; Pace, N.R. Identification by modification-interference of purine N-7 and ribose 2′-OH groups critical for catalysis by bacterial Ribonuclease P. RNA 1998, 4, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Warnecke, J.M.; Held, R.; Busch, S.; Hartmann, R.K. Role of metal ions in the hydrolysis reaction catalyzed by RNase P RNA from Bacillus subtilis. J. Mol. Biol. 1999, 290, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, T.; Tekos, A.; Warnecke, J.M.; Drainas, D.; Engelke, D.R.; Séraphin, B.; Hartmann, R.K. Effects of phosphorothioate modifications on precursor tRNA processing by eukaryotic RNase P enzymes. J. Mol. Biol. 2000, 298, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Christian, E.L.; Kaye, N.M.; Harris, M.E. Evidence for a polynuclear metal ion binding site in the catalytic domain of Ribonuclease P RNA. EMBO J. 2002, 21, 2253–2262. [Google Scholar] [CrossRef] [PubMed]
- Crary, S.M.; Kurz, J.C.; Fierke, C.A. Specific phosphorothioate substitutions probe the active site of Bacillus subtilis Ribonuclease P. RNA 2002, 8, 933–947. [Google Scholar] [CrossRef] [PubMed]
- Persson, T.; Cuzic, S.; Hartmann, R.K. Catalysis by RNase P RNA: unique features and unprecedented active site plasticity. J. Biol. Chem. 2003, 278, 43394–43401. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, Y.; Fierke, C.A.; Department of Chemistry, University of Michigan, Ann Arbor, MI, USA. Unpublished data. 2016.
- Cassano, A.G.; Anderson, V.E.; Harris, M.E. Analysis of solvent nucleophile isotope effects: evidence for concerted mechanisms and nucleophilic activation by metal coordination in nonenzymatic and ribozyme-catalyzed phosphodiester hydrolysis. Biochemistry 2004, 43, 10547–10559. [Google Scholar] [CrossRef] [PubMed]
- Cassano, A.G.; Anderson, V.E.; Harris, M.E. Understanding the transition states of phosphodiester bond cleavage: insights from heavy atom isotope effects. Biopolymers 2004, 73, 110–129. [Google Scholar] [CrossRef] [PubMed]
- Steitz, T.A.; Steitz, J.A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. USA 1993, 90, 6498–6502. [Google Scholar] [CrossRef] [PubMed]
- Brautigam, C.A.; Sun, S.; Piccirilli, J.A.; Steitz, T.A. Structures of normal single-stranded DNA and deoxyribo-3′-S-phosphorothiolates bound to the 3′-5′ exonucleolytic active site of DNA polymerase I from Escherichia coli. Biochemistry 1999, 38, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Sosunov, V.; Sosunova, E.; Mustaev, A.; Bass, I.; Nikiforov, V.; Goldfarb, A. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J. 2003, 22, 2234–2244. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Harris, M.E. Evidence that binding of C5 protein to P RNA enhances ribozyme catalysis by influencing active site metal ion affinity. RNA 2007, 13, 1505–1515. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, L.V.; Gößringer, M.; Weber, C.; Buzet, A.; Rossmanith, W.; Hartmann, R.K. tRNA processing by protein-only versus RNA-based RNase P: kinetic analysis reveals mechanistic differences. ChemBioChem 2012, 13, 2270–2276. [Google Scholar] [CrossRef] [PubMed]
- Thomas, B.C.; Li, X.; Gegenheimer, P. Chloroplast Ribonuclease P does not utilize the ribozyme-type pre-tRNA cleavage mechanism. RNA 2000, 6, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Westhof, E.; Dumas, P.; Moras, D. Restrained refinement of two crystalline forms of yeast aspartic acid and phenylalanine transfer RNA crystals. Acta Crystallogr. Sect. A: Found Adv. 1988, 44 (Pt 2), 112–123. [Google Scholar] [CrossRef]
- Orans, J.; McSweeney, E.A.; Iyer, R.R.; Hast, M.A.; Hellinga, H.W.; Modrich, P.; Beese, L.S. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell 2011, 145, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Brillante, N.; Gößringer, M.; Lindenhofer, D.; Toth, U.; Rossmanith, W.; Hartmann, R.K. Substrate recognition and cleavage-site selection by a single-subunit protein-only RNase P. Nucleic Acids Res. 2016, 44, 2323–2336. [Google Scholar] [CrossRef] [PubMed]
- Howard, M.J.; Karasik, A.; Klemm, B.P.; Mei, C.; Shanmuganathan, A.; Fierke, C.A.; Koutmos, M. Differential substrate recognition by isozymes of plant protein-only Ribonuclease P. RNA 2016, 22, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.; Walker, S.C.; Fierke, C.A.; Engelke, D.R. Pre-tRNA turnover catalyzed by the yeast nuclear RNase P holoenzyme is limited by product release. RNA 2009, 15, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Dirheimer, G.; Keith, G.; Dumas, P.; Westhof, E. Primary, Secondary, and Tertiary Structures of tRNAs. In tRNA: Structure, Biosynthesis, and Function, 1st ed.; Söll, D., RajBhandary, U.L., Eds.; The American Society for Microbiology Press: Washington, DC, USA, 1995; Chapter 8; pp. 93–126. [Google Scholar]
- Suzuki, T.; Nagao, A.; Suzuki, T. Human mitochondrial tRNAs: Biogenesis, function, structural aspects, and diseases. Annu. Revf. Genet. 2011, 45, 299–329. [Google Scholar] [CrossRef] [PubMed]
- Rossmanith, W. Of P and Z: Mitochondrial tRNA processing enzymes. Biochim. Biophys. Acta, Gene Regul. Mech. 2012, 1819, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Byrne, R.T.; Konevega, A.L.; Rodnina, M.V.; Antson, A.A. The crystal structure of unmodified tRNAPhe from Escherichia coli. Nucleic Acids Res. 2010, 38, 4154–4162. [Google Scholar] [CrossRef] [PubMed]
- Yandek, L.E.; Lin, H.C.; Harris, M.E. Alternative substrate kinetics of Escherichia coli Ribonuclease P: determination of relative rate constants by internal competition. J. Biol. Chem. 2013, 288, 8342–8354. [Google Scholar] [CrossRef] [PubMed]
- Tallsjo, A.; Kirsebom, L.A. Product release is a rate-limiting step during cleavage by the catalytic RNA subunit of Escherichia coli RNase P. Nucleic Acids Res. 1993, 21, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Oh, B.K.; Pace, N.R. Interaction of the 3′-end of tRNA with Ribonuclease P RNA. Nucleic Acids Res. 1994, 22, 4087–4094. [Google Scholar] [CrossRef] [PubMed]
- Christian, E.L.; Zahler, N.H.; Kaye, N.M.; Harris, M.E. Analysis of substrate recognition by the ribonucleoprotein endonuclease RNase P. Methods 2002, 28, 307–322. [Google Scholar] [CrossRef]
- Wegscheid, B.; Hartmann, R.K. The precursor tRNA 3’-CCA interaction with Escherichia coli RNase P RNA is essential for catalysis by RNase P in vivo. RNA 2006, 12, 2135–2148. [Google Scholar] [CrossRef] [PubMed]
- Wegscheid, B.; Hartmann, R.K. In vivo and in vitro investigation of bacterial type B RNase P interaction with tRNA 3’-CCA. Nucleic Acids Res. 2007, 35, 2060–2073. [Google Scholar] [CrossRef] [PubMed]
- Koutmou, K.S.; Zahler, N.H.; Kurz, J.C.; Campbell, F.E.; Harris, M.E.; Fierke, C.A. Protein–precursor tRNA contact leads to sequence-specific recognition of 5′ leaders by bacterial Ribonuclease P. J. Mol. Biol. 2010, 396, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Kirsebom, L.A.; Svard, S.G. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 1994, 13, 4870–4876. [Google Scholar] [PubMed]
- Oh, B.K.; Frank, D.N.; Pace, N.R. Participation of the 3′-CCA of tRNA in the binding of catalytic Mg2+ ions by Ribonuclease P. Biochemistry 1998, 37, 7277–7283. [Google Scholar] [CrossRef] [PubMed]
- Heide, C.; Pfeiffer, T.; Nolan, J.M.; Hartmann, R.K. Guanosine 2-NH2 groups of Escherichia coli RNase P RNA involved in intramolecular tertiary contacts and direct interactions with tRNA. RNA 1999, 5, 102–116. [Google Scholar] [CrossRef] [PubMed]
- Busch, S.; Kirsebom, L.A.; Notbohm, H.; Hartmann, R.K. Differential role of the intramolecular base-pairs G292-C75 and G293-C74 in the reaction catalyzed by Escherichia coli RNase P RNA. J. Mol. Biol. 2000, 299, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Heide, C.; Busch, S.; Feltens, R.; Hartmann, R.K. Distinct modes of mature and precursor tRNA binding to Escherichia coli RNase P RNA revealed by NAIM analyses. RNA 2001, 7, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, R.; Gößringer, M.; Späth, B.; Fischer, S.; Marchfelder, A. The making of tRNAs and more – RNase P and tRNase Z. Prog. Mol. Biol. Transl. Sci. 2009, 85, 319–368. [Google Scholar] [PubMed]
- Zahler, N.H.; Sun, L.; Christian, E.L.; Harris, M.E. The pre-tRNA nucleotide base and 2′-hydroxyl at N(-1) contribute to fidelity in tRNA processing by RNase P. J. Mol. Biol. 2005, 345, 969–985. [Google Scholar] [CrossRef] [PubMed]
- Guenther, U.-P.; Yandek, L.E.; Niland, C.N.; Campbell, F.E.; Anderson, D.; Anderson, V.E.; Harris, M.E.; Jankowsky, E. Hidden specificity in an apparently nonspecific RNA-binding protein. Nature 2013, 502, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Brännvall, M.; Mattsson, J.G.; Svard, S.G.; Kirsebom, L.A. RNase P RNA structure and cleavage reflect the primary structure of tRNA genes. J. Mol. Biol. 1998, 283, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Y.; Singh, D.; Lai, L.B.; Stiffler, M.A.; Lai, H.D.; Foster, M.P.; Gopalan, V. Fidelity of tRNA 5′-maturation: a possible basis for the functional dependence of archaeal and eukaryal RNase P on multiple protein cofactors. Nucleic Acids Res. 2012, 40, 4666–4680. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Altman, S. Substrate recognition by human RNase P: identification of small, model substrates for the enzyme. EMBO J. 1995, 14, 159–168. [Google Scholar] [PubMed]
- Marvin, M.C.; Clauder-Münster, S.; Walker, S.C.; Sarkeshik, A.; Yates III, J.R.; Steinmetz, L.M.; Engelke, D.R. Accumulation of noncoding RNA due to an RNase P defect in Saccharomyces cerevisiae. RNA 2011, 17, 1441–1450. [Google Scholar] [CrossRef] [PubMed]
- Marvin, M.C.; Walker, S.C.; Fierke, C.A.; Engelke, D.R. Binding and cleavage of unstructured RNA by nuclear RNase P. RNA 2011, 17, 1429–1440. [Google Scholar] [CrossRef] [PubMed]
- Maréchal-Drouard, L.; Weil, J.H.; Dietrich, A. Transfer RNAs and transfer RNA genes in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993, 44, 13–32. [Google Scholar] [CrossRef]
- Cognat, V.; Pawlak, G.; Duchêne, A.M.; Daujat, M.; Gigant, A.; Salinas, T.; Michaud, M.; Gutmann, B.; Giegé, P.; Gobert, A.; et al. PlantRNA, a database for tRNAs of photosynthetic eukaryotes. Nucleic Acids Res. 2013, 41, D273–D279. [Google Scholar] [CrossRef] [PubMed]
- Gobert, A.; Pinker, F.; Fuchsbauer, O.; Gutmann, B.; Boutin, R.; Roblin, P.; Sauter, C.; Giegé, P. Structural insights into protein-only RNase P complexed with tRNA. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A.; Rojas, M.; Fujii, S.; Yap, A.; Chong, Y.S.; Bond, C.S.; Small, I. A combinatorial amino acid code for rna recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012, 8, e1002910. [Google Scholar] [CrossRef] [PubMed]
- Yagi, Y.; Hayashi, S.; Kobayashi, K.; Hirayama, T.; Nakamura, T. Elucidation of the NA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS ONE 2013, 8, e57286. [Google Scholar] [CrossRef] [PubMed]
- Placido, A.; Sieber, F.; Gobert, A.; Gallerani, R.; Giegé, P.; Maréchal-Drouard, L. Plant mitochondria use two pathways for the biogenesis of tRNAHis. Nucleic Acids Res. 2010, 38, 7711–7717. [Google Scholar] [CrossRef] [PubMed]
- Rossmanith, W.; Tullo, A.; Potuschak, T.; Karwan, R.; Sbisà, E. Human mitochondrial tRNA processing. J. Biol. Chem. 1995, 270, 12885–12891. [Google Scholar] [PubMed]
- Rackham, O.; Shearwood, A.M.; Mercer, T.R.; Davies, S.M.; Mattick, J.S.; Filipovska, A. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 2011, 17, 2085–2093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.H.; Suddath, F.L.; Quigley, G.J.; McPherson, A.; Sussman, J.L.; Wang, A.H.; Seeman, N.C.; Rich, A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 1974, 185, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Moore, P.B. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA 2000, 6, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Giegé, R.; Jühling, F.; Pütz, J.; Stadler, P.; Sauter, C.; Florentz, C. Structure of transfer RNAs: Similarity and variability. Wiley Interdiscip. Rev. RNA 2012, 3, 37–61. [Google Scholar] [CrossRef] [PubMed]
- Voigts-Hoffmann, F.; Hengesbach, M.; Kobitski, A.Y.; van Aerschot, A.; Herdewijn, P.; Nienhaus, G.U.; Helm, M. A methyl group controls conformational equilibrium in human mitochondrial tRNALys. J. Am. Chem. Soc. 2007, 129, 13382–13383. [Google Scholar] [CrossRef] [PubMed]
- Dammertz, K.; Hengesbach, M.; Helm, M.; Nienhaus, G.U.; Kobitski, A.Y. Single-molecule FRET studies of counterion effects on the free energy landscape of human mitochondrial lysine tRNA. Biochemistry 2011, 50, 3107–3115. [Google Scholar] [CrossRef] [PubMed]
- Kobitski, A.Y.; Hengesbach, M.; Seidu-Larry, S.; Dammertz, K.; Chow, C.S.; van Aerschot, A.; Nienhaus, G.U.; Helm, M. Single-molecule FRET reveals a cooperative effect of two methyl group modifications in the folding of human mitochondrial tRNALys. Chem. Biol. 2011, 18, 928–936. [Google Scholar] [CrossRef] [PubMed]
- Ishitani, R.; Nureki, O.; Nameki, N.; Okada, N.; Nishimura, S.; Yokoyama, S. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell 2003, 113, 383–394. [Google Scholar] [CrossRef]
- Holzmann, J.; Rossmanith, W. tRNA recognition, processing, and disease: hypotheses around an unorthodox type of RNase P in human mitochondria. Mitochondrion 2009, 9, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Bothwell, A.L.; Garber, R.L.; Altman, S. Nucleotide sequence and in vitro processing of a precursor molecule to Escherichia coli 4.5 S RNA. J. Biol. Chem. 1976, 251, 7709–7716. [Google Scholar] [PubMed]
- Peck-Miller, K.A.; Altman, S. Kinetics of the processing of the precursor to 4.5 S RNA, a naturally occurring substrate for RNase P from Escherichia coli. J. Mol. Biol. 1991, 221, 1–5. [Google Scholar] [CrossRef]
- Komine, Y.; Kitabatake, M.; Yokogawa, T.; Nishikawa, K.; Inokuchi, H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl. Acad. Sci. USA 1994, 91, 9223–9227. [Google Scholar] [CrossRef] [PubMed]
- Alifano, P.; Rivellini, F.; Piscitelli, C.; Arraiano, C.M.; Bruni, C.B.; Carlomagno, M.S. Ribonuclease E provides substrates for Ribonuclease P-dependent processing of a polycistronic mRNA. Genes Dev. 1994, 8, 3021–3031. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Altman, S. A specific endoribonuclease, RNase P, affects gene expression of polycistronic operon mRNAs. Proc. Natl. Acad. Sci. USA 2003, 100, 13213–13218. [Google Scholar] [CrossRef] [PubMed]
- Altman, S.; Wesolowski, D.; Guerrier-Takada, C.; Li, Y. RNase P cleaves transient structures in some riboswitches. Proc. Natl. Acad. Sci. USA 2005, 102, 11284–11289. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Altman, S. Polarity effects in the lactose operon of Escherichia coli. J. Mol. Biol. 2004, 339, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.H.; Altman, S. OLE RNA, an RNA motif that is highly conserved in several extremophilic bacteria, is a substrate for and can be regulated by RNase P RNA. Proc. Natl. Acad. Sci. USA 2007, 104, 7815–7820. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, R.K.; Heinrich, J.; Schlegl, J.; Schuster, H. Precursor of c4 antisense rna of bacteriophages p1 and p7 is a substrate for rnase p of escherichia coli. Proc. Natl. Acad. Sci. USA 1995, 92, 5822–5826. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, J.R.; Pagan, R.; Kindelberger, D.W.; Engelke, D.R. An RNase P RNA subunit mutation affects ribosomal RNA processing. Nucleic Acids Res. 1996, 24, 3158–3166. [Google Scholar] [CrossRef] [PubMed]
- Gimple, O.; Schön, A. In vitro and in vivo processing of cyanelle tmRNA by RNase P. Biol. Chem. 2001, 382, 1421–1429. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Altman, S. A noncoding RNA in Saccharomyces cerevisiae is an RNase P substrate. RNA 2007, 13, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E.; Freier, S.M.; Spector, D.L. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 2008, 135, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Sunwoo, H.; Dinger, M.E.; Wilusz, J.E.; Amaral, P.P.; Mattick, J.S.; Spector, D.L. MEN ε/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009, 19, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Toledano, R.; Gómez, J. Messenger RNAs bearing tRNA-like features exemplified by interferon alfa 5 mRNA. Cell Mol. Life Sci. 2015, 72, 3747–3768. [Google Scholar] [CrossRef] [PubMed]
- Reiner, R.; Ben-Asouli, Y.; Krilovetzky, I.; Jarrous, N. A role for the catalytic ribonucleoprotein RNase P in RNA polymerase III transcription. Genes Dev. 2006, 20, 1621–1635. [Google Scholar] [CrossRef] [PubMed]
- Jarrous, N.; Reiner, R. Human RNase P: A tRNA-processing enzyme and transcription factor. Nucleic Acids Res. 2007, 35, 3519–3524. [Google Scholar] [CrossRef] [PubMed]
- Reiner, R.; Krasnov-Yoeli, N.; Dehtiar, Y.; Jarrous, N. Function and assembly of a chromatin-associated RNase P that is required for efficient transcription by RNA polymerase I. PLoS ONE 2008, 3, e4072. [Google Scholar] [CrossRef] [PubMed]
- Serruya, R.; Orlovetskie, N.; Reiner, R.; Dehtiar-Zilber, Y.; Wesolowski, D.; Altman, S.; Jarrous, N. Human RNase P ribonucleoprotein is required for formation of initiation complexes of RNA polymerase III. Nucleic Acids Res. 2015, 43, 5442–5450. [Google Scholar] [CrossRef] [PubMed]
- Newhart, A.; Powers, S.L.; Shastrula, P.K.; Sierra, I.; Joo, L.M.; Hayden, J.E.; Cohen, A.R.; Janicki, S.M. RNase P protein subunit, Rpp29, represses histone H3.3 nucleosome deposition. Mol. Biol. Cell 2016, 27, 1154–1169. [Google Scholar] [CrossRef] [PubMed]



| Bacteria | Archaea 1 | Eukarya | |||
|---|---|---|---|---|---|
| RNA-based | PRORP | ||||
| Yeast | Human | Human | Plant | ||
| RnpA | |||||
| Mth687p | POP5 | hPOP5 | |||
| Mth11p | POP4 | RPP29 | |||
| Mth688p | RPP1 | RPP30 | |||
| Mth1618p | RPR2 | RPP21 | |||
| L7Ae 2 | POP3 | RPP38 | |||
| POP1 | hPOP1 | ||||
| POP6 | RPP25 | ||||
| POP7 | RPP20 | ||||
| POP8 | |||||
| RPP14 | |||||
| RPP40 | |||||
| MRPP1 | |||||
| MRPP2 | |||||
| MRPP3 | PRORP1–3 | ||||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klemm, B.P.; Wu, N.; Chen, Y.; Liu, X.; Kaitany, K.J.; Howard, M.J.; Fierke, C.A. The Diversity of Ribonuclease P: Protein and RNA Catalysts with Analogous Biological Functions. Biomolecules 2016, 6, 27. https://doi.org/10.3390/biom6020027
Klemm BP, Wu N, Chen Y, Liu X, Kaitany KJ, Howard MJ, Fierke CA. The Diversity of Ribonuclease P: Protein and RNA Catalysts with Analogous Biological Functions. Biomolecules. 2016; 6(2):27. https://doi.org/10.3390/biom6020027
Chicago/Turabian StyleKlemm, Bradley P., Nancy Wu, Yu Chen, Xin Liu, Kipchumba J. Kaitany, Michael J. Howard, and Carol A. Fierke. 2016. "The Diversity of Ribonuclease P: Protein and RNA Catalysts with Analogous Biological Functions" Biomolecules 6, no. 2: 27. https://doi.org/10.3390/biom6020027