Sequence Analysis and Comparative Study of the Protein Subunits of Archaeal RNase P
Abstract
:1. Introduction
2. Results
2.1. Inventory of Archaeal RPPs
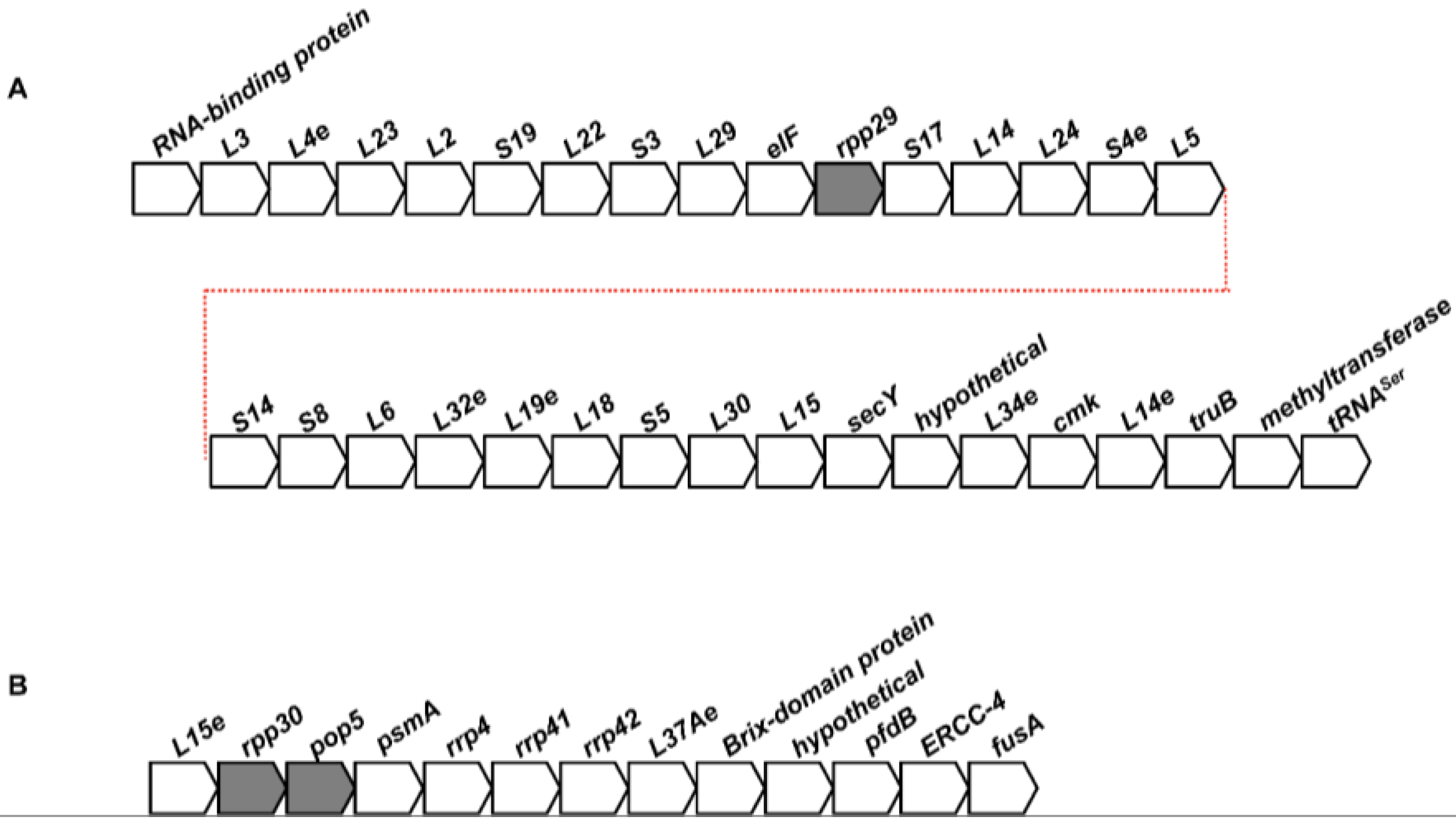
2.2. Conservation of the Genomic Neighborhoods of Archaeal RPPs
2.3. Structure-Function Analyses of Archaeal RPPs
2.3.1. RPP21
2.3.2. RPP29
2.3.3. RPP21•RPP29
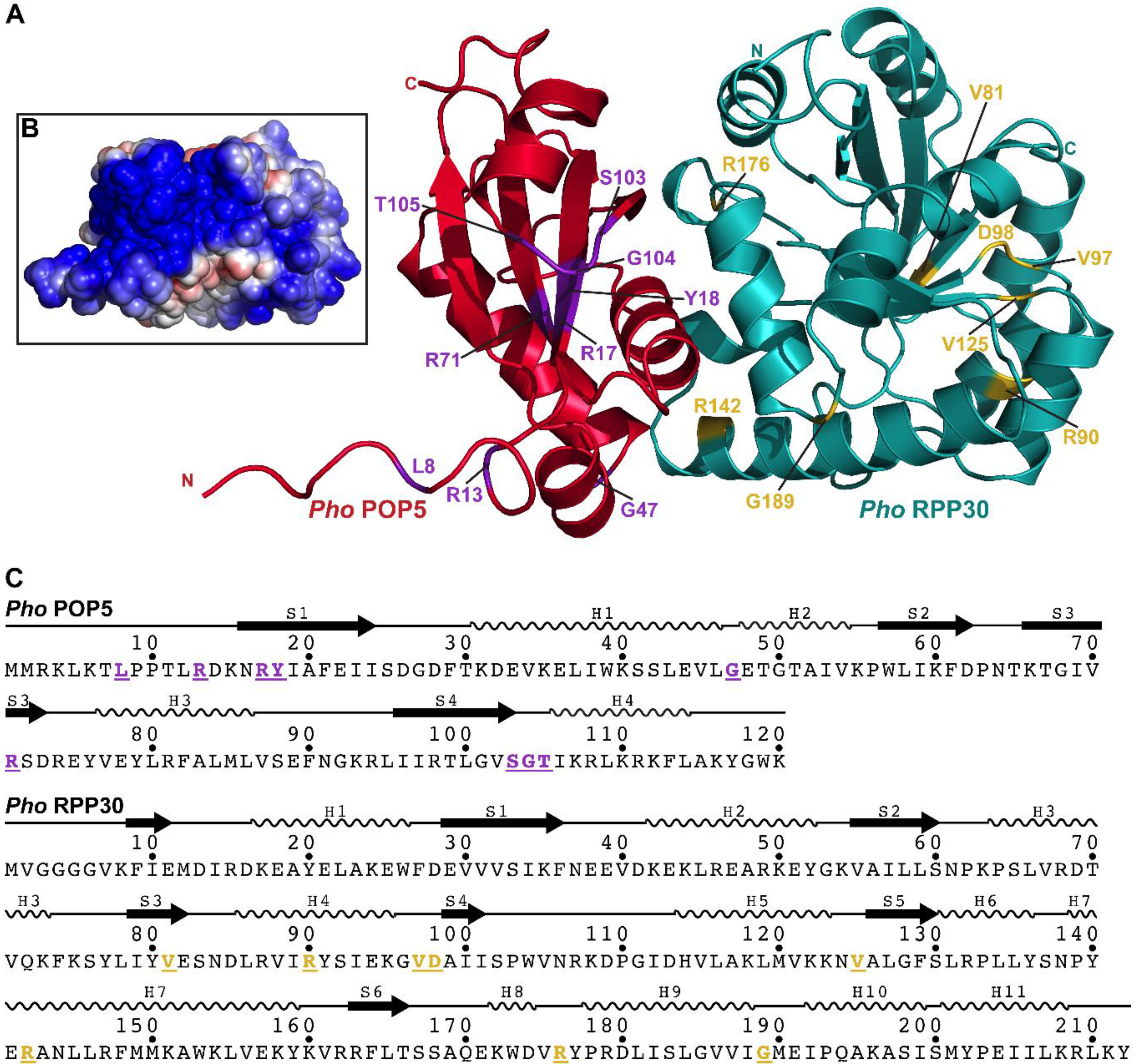
2.3.4. POP5
2.3.5. RPP30
2.3.6. POP5•RPP30
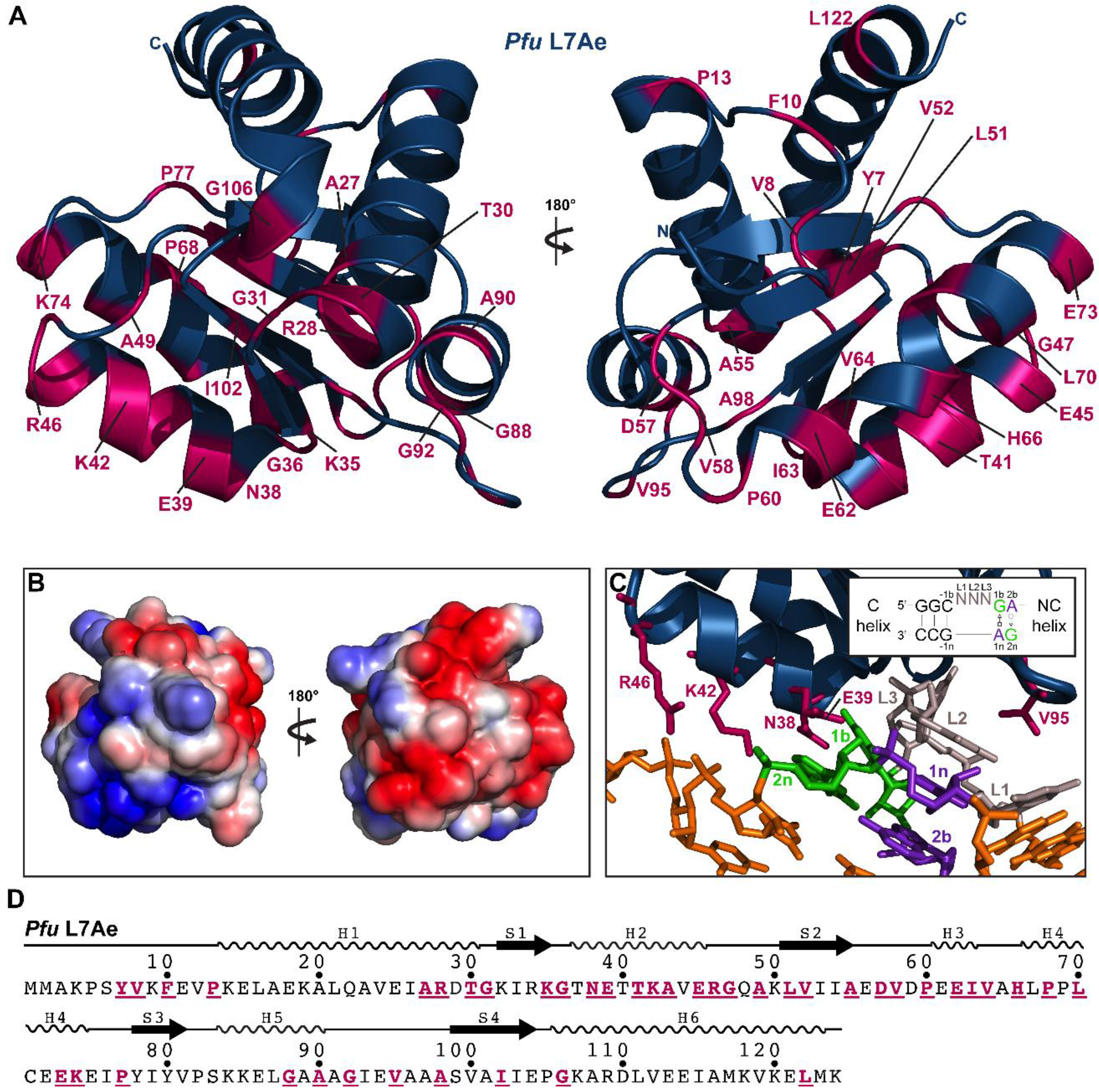
2.3.7. L7Ae
2.3.8. General Remarks
3. Discussion
3.1. Divergence of Bacterial and Archaeal RNase P
3.2. Coordinate Regulation of RNase P and Other Cellular Machineries
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| MEME | Multiple EM for Motif Elucidation |
| NCBI | National Center for Biotechnology Information |
| RPP | RNase P protein |
| RPR | RNase P RNA |
| pre-tRNA | precursor tRNA |
| tRNA | transfer RNA |
References
- Altman, S. A view of RNase P. Mol. Biosyst. 2007, 3, 604–607. [Google Scholar] [CrossRef] [PubMed]
- Esakova, O.; Krasilnikov, A.S. Of proteins and RNA: The RNase P/MRP family. RNA 2010, 16, 1725–1747. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.; Marquez, S.M.; Pace, N.R. RNase P: Interface of the RNA and protein worlds. Trends Biochem. Sci. 2006, 31, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.B.; Vioque, A.; Kirsebom, L.A.; Gopalan, V. Unexpected diversity of RNase P, an ancient tRNA processing enzyme: Challenges and prospects. FEBS Lett. 2010, 584, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Van Eenennaam, H.; Jarrous, N.; van Venrooij, W.J.; Pruijn, G.J. Architecture and function of the human endonucleases RNase P and RNase MRP. IUBMB Life 2000, 49, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.C.; Engelke, D.R. Ribonuclease P: The evolution of an ancient RNA enzyme. Crit Rev. Biochem. Mol. Biol. 2006, 41, 77–102. [Google Scholar] [CrossRef] [PubMed]
- Jarrous, N.; Gopalan, V. Archaeal/eukaryal RNase P: Subunits, functions and RNA diversification. Nucleic Acids Res. 2010, 38, 7885–7894. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.; Rossmanith, W.; Hartmann, R.K.; Tholken, C.; Gutmann, B.; Giege, P.; Gobert, A. Distribution of Ribonucleoprotein and Protein-Only RNase P in Eukarya. Mol. Biol. Evol. 2015, 32, 3186–3193. [Google Scholar] [CrossRef] [PubMed]
- Holzmann, J.; Frank, P.; Loffler, E.; Bennett, K.L.; Gerner, C.; Rossmanith, W. RNase P without RNA: Identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 2008, 135, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, J.R.; Lee, Y.; Lane, W.S.; Engelke, D.R. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998, 12, 1678–1690. [Google Scholar] [CrossRef] [PubMed]
- Gossringer, M.; Kretschmer-Kazemi Far, R.; Hartmann, R.K. Analysis of RNase P protein (rnpA) expression in Bacillus subtilis utilizing strains with suppressible rnpA expression. J. Bacteriol. 2006, 188, 6816–6823. [Google Scholar] [CrossRef] [PubMed]
- Schedl, P.; Primakoff, P. Mutants of Escherichia coli thermosensitive for the synthesis of transfer RNA. Proc. Natl. Acad. Sci. USA 1973, 70, 2091–2095. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, E.; Hartmann, R.K. The enigma of ribonuclease P evolution. Trends Genet. 2003, 19, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Brochier-Armanet, C.; Forterre, P.; Gribaldo, S. Phylogeny and evolution of the Archaea: One hundred genomes later. Curr. Opin. Microbiol. 2011, 14, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.B.; Cho, I.-M.; Chen, W.-Y.; Gopalan, V. Ribonuclease P.; Liu, F., Altman, S., Eds.; Springer Verlag: New York, NY, USA, 2010; pp. 153–172. [Google Scholar]
- Randau, L. Evolution of small guide RNA genes in hyperthermophilic archaea. Ann. N. Y. Acad. Sci. 2015, 1341, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. Origin of eukaryotes from within archaea, archaeal eukaryome and bursts of gene gain: Eukaryogenesis just made easier? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A.; Brown, J.W. The ribonuclease P family. Methods Enzymol. 2001, 341, 56–77. [Google Scholar] [PubMed]
- Hall, T.A.; Brown, J.W. Archaeal RNase P has multiple protein subunits homologous to eukaryotic nuclear RNase P proteins. RNA 2002, 8, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Amero, C.D.; Boomershine, W.P.; Xu, Y.; Foster, M. Solution structure of Pyrococcus furiosus RPP21, a component of the archaeal RNase P holoenzyme, and interactions with its RPP29 protein partner. Biochemistry 2008, 47, 11704–11710. [Google Scholar] [CrossRef] [PubMed]
- Boomershine, W.P.; McElroy, C.A.; Tsai, H.Y.; Wilson, R.C.; Gopalan, V.; Foster, M.P. Structure of Mth11/Mth Rpp29, an essential protein subunit of archaeal and eukaryotic RNase P. Proc. Natl. Acad. Sci. USA 2003, 100, 15398–15403. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Y.; Pulukkunat, D.K.; Cho, I.M.; Tsai, H.Y.; Gopalan, V. Dissecting functional cooperation among protein subunits in archaeal RNase P, a catalytic ribonucleoprotein complex. Nucleic Acids Res. 2010, 38, 8316–8327. [Google Scholar] [CrossRef] [PubMed]
- Kawano, S.; Nakashima, T.; Kakuta, Y.; Tanaka, I.; Kimura, M. Crystal structure of protein Ph1481p in complex with protein Ph1877p of archaeal RNase P from Pyrococcus horikoshii OT3: Implication of dimer formation of the holoenzyme. J. Mol. Biol. 2006, 357, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Kouzuma, Y.; Mizoguchi, M.; Takagi, H.; Fukuhara, H.; Tsukamoto, M.; Numata, T.; Kimura, M. Reconstitution of archaeal ribonuclease P from RNA and four protein components. Biochem. Biophys. Res. Commun. 2003, 306, 666–673. [Google Scholar] [CrossRef]
- Pulukkunat, D.K.; Gopalan, V. Studies on Methanocaldococcus jannaschii RNase P reveal insights into the roles of RNA and protein cofactors in RNase P catalysis. Nucleic Acids Res. 2008, 36, 4172–4180. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.Y.; Pulukkunat, D.K.; Woznick, W.K.; Gopalan, V. Functional reconstitution and characterization of Pyrococcus furiosus RNase P. Proc. Natl. Acad. Sci. USA 2006, 103, 16147–16152. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.C.; Bohlen, C.J.; Foster, M.P.; Bell, C.E. Structure of Pfu Pop5, an archaeal RNase P protein. Proc. Natl. Acad. Sci. USA 2006, 103, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Amero, C.D.; Pulukkunat, D.K.; Gopalan, V.; Foster, M.P. Solution structure of an archaeal RNase P binary protein complex: Formation of the 30-kDa complex between Pyrococcus furiosus RPP21 and RPP29 is accompanied by coupled protein folding and highlights critical features for protein-protein and protein-RNA interactions. J. Mol. Biol. 2009, 393, 1043–1055. [Google Scholar] [PubMed]
- Honda, T.; Kakuta, Y.; Kimura, K.; Saho, J.; Kimura, M. Structure of an archaeal homolog of the human protein complex Rpp21-Rpp29 that is a key core component for the assembly of active ribonuclease P. J. Mol. Biol. 2008, 384, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Numata, T.; Ishimatsu, I.; Kakuta, Y.; Tanaka, I.; Kimura, M. Crystal structure of archaeal ribonuclease P protein Ph1771p from Pyrococcus horikoshii OT3: An archaeal homolog of eukaryotic ribonuclease P protein Rpp29. RNA 2004, 10, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Sidote, D.J.; Heideker, J.; Hoffman, D.W. Crystal structure of archaeal ribonuclease P protein aRpp29 from Archaeoglobus fulgidus. Biochemistry 2004, 43, 14128–14138. [Google Scholar] [CrossRef] [PubMed]
- Sidote, D.J.; Hoffman, D.W. NMR structure of an archaeal homologue of ribonuclease P protein Rpp29. Biochemistry 2003, 42, 13541–13550. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Watanabe, M.; Kakuta, Y.; Kamachi, R.; Numata, T.; Tanaka, I.; Kimura, M. Crystal structure of the ribonuclease P protein Ph1877p from hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biochem. Biophys. Res. Commun. 2004, 319, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Kakuta, Y.; Ishimatsu, I.; Numata, T.; Kimura, K.; Yao, M.; Tanaka, I.; Kimura, M. Crystal structure of a ribonuclease P protein Ph1601p from Pyrococcus horikoshii OT3: An archaeal homologue of human nuclear ribonuclease P protein Rpp21. Biochemistry 2005, 44, 12086–12093. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.M.; Lai, L.B.; Susanti, D.; Mukhopadhyay, B.; Gopalan, V. Ribosomal protein L7Ae is a subunit of archaeal RNase P. Proc. Natl. Acad. Sci. USA 2010, 107, 14573–14578. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Y.; Xu, Y.; Cho, I.M.; Oruganti, S.V.; Foster, M.P.; Gopalan, V. Cooperative RNP Assembly: Complementary Rescue of Structural Defects by Protein and RNA Subunits of Archaeal RNase P. J. Mol. Biol. 2011, 411, 368–383. [Google Scholar] [CrossRef] [PubMed]
- Eder, P.S.; Kekuda, R.; Stolc, V.; Altman, S. Characterization of two scleroderma autoimmune antigens that copurify with human ribonuclease P. Proc. Natl. Acad. Sci. USA 1997, 94, 1101–1106. [Google Scholar] [CrossRef] [PubMed]
- Jarrous, N. Human ribonuclease P: Subunits, function, and intranuclear localization. RNA 2002, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Fukuhara, H.; Kifusa, M.; Watanabe, M.; Terada, A.; Honda, T.; Numata, T.; Kakuta, Y.; Kimura, M. A fifth protein subunit Ph1496p elevates the optimum temperature for the ribonuclease P activity from Pyrococcus horikoshii OT3. Biochem. Biophys. Res. Commun. 2006, 343, 956–964. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.M.; Lai, L.B.; Foster, M.P.; Gopalan, V. The L7Ae protein binds to two kink-turns in the Pyrococcus furiosus RNase P RNA. Nucleic Acids Res. 2014, 42, 13328–13338. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Dasgupta, I.; Fox, G.E. Many nonuniversal archaeal ribosomal proteins are found in conserved gene clusters. Archaea 2009, 2, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Gil, M.A.; Sherwood, K.E.; Maupin-Furlow, J.A. Transcriptional linkage of Haloferax volcanii proteasomal genes with non-proteasomal gene neighbours including RNase P, MOSC domain and SAM-methyltransferase homologues. Microbiology 2007, 153, 3009–3022. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Wolf, Y.I.; Aravind, L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 2001, 11, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Crowe, B.L.; Bohlen, C.J.; Wilson, R.C.; Gopalan, V.; Foster, M.P. Assembly of the complex between archaeal RNase P proteins RPP30 and Pop5. Archaea 2011. [Google Scholar] [CrossRef] [PubMed]
- Hamma, T.; Ferré-D'Amaré, A.R. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 Å resolution. Structure 2004, 12, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Lilley, D.M.J. The molecular recognition of kink-turn structure by the L7Ae class of proteins. RNA 2013, 19, 1703–1710. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.F.; Tran, E.J.; Maxwell, E.S. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5 kD/Snu13p snoRNP core protein. Nucleic Acids Res. 2002, 30, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Lilley, D.M. The K-turn motif in riboswitches and other RNA species. Biochim. Biophys. Acta 2014, 1839, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Moore, T.; Zhang, Y.; Fenley, M.O.; Li, H. Molecular basis of box C/D RNA-protein interactions: Cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure 2004, 12, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Moschen, T.; Wunderlich, C.; Kreutz, C.; Tollinger, M. NMR resonance assignments of the archaeal ribosomal protein L7Ae in the apo form and bound to a 25 nt RNA. Biomol. NMR Assign. 2015, 9, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Rozhdestvensky, T.S.; Tang, T.H.; Tchirkova, I.V.; Brosius, J.; Bachellerie, J.-P.; Hüttenhofer, A. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: A shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 2003, 31, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Szewczak, L.B.W.; DeGregorio, S.J.; Strobel, S.A.; Steitz, J.A. Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol. 2002, 9, 1095–1107. [Google Scholar] [CrossRef]
- Vidovic, I.; Nottrott, S.; Hartmuth, K.; Lührmann, R.; Ficner, R. Crystal structure of the spliceosomal 15.5 kD protein bound to a U4 snRNA fragment. Mol. Cell 2000, 6, 1331–1342. [Google Scholar] [CrossRef]
- Xue, S.; Wang, R.; Yang, F.; Terns, R.M.; Terns, M.P.; Zhang, X.; Maxwell, E.S.; Li, H. Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle. Mol. Cell 2010, 39, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Charron, C.; Manival, X.; Cléry, A.; Senty-Ségault, V.; Charpentier, B.; Marmier-Gourrier, N.; Branlant, C.; Aubry, A. The archaeal sRNA binding protein L7Ae has a 3D structure very similar to that of its eukaryal counterpart while having a broader RNA-binding specificity. J. Mol. Biol. 2004, 342, 757–773. [Google Scholar] [CrossRef] [PubMed]
- Kalavrizioti, D.; Vourekas, A.; Drainas, D. DRpp20 and DRpp40: Two protein subunits involved in Dictyostelium discoideum ribonuclease P holoenzyme assembly. Gene 2007, 400, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Stamatopoulou, V.; Toumpeki, C.; Tzakos, A.; Vourekas, A.; Drainas, D. Domain architecture of the DRpp29 protein and its interaction with the RNA subunit of Dictyostelium discoideum RNase P. Biochemistry 2010, 49, 10714–10727. [Google Scholar] [CrossRef] [PubMed]
- Vourekas, A.; Kalavrizioti, D.; Zarkadis, I.K.; Spyroulias, G.A.; Stathopoulos, C.; Drainas, D. A 40.7 kDa Rpp30/Rpp1 homologue is a protein subunit of Dictyostelium discoideum RNase P holoenzyme. Biochimie 2007, 89, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Elkan, C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995, 3, 21–29. [Google Scholar] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Jarrous, N.; Reiner, R.; Wesolowski, D.; Mann, H.; Guerrier-Takada, C.; Altman, S. Function and subnuclear distribution of Rpp21, a protein subunit of the human ribonucleoprotein ribonuclease P. RNA 2001, 7, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Niranjanakumari, S.; Stams, T.; Crary, S.M.; Christianson, D.W.; Fierke, C.A. Protein component of the ribozyme ribonuclease P alters substrate recognition by directly contacting precursor tRNA. Proc. Natl. Acad. Sci. USA 1998, 95, 15212–15217. [Google Scholar] [CrossRef] [PubMed]
- Reiter, N.J.; Osterman, A.; Torres-Larios, A.; Swinger, K.K.; Pan, T.; Mondragon, A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 2010, 468, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Gopalan, V. Uniformity amid diversity in RNase P. Proc. Natl. Acad. Sci. USA 2007, 104, 2031–2032. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.B.; Chan, P.P.; Cozen, A.E.; Bernick, D.L.; Brown, J.W.; Gopalan, V.; Lowe, T.M. Discovery of a minimal form of RNase P in Pyrobaculum. Proc. Natl. Acad. Sci. USA 2010, 107, 22493–22498. [Google Scholar] [CrossRef] [PubMed]
- Podar, M.; Makarova, K.S.; Graham, D.E.; Wolf, Y.I.; Koonin, E.V.; Reysenbach, A.L. Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park. Biol. Direct. 2013. [Google Scholar] [CrossRef] [PubMed]
- Coughlin, D.J.; Pleiss, J.A.; Walker, S.C.; Whitworth, G.B.; Engelke, D.R. Genome-wide search for yeast RNase P substrates reveals role in maturation of intron-encoded box C/D small nucleolar RNAs. Proc. Natl. Acad. Sci. USA 2008, 105, 12218–12223. [Google Scholar] [CrossRef] [PubMed]
- Samanta, M.P.; Tongprasit, W.; Sethi, H.; Chin, C.S.; Stolc, V. Global identification of noncoding RNAs in Saccharomyces cerevisiae by modulating an essential RNA processing pathway. Proc. Natl. Acad. Sci. USA 2006, 103, 4192–4197. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samanta, M.P.; Lai, S.M.; Daniels, C.J.; Gopalan, V. Sequence Analysis and Comparative Study of the Protein Subunits of Archaeal RNase P. Biomolecules 2016, 6, 22. https://doi.org/10.3390/biom6020022
Samanta MP, Lai SM, Daniels CJ, Gopalan V. Sequence Analysis and Comparative Study of the Protein Subunits of Archaeal RNase P. Biomolecules. 2016; 6(2):22. https://doi.org/10.3390/biom6020022
Chicago/Turabian StyleSamanta, Manoj P., Stella M. Lai, Charles J. Daniels, and Venkat Gopalan. 2016. "Sequence Analysis and Comparative Study of the Protein Subunits of Archaeal RNase P" Biomolecules 6, no. 2: 22. https://doi.org/10.3390/biom6020022





