Identification of Cellular Proteins that Interact with Human Cytomegalovirus Immediate-Early Protein 1 by Protein Array Assay
Abstract
:1. Introduction
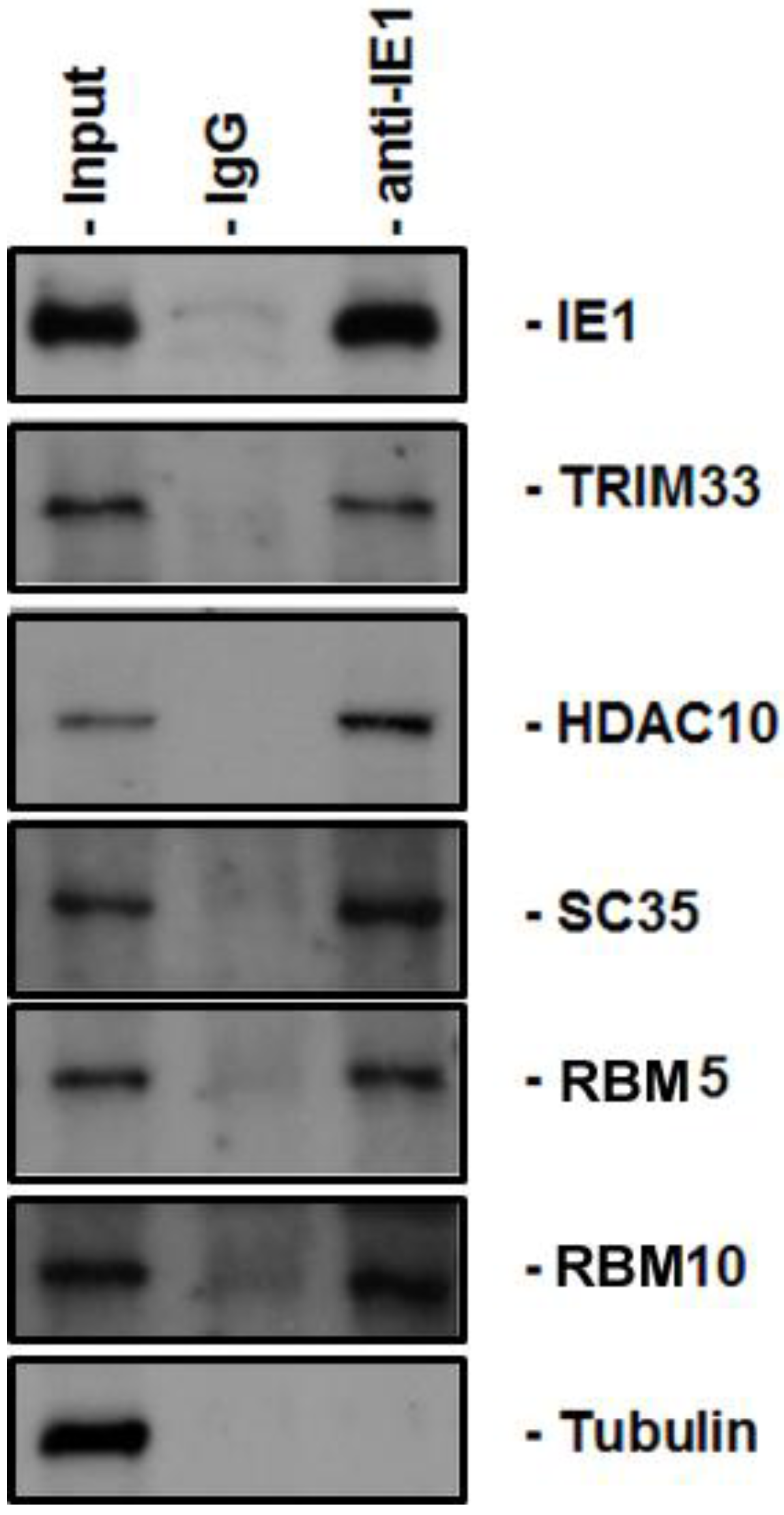
2. Results
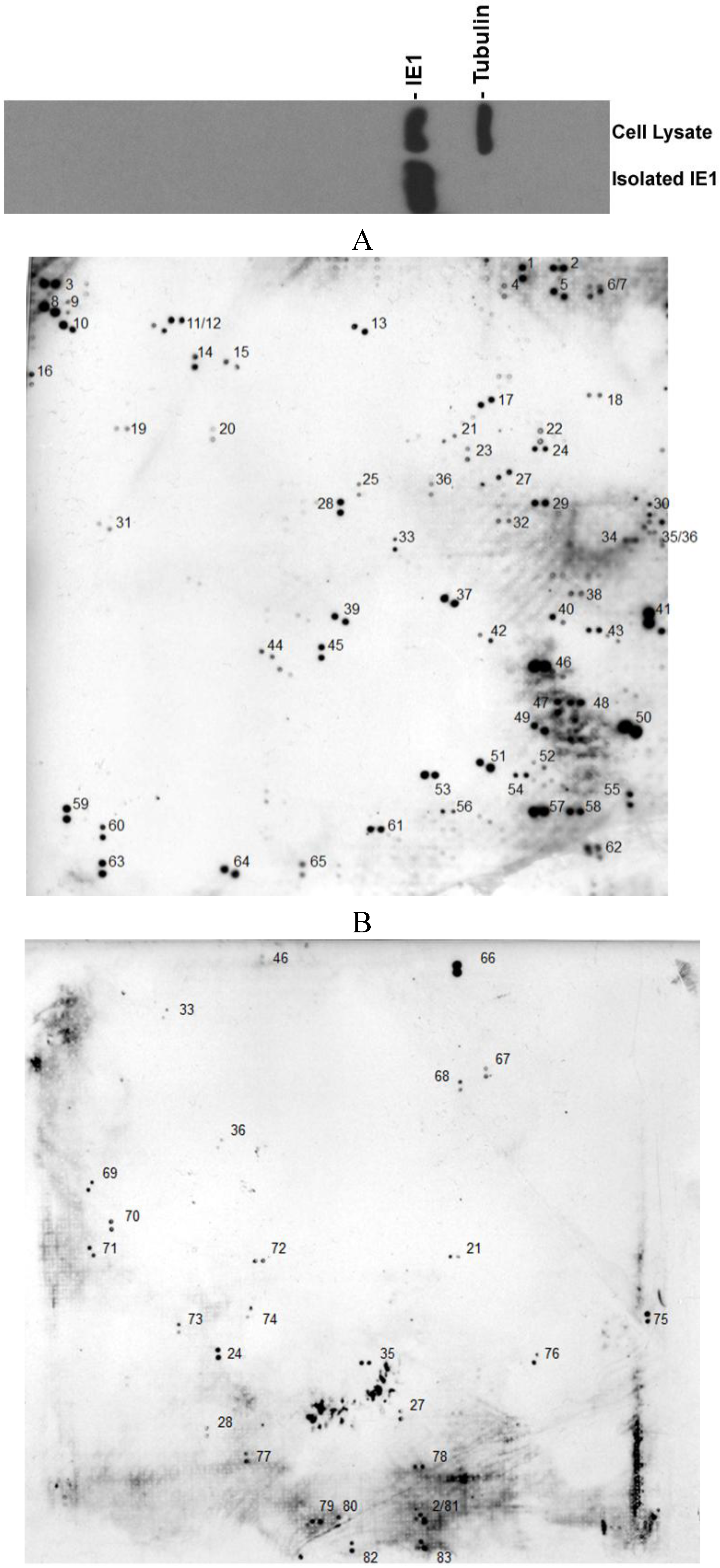
| GenBank entry# | Name of the protein | Function | Ref. |
|---|---|---|---|
| AF217982 | CDK5 regulatory subunit associated protein | Tumorigenesis and metastasis | [35] |
| D84294 | TPRDI (tetratricopeptide repeat) | Down syndrome | [36] |
| BC069268 | Golgi autoantigen, golgin subfamily a | Tentacular matrix | [37] |
| NM_007144 | Polycomb group ring Finger 2 (PCGF2) | Embryogenesis and tumorigenesis | [38] |
| NM_005676 | RNA binding motif protein 10 (RBM10) | RNA-related apoptosis | [39] |
| Z11584 | NuMA protein | Spindle orientation | [40] |
| BC022880 | Breast carcinoma amplified sequence 2 | Negative regulator of p53 | [41] |
| BC057387 | LSM14B, SCD6 homolog B | Regulation of translation | [42] |
| BG354577 | CDCA4 Cell division cycle associated gene 4 | Transcription regulation | [43] |
| NM_001789 | Cell division cycle 25 homolog A (CDC25A) | Cell cycle regulation | [44] |
| NM_003026 | SH3-domain GRB2-like 2 (SH3GL2) | Tumorigenesis | [45] |
| NM_203505 | GTPase activating protein binding protein 2 | Stress granule formation | [46] |
| XM_001128623 | Transcriptional regulator ATRX | Transcriptional repression | [47] |
| NM_153273 | Inositol hexaphosphate kinase 1 (IHPK1) | Type 2 diabetes | [48] |
| NM_032019 | Histone deacetylase 10 (HDAC10) | Cancer metastasis | [49] |
| NM_001040653 | ZXD family zinc finger C (ZXDC) | MHC gene transcription | [50] |
| NM_138383 | Actin-bundling protein with BAIAP2 homology (ABBA-1) | unknown | [51] |
| NM_001082486 | Adrenocortical dysplasia homolog (mouse) (ACD), transcript variant 1 | Tumorigenesis | [52] |
| AK122898 | ADP-ribosylation factor binding protein GGA1 | Trans-olgi network | [53] |
| AB208813 | RNA binding motif protein 5 | Gene splicing factor | [39] |
| NM_170677 | Meis homeobox 2 (MEIS2) | Unknown | [54] |
| NM_000038 | Adenomatosis polyposis coli (APC) | Tumor supresor | [55] |
| NM_003660 | Protein tyrosine phosphatase, receptor type f polypeptide (PTPRF), interacting protein (liprin), alpha 3 (PPFIA3) | Unknown | [56] |
| NM_012398 | Phosphatidylinositol-4-phosphate 5-kinase, type I, gamma (PIP5K1C) | Lethal contractural syndrome | [57] |
| NM_003861 | WD repeat domain 22 (WDR22) | Unknown | N/A |
| NM_017453 | Staufen, RNA binding protein | mRNA traffic | [58] |
| BC017222 | Sequestosome 1 | Signal transduction | [59] |
| NM_152586 | Ubiquitin specific peptidase 54 (USP54) | Unknown | [60] |
| NM_014868 | Ring finger protein 10 (RNF10) | Type 2 diabetes | [61] |
| BC041897 | SplA/ryanodine receptor domain and SOCS box containing 3 | Inflammation | [62] |
| AB209534 | Tumor rejection antigen (gp96) 1 | Unknown | N/A |
| NM_001080424 | Jumonji domain containing 3 (JMJD3) | Histone demethylation | [63] |
| NM_006312 | Nuclear receptor co-repressor 2 (NCOR2) | Gene suppressor | [64] |
| AK124656 | Gamma enolase (EC 4.2.1.11) | Neural tissue development | [65] |
| AK123065 | Sperm acrosomal protein | Motility of the spermatozoon | [66] |
| NM_001098800 | Melanoma antigen family D 4 (MAGED4) | Renal cell carcinoma | [67] |
| NM_021098 | Calcium channel, voltage-dependent, T type, alpha 1H subunit (CACNA1H) | T-type Ca(2+) channel activity | [68] |
| NM_001417 | Eukaryotic translation initiation factor 4B (EIF4B) | Translation control | [69] |
| NM_002973 | Ataxin 2 (ATXN2) | Spinocerebellar ataxia type 2 | [70] |
| BC070086 | Splicing factor, arginine/serine-rich 2 | Gene splicing | [18] |
| NM_001675 | Activating transcription factor 4 (ATF4) | Gene transcription | [71] |
| AB209149 | Phenol-sulfating phenol sulfotransferase 1 | Transfer of a sulfonate moiety | [72] |
| AY509035 | Roundabout-like protein 3 (ROBO3) | Horizontal gaze palsy | [73] |
| NM_014751 | Metastasis suppressor 1 (MTSS1) | Metastasis | [74] |
| AY335491 | GON4L isoform C (GON4L) | Hematopoiesis | [75] |
| NM_014739 | BCL2-associated transcription factor 1 | Transcriptional repression | [76] |
| AB067518 | KIAA1931 protein | Unknown | N/A |
| NM_003200 | Transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) (TCF3) | Transcription regulation | [77] |
| AB209197 | Protein phosphatase 1 (PP1) | Multiple functions | [78] |
| NM_020226 | PR domain containing 8 (PRDM8) | Gene repressor | [79] |
| XM_166659 | OTU domain containing 1 (OTUD1) | Unknown | N/A |
| NM_001098208 | Heterogeneous nuclear ribonucleoprotein F (HNRPF) | Gene splicing | [80] |
| AB209441 | Fibroblast growth factor receptor 3 isoform 1 precursor | Development | [81] |
| NM_152643 | Kinase non-catalytic C-lobe domain (KIND) containing 1 (KNDC1) | Unknown | N/A |
| NM_001982 | V-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (ERBB3) | Cell proliferation or differentiation | [82] |
| NM_015695 | Bromodomain and PHD finger containing, 3 (BRPF3) | Fetal liver erythropoiesis | [83] |
| NM_015906 | Tripartite motif-containing 33 (TRIM33) | Tumor suppressors | [84] |
| NM_032127 | Chromosome 11 open reading frame 56 (C11orf56) | FTS and Hook-interacting protein | [85] |
| NM_002904 | RD RNA binding protein (RDBP) | Repress RNA polymerase II | N/A |
| NM_012272 | PRP40 pre-mRNA processing factor 40 B (PRPF40B) | Gene splicing | [86] |
| NM_020967 | Nuclear receptor coactivator 5 (NCOA5) | Gene regulator | [87] |
| NM_001080495 | KIAA1856 protein (KIAA1856) | Unknown | N/A |
| NM_022748 | Tensin 3 (TNS3) | Signal transduction | [88] |
| BC063642 | Phosphodiesterase 4D interacting protein (myomegalin) | Control microtubules | [89] |
| AB209493 | Death-associated protein 6 (DAXX) | Development and Cancer | [90] |
| NM_003482 | Myeloid/lymphoid or mixed-lineage leukemia 2 (MLL2) | Lymphomagenesis | [91] |
| NM_033396 | Tankyrase 1 binding protein 1, (TNKS1BP1) | Unknown | [92] |
| AY729650 | Intersex-like protein | Unknown | [93] |
| BC110647 | Immediate-early response 2 | Unknown | N/A |
| AB014581 | KIAA0681 protein | Unknown | N/A |
| NM_004235 | Kruppel-like factor 4 (gut) (KLF4) | Transactivation and growth suppression | [94] |
| AB051455 | KIAA1668 protein | Unknown | N/A |
| AF045458 | Serine/threonine kinase ULK1 (ULK1) | Autophagy activation | [95] |
| NM_001003694 | Bromodomain and PHD finger containing, 1 (BRPF1) | Unknown | N/A |
| NM_003626 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 (PPFIA1) | Unknown | N/A |
| NM_006291 | tumor necrosis factor, alpha-induced protein 2 (TNFAIP2) | Unknown | N/A |
| AB209643 | smoothelin isoform b | Unknown | N/A |
| NM_002857 | peroxisomal biogenesis factor 19 (PEX19) | Peroxisomal assembly | [96] |
| AB061669 | receptor for advanced glycation end-products | Signaling and inflammation | [97] |
| NM_014678 | SAPS domain family, member 2 (SAPS2) | Unknown | N/A |
| NM_006887 | zinc finger protein 36, C3H type-like 2 (ZFP36L2) | Unknown | N/A |
| AB208876 | axin 1 isoform | Wnt signaling pathway | [98] |
| NM_004530 | matrix metallopeptidase 2 (MMP2) | Metastasis and inflammation | [99] |
3. Discussion
4. Experimental
4.1. Tissue Culture and Viruses
4.2. Antibodies
4.3. Preparation of Nuclear Extracts
4.4. Coimmunoprecipitation
4.5. Immunoblot Analysis
5. Conclusions
Acknowledgements
Conflicts of Interest
References and Notes
- Sweet, C. The pathogenicity of cytomegalovirus. FEMS Microbiol. Rev. 1999, 23, 457–482. [Google Scholar] [CrossRef]
- Landolfo, S.; Gariglio, M.; Gribaudo, G.; Lembo, D. The human cytomegalovirus. Pharmacol. Ther. 2003, 98, 269–297. [Google Scholar] [CrossRef]
- Mocarski, E.S., Jr.; Shenk, T.; Pass, R.F. Cytomegaloviruses, 5th ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2006. [Google Scholar]
- Tang, Q.; Li, L.; Maul, G.G. Mouse cytomegalovirus early M112/113 proteins control the repressive effect of IE3 on the major immediate-early promoter. J. Virol. 2005, 79, 257–263. [Google Scholar] [CrossRef]
- Tang, Q.; Maul, G. Immediate early interactions and epigenetic defense mechanisms. In In Cytomegaloviruses: Molecular Biology and Immunology; Reddehase, M.J., Ed.; Hethersett, Horizon Scientific Press: Norwich, UK, 2005. [Google Scholar]
- Hagemeier, C.; Walker, S.M.; Sissons, P.J.; Sinclair, J.H. The 72K IE1 and 80K IE2 proteins of human cytomegalovirus independently trans-activate the c-Fos, c-Myc and hsp70 promoters via basal promoter elements. J. Gen. Virol. 1992, 73, 2385–2393. [Google Scholar] [CrossRef]
- Liu, B.; Hermiston, T.W.; Stinski, M.F. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J. Virol. 1991, 65, 897–903. [Google Scholar]
- Scully, A.L.; Sommer, M.H.; Schwartz, R.; Spector, D.H. The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J. Virol. 1995, 69, 6533–6540. [Google Scholar]
- Awasthi, S.; Isler, J.A.; Alwine, J.C. Analysis of splice variants of the immediate-early 1 region of human cytomegalovirus. J. Virol. 2004, 78, 8191–8200. [Google Scholar] [CrossRef]
- Sadanari, H.; Yamada, R.; Yamagoshi, T.; Ohnishi, K.; Matsubara, K.; Fukuda, S.; Tanaka, J. The major immediate-early genes of human cytomegalovirus induce two novel proteins with molecular weights of 91 and 102 kilodaltons. Arch. Virol. 2000, 145, 1257–1266. [Google Scholar] [CrossRef]
- Ahn, J.H.; Hayward, G.S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J. Virol. 1997, 71, 4599–4613. [Google Scholar]
- Meier, J.L.; Stinski, M.F. Effect of a modulator deletion on transcription of the human cytomegalovirus major immediate-early genes in infected undifferentiated and differentiated cells. J. Virol. 1997, 71, 1246–1255. [Google Scholar]
- Stenberg, R.M. The human cytomegalovirus major immediate-early gene. Intervirology 1996, 39, 343–349. [Google Scholar]
- Stenberg, R.M.; Thomsen, D.R.; Stinski, M.F. Structural analysis of the major immediate early gene of human cytomegalovirus. J. Virol. 1984, 49, 190–199. [Google Scholar]
- Tang, Q.; Maul, G.G. Mouse cytomegalovirus immediate-early protein 1 binds with host cell repressors to relieve suppressive effects on viral transcription and replication during lytic infection. J. Virol. 2003, 77, 1357–1367. [Google Scholar] [CrossRef]
- Lee, H.R.; Kim, D.J.; Lee, J.M.; Choi, C.Y.; Ahn, B.Y.; Hayward, G.S.; Ahn, J.H. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol. 2004, 78, 6527–6542. [Google Scholar]
- Nevels, M.; Paulus, C.; Shenk, T. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc. Natl. Acad. Sci. USA 2004, 101, 17234–17239. [Google Scholar] [CrossRef]
- Ahn, J.H.; Hayward, G.S. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology 2000, 274, 39–55. [Google Scholar] [CrossRef]
- Ishov, A.M.; Stenberg, R.M.; Maul, G.G. Human cytomegalovirus immediate early interaction with host nuclear structures: Definition of an immediate transcript environment. J. Cell Biol. 1997, 138, 5–16. [Google Scholar] [CrossRef]
- Lee, H.R.; Huh, Y.H.; Kim, Y.E.; Lee, K.; Kim, S.; Ahn, J.H. N-Terminal determinants of human cytomegalovirus IE1 protein in nuclear targeting and disrupting PML-associated subnuclear structures. Biochem. Biophys. Res. Commun. 2007, 356, 499–504. [Google Scholar] [CrossRef]
- Maul, G.G.; Negorev, D.; Bell, P.; Ishov, A.M. Review: Properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 2000, 129, 278–287. [Google Scholar] [CrossRef]
- Paulus, C.; Krauss, S.; Nevels, M. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 3840–3845. [Google Scholar] [CrossRef]
- Huh, Y.H.; Kim, Y.E.; Kim, E.T.; Park, J.J.; Song, M.J.; Zhu, H.; Hayward, G.S.; Ahn, J.H. Binding STAT2 by the acidic domain of human cytomegalovirus IE1 promotes viral growth and is negatively regulated by SUMO. J. Virol. 2008, 82, 10444–10454. [Google Scholar] [CrossRef]
- Krauss, S.; Kaps, J.; Czech, N.; Paulus, C.; Nevels, M. Physical requirements and functional consequences of complex formation between the cytomegalovirus IE1 protein and human STAT2. J. Virol. 2009, 83, 12854–12870. [Google Scholar] [CrossRef]
- Knoblach, T.; Grandel, B.; Seiler, J.; Nevels, M.; Paulus, C. Human cytomegalovirus IE1 protein elicits a type II interferon-like host cell response that depends on activated STAT1 but not interferon-gamma. PLoS Pathog. 2011, 7, e1002016. [Google Scholar] [CrossRef]
- Zhang, Z.; Huong, S.M.; Wang, X.; Huang, D.Y.; Huang, E.S. Interactions between human cytomegalovirus IE1-72 and cellular p107: Functional domains and mechanisms of up-regulation of cyclin E/cdk2 kinase activity. J. Virol. 2003, 77, 12660–12670. [Google Scholar] [CrossRef]
- Castillo, J.P.; Frame, F.M.; Rogoff, H.A.; Pickering, M.T.; Yurochko, A.D.; Kowalik, T.F. Human cytomegalovirus IE1-72 activates ataxia telangiectasia mutated kinase and a p53/p21-mediated growth arrest response. J. Virol. 2005, 79, 11467–11475. [Google Scholar] [CrossRef]
- Hu, S.; Xie, Z.; Onishi, A.; Yu, X.; Jiang, L.; Lin, J.; Rho, H.S.; Woodard, C.; Wang, H.; Jeong, J.S.; et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell 2009, 139, 610–622. [Google Scholar] [CrossRef]
- Tang, Q.; Maul, G.G. Mouse cytomegalovirus crosses the species barrier with help from a few human cytomegalovirus proteins. J. Virol. 2006, 80, 7510–7521. [Google Scholar] [CrossRef]
- Reeves, M.; Woodhall, D.; Compton, T.; Sinclair, J. Human cytomegalovirus IE72 protein interacts with the transcriptional repressor hDaxx to regulate LUNA gene expression during lytic infection. J. Virol. 2010, 84, 7185–7194. [Google Scholar] [CrossRef]
- Gaspar, M.; Shenk, T. Human cytomegalovirus inhibits a DNA damage response by mislocalizing checkpoint proteins. Proc. Natl. Acad. Sci. USA 2006, 103, 2821–2826. [Google Scholar] [CrossRef]
- Xuan, B.; Qian, Z.; Torigoi, E.; Yu, D. Human cytomegalovirus protein pUL38 induces ATF4 expression, inhibits persistent JNK phosphorylation, and suppresses endoplasmic reticulum stress-induced cell death. J. Virol. 2009, 83, 3463–3474. [Google Scholar] [CrossRef]
- Andoniou, C.E.; Andrews, D.M.; Manzur, M.; Ricciardi-Castagnoli, P.; Degli-Esposti, M.A. A novelcheckpoint in the Bcl-2-regulated apoptotic pathway revealed by murine cytomegalovirus infection of dendritic cells. J. Cell Biol. 2004, 166, 827–837. [Google Scholar] [CrossRef]
- Tower, C.; Fu, L.; Gill, R.; Prichard, M.; Lesort, M.; Sztul, E. Human cytomegalovirus UL97 kinase prevents the deposition of mutant protein aggregates in cellular models of Huntington’s disease and ataxia. Neurobiol. Dis. 2011, 41, 11–22. [Google Scholar] [CrossRef]
- Mak, G.W.; Lai, W.L.; Zhou, Y.; Li, M.; Ng, I.O.; Ching, Y.P. CDK5RAP3 is a novel repressor of p14ARF in hepatocellular carcinoma cells. PLoS One 2012, 7, e42210. [Google Scholar]
- Tsukahara, F.; Hattori, M.; Muraki, T.; Sakaki, Y. Identification and cloning of a novel cDNA belonging to tetratricopeptide repeat gene family from Down syndrome-critical region 21q22.2. J. Biochem. 1996, 120, 820–827. [Google Scholar] [CrossRef]
- Munro, S. The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef]
- Hennig, L.; Derkacheva, M. Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet. 2009, 25, 414–423. [Google Scholar] [CrossRef]
- Sutherland, L.C.; Rintala-Maki, N.D.; White, R.D.; Morin, C.D. RNA binding motif (RBM) proteins: A novel family of apoptosis modulators? J. Cell Biochem. 2005, 94, 5–24. [Google Scholar] [CrossRef]
- Seldin, L.; Poulson, N.D.; Foote, H.P.; Lechler, T. NuMA localization, stability and function in spindle orientation involves 4.1 and Cdk1 interactions. Mol. Biol. Cell 2013, 24, 3651–3662. [Google Scholar] [CrossRef]
- Kuo, P.C.; Tsao, Y.P.; Chang, H.W.; Chen, P.H.; Huang, C.W.; Lin, S.T.; Weng, Y.T.; Tsai, T.C.; Shieh, S.Y.; Chen, S.L. Breast cancer amplified sequence 2, a novel negative regulator of the p53 tumor suppressor. Cancer Res. 2009, 69, 8877–8885. [Google Scholar] [CrossRef]
- Huang, Y.C.; Schmitt, M.; Yang, Z.; Que, L.G.; Stewart, J.C.; Frampton, M.W.; Devlin, R.B. Gene expression profile in circulating mononuclear cells after exposure to ultrafine carbon particles. Inhal. Toxicol. 2010, 22, 835–846. [Google Scholar] [CrossRef]
- Tategu, M.; Nakagawa, H.; Hayashi, R.; Yoshida, K. Transcriptional co-factor CDCA4 participates in the regulation of JUN oncogene expression. Biochimie 2008, 90, 1515–1522. [Google Scholar] [CrossRef]
- Shen, T.; Huang, S. The role of Cdc25A in the regulation of cell proliferation and apoptosis. Anticancer Agents Med. Chem. 2012, 12, 631–639. [Google Scholar] [CrossRef]
- Trub, T.; Frantz, J.D.; Miyazaki, M.; Band, H.; Shoelson, S.E. The role of a lymphoid-restricted, Grb2-like SH3-SH2-SH3 protein in T cell receptor signaling. J. Biol. Chem. 1997, 272, 894–902. [Google Scholar] [CrossRef]
- Matsuki, H.; Takahashi, M.; Higuchi, M.; Makokha, G.N.; Oie, M.; Fujii, M. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells 2013, 18, 135–146. [Google Scholar] [CrossRef]
- Salomoni, P.; Khelifi, A.F. DAXX: Death or survival protein? Trends Cell Biol. 2006, 16, 97–104. [Google Scholar] [CrossRef]
- Kamimura, J.; Wakui, K.; Kadowaki, H.; Watanabe, Y.; Miyake, K.; Harada, N.; Sakamoto, M.; Kinoshita, A.; Yoshiura, K.; Ohta, T.; et al. The IHPK1 gene is disrupted at the 3p21.31 breakpoint of t(3;9) in a family with type 2 diabetes mellitus. J. Hum. Genet. 2004, 49, 360–365. [Google Scholar]
- Song, C.; Zhu, S.; Wu, C.; Kang, J. Histone Deacetylase (HDAC) 10 suppresses cervical cancer metastasis through inhibition of matrix metalloproteinase (MMP) 2 and 9 Expression. J. Biol. Chem. 2013, 288, 28021–28033. [Google Scholar] [CrossRef]
- Al-Kandari, W.; Jambunathan, S.; Navalgund, V.; Koneni, R.; Freer, M.; Parimi, N.; Mudhasani, R.; Fontes, J.D. ZXDC, a novel zinc finger protein that binds CIITA and activates MHC gene transcription. Mol. Immunol. 2007, 44, 311–321. [Google Scholar] [CrossRef]
- Machesky, L.M.; Johnston, S.A. MIM: A multifunctional scaffold protein. J. Mol. Med. 2007, 85, 569–576. [Google Scholar] [CrossRef]
- Else, T.; Trovato, A.; Kim, A.C.; Wu, Y.; Ferguson, D.O.; Kuick, R.D.; Lucas, P.C.; Hammer, G.D. Genetic p53 deficiency partially rescues the adrenocortical dysplasia phenotype at the expense of increased tumorigenesis. Cancer Cell 2009, 15, 465–476. [Google Scholar] [CrossRef]
- Boman, A.L.; Zhang, C.; Zhu, X.; Kahn, R.A. A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol. Biol. Cell 2000, 11, 1241–1255. [Google Scholar] [CrossRef]
- Nakamura, T.; Jenkins, N.A.; Copeland, N.G. Identification of a new family of Pbx-related homeobox genes. Oncogene 1996, 13, 2235–2242. [Google Scholar]
- Wong, M.H.; Hermiston, M.L.; Syder, A.J.; Gordon, J.I. Forced expression of the tumor suppressor adenomatosis polyposis coli protein induces disordered cell migration in the intestinal epithelium. Proc. Natl. Acad. Sci. USA 1996, 93, 9588–9593. [Google Scholar] [CrossRef]
- Katoh, M. Identification and characterization of mouse Ppfia1 gene in silico. Int. J. Mol. Med. 2003, 12, 263–267. [Google Scholar]
- Narkis, G.; Ofir, R.; Landau, D.; Manor, E.; Volokita, M.; Hershkowitz, R.; Elbedour, K.; Birk, O.S. Lethal contractural syndrome type 3 (LCCS3) is caused by a mutation in PIP5K1C, which encodes PIPKI gamma of the phophatidylinsitol pathway. Am. J. Hum. Genet. 2007, 81, 530–539. [Google Scholar] [CrossRef]
- Miki, T.; Takano, K.; Yoneda, Y. The role of mammalian Staufen on mRNA traffic: A view from its nucleocytoplasmic shuttling function. Cell Struct. Funct. 2005, 30, 51–56. [Google Scholar] [CrossRef]
- Geetha, T.; Vishwaprakash, N.; Sycheva, M.; Babu, J.R. Sequestosome 1/p62: Across diseases. Biomarkers 2012, 17, 99–103. [Google Scholar] [CrossRef]
- Rigden, D.J.; Liu, H.; Hayes, S.D.; Urbe, S.; Clague, M.J. Ab initio protein modelling reveals novel human MIT domains. FEBS Lett. 2009, 583, 872–878. [Google Scholar] [CrossRef]
- Huang, K.; Nair, A.K.; Muller, Y.L.; Piaggi, P.; Bian, L.; del Rosario, M.; Knowler, W.C.; Kobes, S.; Hanson, R.L.; Bogardus, C.; et al. Whole exome sequencing identifies variation in CYB5A and RNF10 associated with adiposity and type 2 diabetes. Obesity 2013. [Google Scholar] [CrossRef]
- Kuang, Z.; Yao, S.; Xu, Y.; Lewis, R.S.; Low, A.; Masters, S.L.; Willson, T.A.; Kolesnik, T.B.; Nicholson, S.E.; Garrett, T.J.; et al. SPRY domain-containing SOCS box protein 2: Crystal structure and residues critical for protein binding. J. Mol. Biol. 2009, 386, 662–674. [Google Scholar] [CrossRef]
- Svotelis, A.; Bianco, S.; Madore, J.; Huppe, G.; Nordell-Markovits, A.; Mes-Masson, A.M.; Gevry, N. H3K27 demethylation by JMJD3 at a poised enhancer of anti-apoptotic gene BCL2 determines ERalpha ligand dependency. EMBO J. 2011, 30, 3947–3961. [Google Scholar] [CrossRef]
- Privalsky, M.L. Regulation of SMRT and N-CoR corepressor function. Curr. Top. Microbiol. Immunol. 2001, 254, 117–136. [Google Scholar]
- Marangos, P.J.; Schmechel, D. The neurobiology of the brain enolases. Essays Neurochem. Neuropharmacol. 1980, 4, 211–247. [Google Scholar]
- Topfer-Petersen, E.; Cechova, D.; Henschen, A.; Steinberger, M.; Friess, A.E.; Zucker, A. Cell biology of acrosomal proteins. Andrologia 1990, 22, 110–121. [Google Scholar]
- Kramer, B.F.; Schoor, O.; Kruger, T.; Reichle, C.; Muller, M.; Weinschenk, T.; Hennenlotter, J.; Stenzl, A.; Rammensee, H.G.; Stevanovic, S. MAGED4-expression in renal cell carcinoma and identification of an HLA-A*25-restricted MHC class I ligand from solid tumor tissue. Cancer Biol. Ther. 2005, 4, 943–948. [Google Scholar] [CrossRef]
- Kuwahara, K.; Takano, M.; Nakao, K. Pathophysiological significance of T-type Ca2+ channels: Transcriptional regulation of T-type Ca2+ channel—Regulation of CACNA1H by neuron-restrictive silencer factor. J. Pharmacol. Sci. 2005, 99, 211–213. [Google Scholar] [CrossRef]
- Shahbazian, D.; Parsyan, A.; Petroulakis, E.; Hershey, J.; Sonenberg, N. eIF4B controls survival and proliferation and is regulated by proto-oncogenic signaling pathways. Cell Cycle 2010, 9, 4106–4109. [Google Scholar] [CrossRef]
- Bonini, N.M.; Gitler, A.D. Model organisms reveal insight into human neurodegenerative disease:Ataxin-2 intermediate-length polyglutamine expansions are a risk factor for ALS. J. Mol. Neurosci. 2011, 45, 676–683. [Google Scholar] [CrossRef]
- Rutkowski, D.T.; Kaufman, R.J. All roads lead to ATF4. Dev. Cell 2003, 4, 442–444. [Google Scholar] [CrossRef]
- Bernier, F.; Soucy, P.; Luu-The, V. Human phenol sulfotransferase gene contains two alternative promoters: Structure and expression of the gene. DNA Cell Biol. 1996, 15, 367–375. [Google Scholar] [CrossRef]
- Bosley, T.M.; Salih, M.A.; Jen, J.C.; Lin, D.D.; Oystreck, D.; Abu-Amero, K.K.; MacDonald, D.B.; al Zayed, Z.; al Dhalaan, H.; Kansu, T.; et al. Neurologic features of horizontal gaze palsy and progressive scoliosis with mutations in ROBO3. Neurology 2005, 64, 1196–1203. [Google Scholar]
- Du, P.; Ye, L.; Li, H.; Yang, Y.; Jiang, W.G. The tumour suppressive role of metastasis suppressor-1, MTSS1, in human kidney cancer, a possible connection with the SHH pathway. J. Exp. Ther. Oncol. 2012, 10, 91–99. [Google Scholar]
- Lu, P.; Hankel, I.L.; Hostager, B.S.; Swartzendruber, J.A.; Friedman, A.D.; Brenton, J.L.; Rothman, P.B.; Colgan, J.D. The developmental regulator protein Gon4l associates with protein YY1, co-repressor Sin3a, and histone deacetylase 1 and mediates transcriptional repression. J. Biol. Chem. 2011, 286, 18311–18319. [Google Scholar] [CrossRef]
- McPherson, J.P.; Sarras, H.; Lemmers, B.; Tamblyn, L.; Migon, E.; Matysiak-Zablocki, E.; Hakem, A.; Azami, S.A.; Cardoso, R.; Fish, J.; et al. Essential role for Bclaf1 in lung development and immune system function. Cell Death Differ. 2009, 16, 331–339. [Google Scholar] [CrossRef]
- Yi, F.; Pereira, L.; Merrill, B.J. Tcf3 functions as a steady-state limiter of transcriptional programs of mouse embryonic stem cell self-renewal. Stem Cells 2008, 26, 1951–1960. [Google Scholar] [CrossRef]
- Fong, N.M.; Jensen, T.C.; Shah, A.S.; Parekh, N.N.; Saltiel, A.R.; Brady, M.J. Identification of binding sites on protein targeting to glycogen for enzymes of glycogen metabolism. J. Biol. Chem. 2000, 275, 35034–35039. [Google Scholar]
- Ross, S.E.; McCord, A.E.; Jung, C.; Atan, D.; Mok, S.I.; Hemberg, M.; Kim, T.K.; Salogiannis, J.; Hu, L.; Cohen, S.; et al. Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron 2012, 73, 292–303. [Google Scholar] [CrossRef]
- Honore, B.; Rasmussen, H.H.; Vorum, H.; Dejgaard, K.; Liu, X.; Gromov, P.; Madsen, P.; Gesser, B.; Tommerup, N.; Celis, J.E. Heterogeneous nuclear ribonucleoproteins H, H’, and F are members of a ubiquitously expressed subfamily of related but distinct proteins encoded by genes mapping to different chromosomes. J. Biol. Chem. 1995, 270, 28780–28789. [Google Scholar] [CrossRef]
- Keegan, K.; Johnson, D.E.; Williams, L.T.; Hayman, M.J. Isolation of an additional member of the fibroblast growth factor receptor family, FGFR-3. Proc. Natl. Acad. Sci. USA 1991, 88, 1095–1099. [Google Scholar] [CrossRef]
- Jaiswal, B.S.; Kljavin, N.M.; Stawiski, E.W.; Chan, E.; Parikh, C.; Durinck, S.; Chaudhuri, S.; Pujara, K.; Guillory, J.; Edgar, K.A.; et al. Oncogenic ERBB3 mutations in human cancers. Cancer Cell 2013, 23, 603–617. [Google Scholar] [CrossRef]
- Mishima, Y.; Miyagi, S.; Saraya, A.; Negishi, M.; Endoh, M.; Endo, T.A.; Toyoda, T.; Shinga, J.; Katsumoto, T.; Chiba, T.; et al. The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood 2011, 118, 2443–2453. [Google Scholar] [CrossRef]
- Hatakeyama, S. TRIM proteins and cancer. Nat. Rev. Cancer 2011, 11, 792–804. [Google Scholar] [CrossRef]
- Wiemann, S.; Weil, B.; Wellenreuther, R.; Gassenhuber, J.; Glassl, S.; Ansorge, W.; Bocher, M.; Blocker, H.; Bauersachs, S.; Blum, H.; et al. Toward a catalog of human genes and proteins: Sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res. 2001, 11, 422–435. [Google Scholar] [CrossRef]
- Maciejewski, J.P.; Padgett, R.A. Defects in spliceosomal machinery: A new pathway of leukaemogenesis. Br. J. Haematol. 2012, 158, 165–173. [Google Scholar] [CrossRef]
- Sauve, F.; McBroom, L.D.; Gallant, J.; Moraitis, A.N.; Labrie, F.; Giguere, V. CIA, a novel estrogen receptor coactivator with a bifunctional nuclear receptor interacting determinant. Mol. Cell Biol. 2001, 21, 343–353. [Google Scholar] [CrossRef]
- Lo, S.H.; Weisberg, E.; Chen, L.B. Tensin: A potential link between the cytoskeleton and signal transduction. Bioessays 1994, 16, 817–823. [Google Scholar] [CrossRef]
- Roubin, R.; Acquaviva, C.; Chevrier, V.; Sedjai, F.; Zyss, D.; Birnbaum, D.; Rosnet, O. Myomegalin is necessary for the formation of centrosomal and Golgi-derived microtubules. Biol. Open 2013, 2, 238–250. [Google Scholar] [CrossRef]
- Ishov, A.M.; Vladimirova, O.V.; Maul, G.G. Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71 at ND10 facilitates initiation of viral infection at these nuclear domains. J. Virol. 2002, 76, 7705–7712. [Google Scholar] [CrossRef]
- Morin, R.D.; Mendez-Lago, M.; Mungall, A.J.; Goya, R.; Mungall, K.L.; Corbett, R.D.; Johnson, N.A.; Severson, T.M.; Chiu, R.; Field, M.; et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 2011, 476, 298–303. [Google Scholar] [CrossRef]
- Seimiya, H.; Smith, S. The telomeric poly(ADP-ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1) and a novel acceptor, 182-kDa tankyrase-binding protein (TAB182). J. Biol. Chem. 2002, 277, 14116–14126. [Google Scholar] [CrossRef]
- Kuuselo, R.; Savinainen, K.; Azorsa, D.O.; Basu, G.D.; Karhu, R.; Tuzmen, S.; Mousses, S.; Kallioniemi, A. Intersex-like (IXL) is a cell survival regulator in pancreatic cancer with 19q13 amplification. Cancer Res. 2007, 67, 1943–1949. [Google Scholar] [CrossRef]
- Geiman, D.E.; Ton-That, H.; Johnson, J.M.; Yang, V.W. Transactivation and growth suppression by the gut-enriched Kruppel-like factor (Kruppel-like factor 4) are dependent on acidic amino acid residues and protein-protein interaction. Nucleic Acids Res. 2000, 28, 1106–1113. [Google Scholar] [CrossRef]
- Wong, P.M.; Puente, C.; Ganley, I.G.; Jiang, X. The ULK1 complex: Sensing nutrient signals for autophagy activation. Autophagy 2013, 9, 124–137. [Google Scholar] [CrossRef]
- Mayerhofer, P.U.; Kattenfeld, T.; Roscher, A.A.; Muntau, A.C. Two splice variants of human PEX19 exhibit distinct functions in peroxisomal assembly. Biochem. Biophys. Res. Commun. 2002, 291, 1180–1186. [Google Scholar] [CrossRef]
- Kierdorf, K.; Fritz, G. RAGE regulation and signaling in inflammation and beyond. J. Leukoc. Biol. 2013, 94, 55–68. [Google Scholar] [CrossRef]
- Kikuchi, A. Roles of Axin in the Wnt signalling pathway. Cell. Signal. 1999, 11, 777–788. [Google Scholar] [CrossRef]
- Martignetti, J.A.; Aqeel, A.A.; Sewairi, W.A.; Boumah, C.E.; Kambouris, M.; Mayouf, S.A.; Sheth, K.V.; Eid, W.A.; Dowling, O.; Harris, J.; et al. Mutation of the matrix metalloproteinase 2 gene (MMP2) causes a multicentric osteolysis and arthritis syndrome. Nat. Genet. 2001, 28, 261–265. [Google Scholar] [CrossRef]
- Stagno, S.; Pass, R.F.; Cloud, G.; Britt, W.J.; Henderson, R.E.; Walton, P.D.; Veren, D.A.; Page, F.; Alford, C.A. Primary cytomegalovirus infection in pregnancy. Incidence, transmission to fetus, and clinical outcome. JAMA 1986, 256, 1904–1908. [Google Scholar] [CrossRef]
- Revello, M.G.; Gerna, G. Diagnosis and management of human cytomegalovirus infection in the mother, fetus, and newborn infant. Clin. Microbiol. Rev. 2002, 15, 80–715. [Google Scholar]
- Revello, M.G.; Zavattoni, M.; Furione, M.; Lilleri, D.; Gorini, G.; Gerna, G. Diagnosis and outcomeof preconceptional and periconceptional primary human cytomegalovirus infections. J. Infect. Dis. 2002, 186, 553–557. [Google Scholar] [CrossRef]
- Ramsay, M.E.; Miller, E.; Peckham, C.S. Outcome of confirmed symptomatic congenital cytomegalovirus infection. Arch. Dis. Child. 1991, 66, 1068–1069. [Google Scholar] [CrossRef]
- Chou, S.; Marousek, G.; Guentzel, S.; Follansbee, S.E.; Poscher, M.E.; Lalezari, J.P.; Miner, R.C.; Drew, W.L. Evolution of mutations conferring multidrug resistance during prophylaxis and therapy for cytomegalovirus disease. J. Infect. Dis. 1997, 176, 786–789. [Google Scholar] [CrossRef]
- Anderson, K.P.; Fox, M.C.; Brown-Driver, V.; Martin, M.J.; Azad, R.F. Inhibition of human cytomegalovirus immediate-early gene expression by an antisense oligonucleotide complementary to immediate-early RNA. Antimicrob. Agents Chemother. 1996, 40, 2004–2011. [Google Scholar]
- Smith, J.A.; Pari, G.S. Expression of human cytomegalovirus UL36 and UL37 genes is required for viral DNA replication. J. Virol. 1995, 69, 1925–1931. [Google Scholar]
- Munch, K.; Messerle, M.; Plachter, B.; Koszinowski, U.H. An acidic region of the 89K murine cytomegalovirus immediate early protein interacts with DNA. J. Gen. Virol. 1992, 73, 499–506. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Martínez, F.P.; Tang, Q. Identification of Cellular Proteins that Interact with Human Cytomegalovirus Immediate-Early Protein 1 by Protein Array Assay. Viruses 2014, 6, 89-105. https://doi.org/10.3390/v6010089
Martínez FP, Tang Q. Identification of Cellular Proteins that Interact with Human Cytomegalovirus Immediate-Early Protein 1 by Protein Array Assay. Viruses. 2014; 6(1):89-105. https://doi.org/10.3390/v6010089
Chicago/Turabian StyleMartínez, Francisco Puerta, and Qiyi Tang. 2014. "Identification of Cellular Proteins that Interact with Human Cytomegalovirus Immediate-Early Protein 1 by Protein Array Assay" Viruses 6, no. 1: 89-105. https://doi.org/10.3390/v6010089




