Proteomic Analyses of Human Cytomegalovirus Strain AD169 Derivatives Reveal Highly Conserved Patterns of Viral and Cellular Proteins in Infected Fibroblasts
Abstract
:1. Introduction
2. Results and Discussion
2.1. Conserved Pattern of Viral Protein Expression in Infected Cells
2.2. Comparable Impact of RV-HB15- and RV-BADwt-Infection on the Cellular Proteome
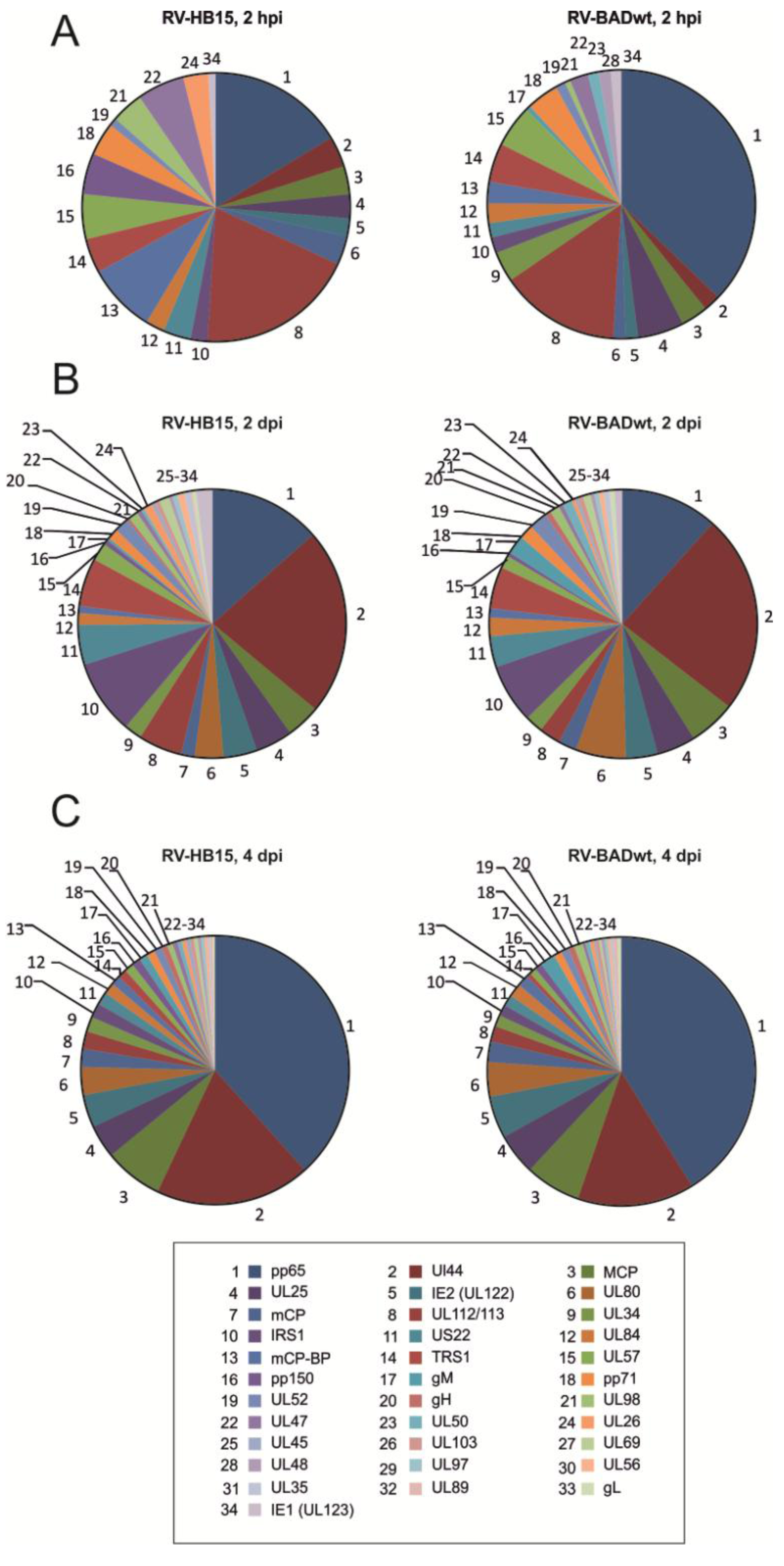

| Capsid proteins | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| HCMV ORF | Synonym | Max score a | Reported peptides b | RV-HB15 | RV-HB15 | RV-HB15 | RV-BADwt | RV-BADwt | RV-BADwt |
| 2 hpi c | 2 dpi c | 4 dpi c | 2 hpi c | 2 dpi c | 4 dpi c | ||||
| UL46 | mCP-BP | 7,323.97 | 9 | 17,316 | 14,320 | 89,186 | 7,064 | 30,425 | 115,613 |
| UL85 | mCP | 11,871.54 | 12 | 7,257 | 27,654 | 173,498 | 4,010 | 58,905 | 211,460 |
| UL86 | MCP | 29,574.99 | 42 | 6,941 | 69,181 | 556,028 | 8,708 | 149,287 | 572,567 |
| UL80 | pUL80 | 19,479.66 | 9 | nd | 57,699 | 267,196 | nd | 160,901 | 354,602 |
| Tegument proteins | |||||||||
| HCMV ORF | Synonym | Max score a | Reported peptides b | RV-HB15 | RV-HB15 | RV-HB15 | RV-BADwt | RV-BADwt | RV-BADwt |
| 2 hdpi | 2 dpi | 4 dpi | 2 hpi | 2 dpi | 4 dpi | ||||
| UL25 | pUL25 | 11,575.85 | 14 | 5,881 | 75,440 | 316,023 | 15,207 | 123,334 | 425,577 |
| UL26 | pUL26 | 2,947.21 | 2 | 6,186 | 17,749 | 39,969 | nd | 11,563 | 39,897 |
| UL32 | pp150 | 1,592.09 | 10 | 10,230 | 10,049 | 84,106 | nd | 12,225 | 76,558 |
| UL35 | pUL35 | 1,480.43 | 3 | nd | 12,827 | 26,106 | nd | 18,355 | 28,489 |
| UL45 | RR1 | 765.52 | 2 | nd | 7,799 | 38,594 | nd | 9,887 | 21,194 |
| UL47 | HMWP-BP | 1,950.86 | 8 | 11,842 | 9,849 | 48,624 | 5,974 | 13,690 | 29,478 |
| UL48 | HMW-P | 934.58 | 6 | nd | 7,967 | 33,958 | 3,938 | 12,393 | 30,863 |
| UL82 | pp71 | 3,682.85 | 9 | 8,167 | 21,851 | 78,681 | 10,465 | 46,063 | 81,361 |
| UL83 | pp65 | 73,413.98 | 27 | 33,740 | 227,127 | 3,026,292 | 100,786 | 309,371 | 3,521,603 |
| UL103 | pUL103 | 2114.97 | 2 | nd | 9,058 | 38,143 | nd | 16,642 | 34,996 |
| US22 | pUS22 | 3681.41 | 15 | 6,956 | 80,747 | 120,418 | 4,665 | 99,484 | 116,925 |
| Envelope proteins/glycoproteins | |||||||||
| HCMV ORF | Synonym | Max score a | Reported peptides b | RV-HB15 | RV-HB15 | RV-HB15 | RV-BADwt | RV-BADwt | RV-BADwt |
| 2 hpi | 2 dpi | 4 dpi | 2 hpi | 2 dpi | 4 dpi | ||||
| UL75 | gH | 848.31 | 4 | nd | 5,856 | 57,842 | nd | 18,448 | 64,162 |
| UL100 | gM | 2,800.15 | 2 | nd | 5,570 | 80,032 | 1,528 | 61,189 | 158,290 |
| UL115 | gL | 3,596.83 | 2 | nd | 8,146 | 19,951 | nd | 15,905 | 32,299 |
| Other virion proteins | |||||||||
| HCMV ORF | Protein | Max score a | Reported peptides b | RV-HB15 | RV-HB15 | RV-HB15 | RV-BADwt | RV-BADwt | RV-BADwt |
| 0 dpi | 2 dpi | 4 dpi | 0 dpi | 2 dpi | 4 dpi | ||||
| IRS1 | pIRS1 | 13,209.55 | 19 | 4,284 | 150,080 | 138,219 | 5,178 | 189,945 | 114,985 |
| TRS1 | pTRS1 | 10,401.20 | 18 | 8,602 | 97,473 | 87,607 | 12,635 | 133,632 | 45,993 |
| UL34 | pUL34 | 2,813.53 | 5 | nd | 35,364 | 142,758 | 9,861 | 56,510 | 117,824 |
| UL44 | pUL44 | 70,382.59 | 20 | 7,621 | 379,249 | 1,477,506 | 5,522 | 631,538 | 1,200,976 |
| UL50 | pUL50 | 1,314.75 | 2 | nd | 6,143 | 40,091 | 3,617 | 31,638 | 45,208 |
| UL52 | pUL52 | 1,623.85 | 11 | 2,043 | 29,069 | 73,220 | 3,252 | 58,778 | 77,154 |
| UL56 | pUL56 | 1,143.55 | 2 | nd | 14,351 | 29,524 | nd | 16,585 | 31,192 |
| UL57 | pUL57 | 2,937.02 | 23 | 11,186 | 36,083 | 84,431 | 13,807 | 40,880 | 69,838 |
| UL69 | pUL69 | 878.59 | 5 | nd | 20,258 | 36,525 | nd | 24,015 | 25,949 |
| UL84 | pUL84 | 8,406.89 | 13 | 4,630 | 23,259 | 108,290 | 6,403 | 59,514 | 137,353 |
| UL89 | pUL89 | 1,075.02 | 2 | nd | 2,815 | 21,271 | nd | nd | 40,538 |
| UL97 | pUL97 | 1,740.41 | 4 | nd | 10,305 | 33,074 | nd | 14,152 | 35,229 |
| UL98 | pUL98 | 2,791.21 | 9 | 7,904 | 18,894 | 53,714 | 1,835 | 31,351 | 89,966 |
| UL112/113 | pUL112/113 | 4,259.03 | 9 | 39,266 | 88,103 | 163,205 | 39,268 | 68,815 | 160,294 |
| UL122 | IE2 | 38,858.88 | 9 | 4,333 | 68,426 | 309,712 | 4,071 | 101,350 | 431,908 |
| UL123 | IE1 | 2,927.93 | 7 | 1,829 | 33,285 | 2,040 | 3,456 | 25,160 | 7404 |
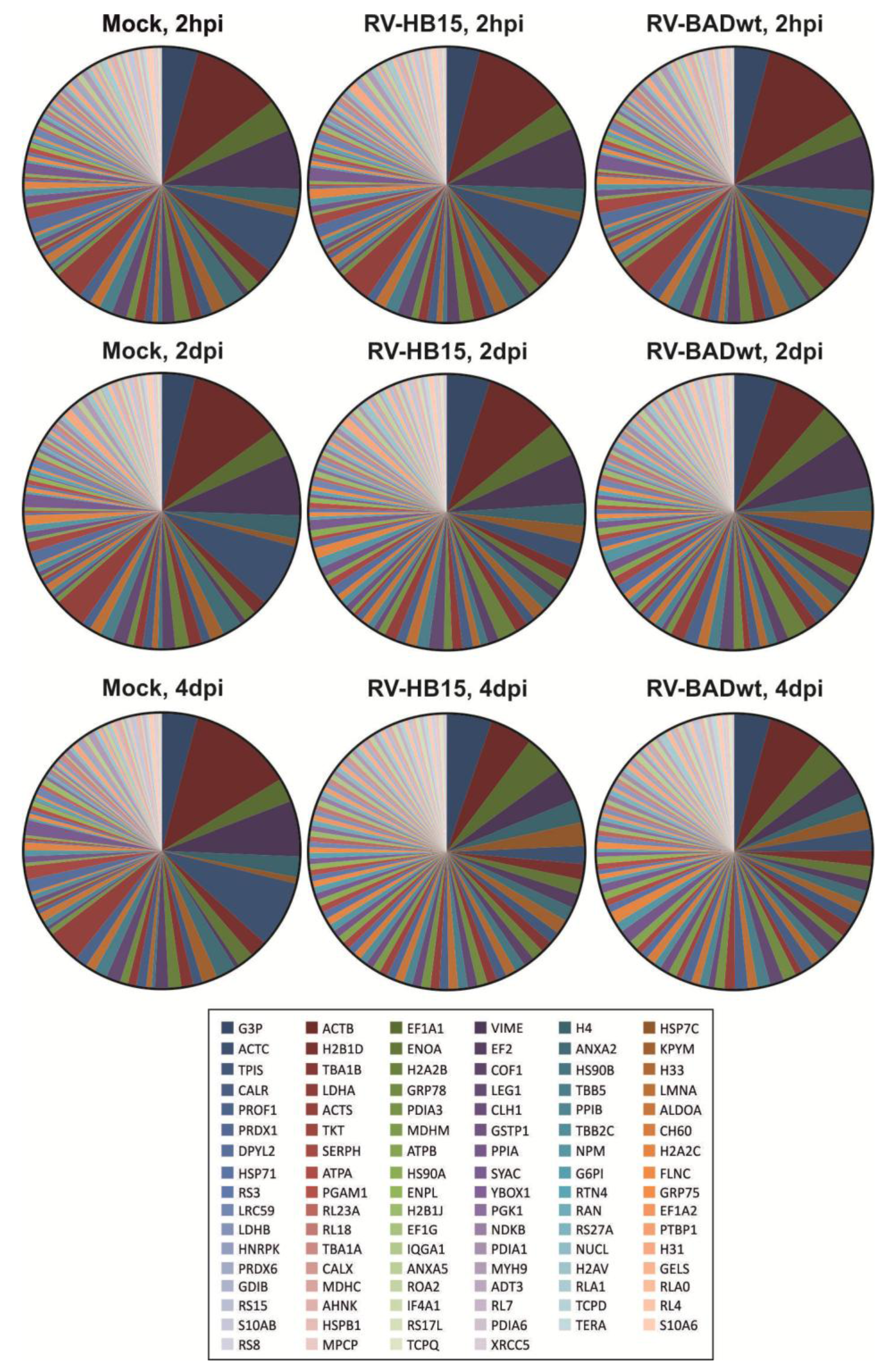
2.3. Conserved Stoichiometry of Viral Proteins in Virions of AD169-Derived Viruses

3. Experimental Section
4. Conclusions
Acknowledgments
Conflicts of Interest
References and Notes
- Mocarski, E.S.; Shenk, T.; Griffiths, P.D.; Pass, R.F. Cytomegaloviruses. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Wolters Kluwer Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 1960–2014. [Google Scholar]
- Gibson, W.; Bogner, E. Morphogenesis of the cytomegalovirus virion and subviral particles. In Cytomegaloviruses: From Molecular Pathogenesis to Intervention, 2th ed.; Reddehase, M.J., Ed.; Caister Academic Press: Norfolk, UK, 2013; pp. 230–246. [Google Scholar]
- Milbradt, J.; Auerochs, S.; Marschall, M. Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J. Gen. Virol. 2007, 88, 2642–2650. [Google Scholar] [CrossRef]
- Milbradt, J.; Auerochs, S.; Sticht, H.; Marschall, M. Cytomegaloviral proteins that associate with the nuclear lamina: Components of a postulated nuclear egress complex. J. Gen. Virol. 2009, 90, 579–590. [Google Scholar] [CrossRef]
- Camozzi, D.; Pignatelli, S.; Valvo, C.; Lattanzi, G.; Capanni, C.; Dal, M.P.; Landini, M.P. Remodelling of the nuclear lamina during human cytomegalovirus infection: Role of the viral proteins pUL50 and pUL53. J. Gen. Virol. 2008, 89, 731–740. [Google Scholar]
- Colberg Poley, A.M.; Williamson, C.A. Intracellular sorting and trafficking of cytomegalovirus proteins during permissive infection. In Cytomegaloviruses From Molecular Pathogenesis to Intervention, 2th ed.; Reddehase, M.J., Ed.; Caister Academic Press: Norfolk, UK, 2013; pp. 196–229. [Google Scholar]
- Marschall, M.; Feichtinger, S.; Milbradt, J. Regulatory roles of protein kinases in cytomegalovirus replication. Adv. Virus Res. 2011, 80, 69–101. [Google Scholar]
- Krosky, P.M.; Baek, M.C.; Coen, D.M. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J. Virol. 2003, 77, 905–914. [Google Scholar] [CrossRef]
- Sanchez, V.; Greis, K.D.; Sztul, E.; Britt, W.J. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: Characterization of a potential site of virus assembly. J. Virol. 2000, 74, 975–986. [Google Scholar] [CrossRef]
- Das, S.; Vasanji, A.; Pellett, P.E. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J. Virol. 2007, 81, 11861–11869. [Google Scholar] [CrossRef]
- Das, S.; Pellett, P.E. Spatial relationships between markers for secretory and endosomal machinery in human cytomegalovirus-infected cells versus those in uninfected cells. J. Virol. 2011, 85, 5864–5879. [Google Scholar] [CrossRef]
- Indran, S.V.; Britt, W.J. A role for the small GTPase Rab6 in assembly of human cytomegalovirus. J. Virol. 2011, 85, 5213–5219. [Google Scholar] [CrossRef]
- Tandon, R.; AuCoin, D.P.; Mocarski, E.S. Human cytomegalovirus exploits ESCRT machinery in the process of virion maturation. J. Virol. 2009, 83, 10797–10807. [Google Scholar] [CrossRef]
- Ahlqvist, J.; Mocarski, E. Cytomegalovirus UL103 controls virion and dense body egress. J. Virol. 2011, 85, 5125–5135. [Google Scholar] [CrossRef]
- Irmiere, A.; Gibson, W. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 1983, 130, 118–133. [Google Scholar] [CrossRef]
- Roby, C.; Gibson, W. Characterization of phosphoproteins and protein kinase activity of virions, noninfectious enveloped particles, and dense bodies of human cytomegalovirus. J. Virol. 1986, 59, 714–727. [Google Scholar]
- Jahn, G.; Scholl, B.C.; Traupe, B.; Fleckenstein, B. The two major structural phosphoproteins (pp65 and pp150) of human cytomegalovirus and their antigenic properties. J. Gen. Virol. 1987, 68, 1327–1337. [Google Scholar] [CrossRef]
- Schmolke, S.; Kern, H.F.; Drescher, P.; Jahn, G.; Plachter, B. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J. Virol. 1995, 69, 5959–5968. [Google Scholar]
- Baldick, C.J., Jr.; Shenk, T. Proteins associated with purified human cytomegalovirus particles. J. Virol. 1996, 70, 6097–6105. [Google Scholar]
- Grundy, J.E.; McKeating, J.A.; Griffiths, P.D. Cytomegalovirus strain AD169 binds beta 2 microglobulin in vitro after release from cells. J. Gen. Virol. 1987, 68, 777–784. [Google Scholar] [CrossRef]
- Stannard, L.M. Beta 2 microglobulin binds to the tegument of cytomegalovirus: An immunogold study. J. Gen. Virol. 1989, 70, 2179–2184. [Google Scholar] [CrossRef]
- Wright, J.F.; Kurosky, A.; Pryzdial, E.L.; Wasi, S. Host cellular annexin II is associated with cytomegalovirus particles isolated from cultured human fibroblasts. J. Virol. 1995, 69, 4784–4791. [Google Scholar]
- Giugni, T.D.; Soderberg, C.; Ham, D.J.; Bautista, R.M.; Hedlund, K.O.; Moller, E.; Zaia, J.A. Neutralization of human cytomegalovirus by human CD13-specific antibodies. J. Infect. Dis. 1996, 173, 1062–1071. [Google Scholar] [CrossRef]
- Michelson, S.; Turowski, P.; Picard, L.; Goris, J.; Landini, M.P.; Topilko, A.; Hemmings, B.; Bessia, C.; Garcia, A.; Virelizier, J.L. Human cytomegalovirus carries serine/threonine protein phosphatases PP1 and a host-cell derived PP2A. J. Virol. 1996, 70, 1415–1423. [Google Scholar]
- Varnum, S.M.; Streblow, D.N.; Monroe, M.E.; Smith, P.; Auberry, K.J.; Pasa-Tolic, L.; Wang, D.; Camp, D.G.; Rodland, K.; Wiley, S.; et al. Identification of proteins in human cytomegalovirus (HCMV) particles: The HCMV proteome. J. Virol. 2004, 78, 10960–10966. [Google Scholar] [CrossRef]
- Caposio, P.; Streblow, D.N.; Nelson, J.A. Cytomegalovirus proteomics. In Cytomegaloviruses From Molecular Pathogenesis to Intervention, 2th ed.; Reddehase, M.J., Ed.; Caister Academic Press: Norfolk, UK, 2013; pp. 86–108. [Google Scholar]
- Kattenhorn, L.M.; Mills, R.; Wagner, M.; Lomsadze, A.; Makeev, V.; Borodovsky, M.; Ploegh, H.L.; Kessler, B.M. Identification of proteins associated with murine cytomegalovirus virions. J. Virol. 2004, 78, 11187–11197. [Google Scholar] [CrossRef]
- Borst, E.M.; Hahn, G.; Koszinowski, U.H.; Messerle, M. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: A new approach for construction of HCMV mutants. J. Virol. 1999, 73, 8320–8329. [Google Scholar]
- Hobom, U.; Brune, W.; Messerle, M.; Hahn, G.; Koszinowski, U.H. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: Mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 2000, 74, 7720–7729. [Google Scholar] [CrossRef]
- Yu, D.; Smith, G.A.; Enquist, L.W.; Shenk, T. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 2002, 76, 2316–2328. [Google Scholar] [CrossRef]
- Besold, K.; Frankenberg, N.; Pepperl-Klindworth, S.; Kuball, J.; Theobald, M.; Hahn, G.; Plachter, B. Processing and MHC class I presentation of human cytomegalovirus pp65-derived peptides persist despite gpUS2–11-mediated immune evasion. J. Gen. Virol. 2007, 88, 1429–1439. [Google Scholar] [CrossRef]
- Hesse, J.; Reyda, S.; Tenzer, S.; Besold, K.; Reuter, N.; Krauter, S.; Büscher, N.; Stamminger, T.; Plachter, B. Human cytomegalovirus pp71 stimulates major histocompatibility complex class i presentation of IE1-derived peptides at immediate early times of infection. J. Virol. 2013, 87, 5229–5238. [Google Scholar] [CrossRef]
- Zhu, H.; Cong, J.P.; Mamtora, G.; Gingeras, T.; Shenk, T. Cellular gene expression altered by human cytomegalovirus: Global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 1998, 95, 14470–14475. [Google Scholar] [CrossRef]
- Rabinowitz, J.D.; Shenk, T. Human cytomegalovirus metabolomics. In Cytomegalovirus from Molecular Pathogenesis to Intervention, 2th ed.; Reddehase, M.J., Ed.; Caister Academic Press: Norfolk, UK, 2013; pp. 59–67. [Google Scholar]
- Protein Lynx Global Server (PLGS) Ion Accounting Search Algorithm. PLGS is available from Waters, Manchester, UK. Available online: http://www.waters.com/ (accessed on 21 October 2013).
- Chevillotte, M.; Landwehr, S.; Linta, L.; Frascaroli, G.; Luske, A.; Buser, C.; Mertens, T.; von Einem, J. Major tegument protein pp65 of human cytomegalovirus is required for the incorporation of pUL69 and pUL97 into the virus particle and for viral growth in macrophages. J. Virol. 2009, 83, 2480–2490. [Google Scholar] [CrossRef]
- To, A.; Bai, Y.; Shen, A.; Gong, H.; Umamoto, S.; Lu, S.; Liu, F. Yeast two hybrid analyses reveal novel binary interactions between human cytomegalovirus-encoded virion proteins. PLoS One 2011, 6, e17796. [Google Scholar]
- Becke, S.; Fabre-Mersseman, V.; Aue, S.; Auerochs, S.; Sedmak, T.; Wolfrum, U.; Strand, D.; Marschall, M.; Plachter, B.; Reyda, S. Modification of the major tegument protein pp65 of human cytomegalovirus inhibits virus growth and leads to the enhancement of a protein complex with pUL69 and pUL97 in infected cells. J. Gen. Virol. 2010, 91, 2531–2541. [Google Scholar] [CrossRef]
- Phillips, S.L.; Bresnahan, W.A. Identification of binary interactions between human cytomegalovirus virion proteins. J. Virol. 2011, 85, 440–447. [Google Scholar] [CrossRef]
- Tenzer, S.; Docter, D.; Rosfa, S.; Wlodarski, A.; Kuharev, J.; Rekik, A.; Knauer, S.K.; Bantz, C.; Nawroth, T.; Bier, C.; et al. Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: a comprehensive quantitative proteomic analysis. ACS Nano. 2011, 5, 7155–7167. [Google Scholar]
- ProteinLynx GlobalSERVER, version 2.5.2; Waters Corporation: Manchester, UK, 2011.
- Patzig, J.; Jahn, O.; Tenzer, S.; Wichert, S.P.; de Monasterio-Schrader, P.; Rosfa, S.; Kuharev, J.; Yan, K.; Bormuth, I.; Bremer, J.; et al. Quantitative and integrative proteome analysis of peripheral nerve myelin identifies novel myelin proteins and candidate neuropathy loci. J. Neurosci. 2011, 31, 16369–16386. [Google Scholar] [CrossRef]
- Silva, J.C.; Gorenstein, M.V.; Li, G.Z.; Vissers, J.P.; Geromanos, S.J. Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Mol. Cell Proteomics. 2006, 5, 144–156. [Google Scholar]
- Distler, U.; Kuharev, J.; Navarro, P.; Levin, Y.; Schild, H.J.; Tenzer, S. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat. Method 2013. [Google Scholar] [CrossRef]
Supplementary Files
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Reyda, S.; Büscher, N.; Tenzer, S.; Plachter, B. Proteomic Analyses of Human Cytomegalovirus Strain AD169 Derivatives Reveal Highly Conserved Patterns of Viral and Cellular Proteins in Infected Fibroblasts. Viruses 2014, 6, 172-188. https://doi.org/10.3390/v6010172
Reyda S, Büscher N, Tenzer S, Plachter B. Proteomic Analyses of Human Cytomegalovirus Strain AD169 Derivatives Reveal Highly Conserved Patterns of Viral and Cellular Proteins in Infected Fibroblasts. Viruses. 2014; 6(1):172-188. https://doi.org/10.3390/v6010172
Chicago/Turabian StyleReyda, Sabine, Nicole Büscher, Stefan Tenzer, and Bodo Plachter. 2014. "Proteomic Analyses of Human Cytomegalovirus Strain AD169 Derivatives Reveal Highly Conserved Patterns of Viral and Cellular Proteins in Infected Fibroblasts" Viruses 6, no. 1: 172-188. https://doi.org/10.3390/v6010172




