Analysis of Essential Viral Gene Functions after Highly Efficient Adenofection of Cells with Cloned Human Cytomegalovirus Genomes
Abstract
:1. Introduction
2. Results and Discussion
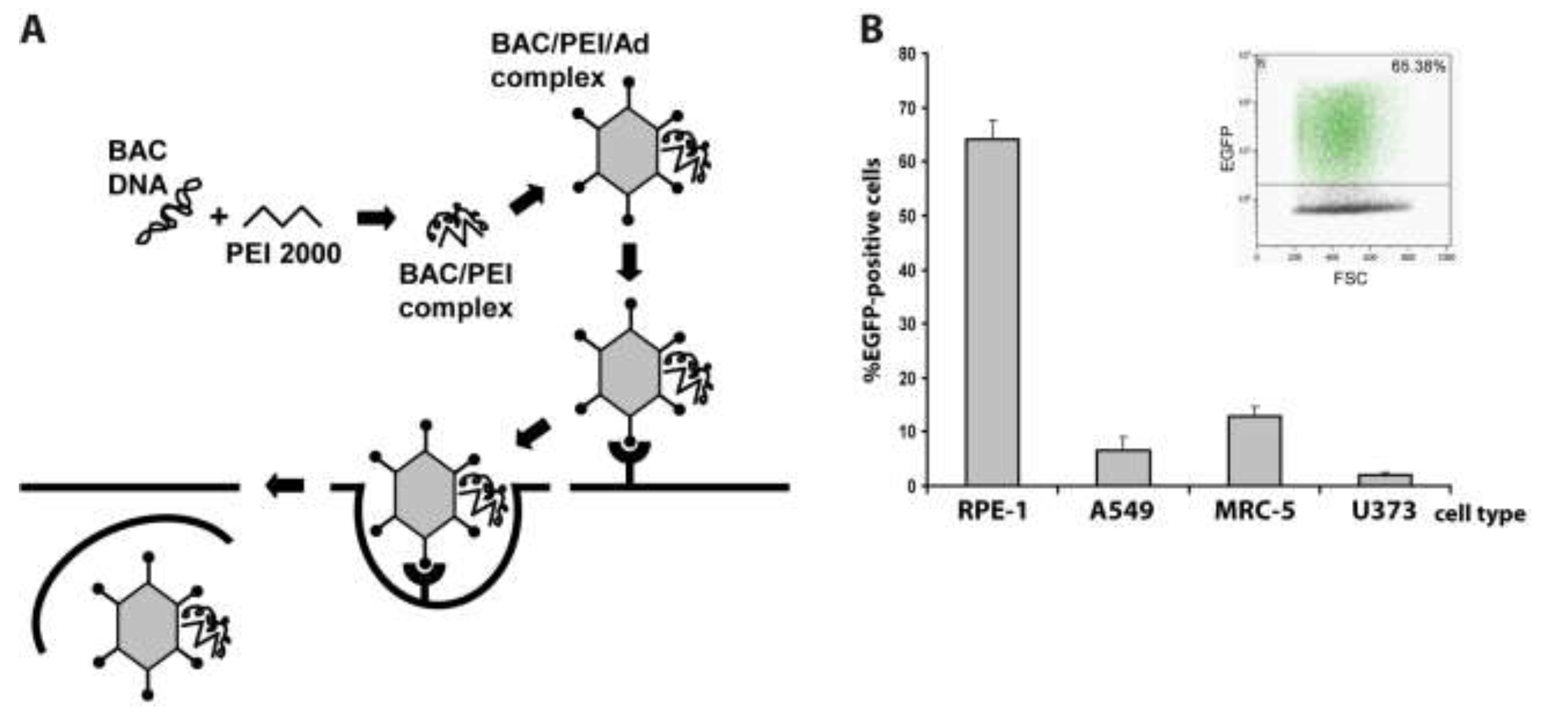
2.1. Adenovirus-Mediated Delivery of HCMV BACs into Different Cell Types and Optimization of the Procedure
| Name of HCMV BAC | Mutation |
|---|---|
| pHG (parental BAC) | EGFP ORF under control of HCMV MIEP deletion of UL1-10 [28] |
| pHG-11 | deletion in UL77 |
| pHG-12 | deletion in UL93 |
| pHG-13 | deletion in UL104 |
| pHG-14 | deletion in UL44 |
| pHG-Δ1.5 | 1.5 kb deletion in oriLyt [28] |
| pHG-Δ3.7 | 3.7 kb deletion in oriLyt [28] |
| pHG-Δ52 | deletion in UL52 [14] |
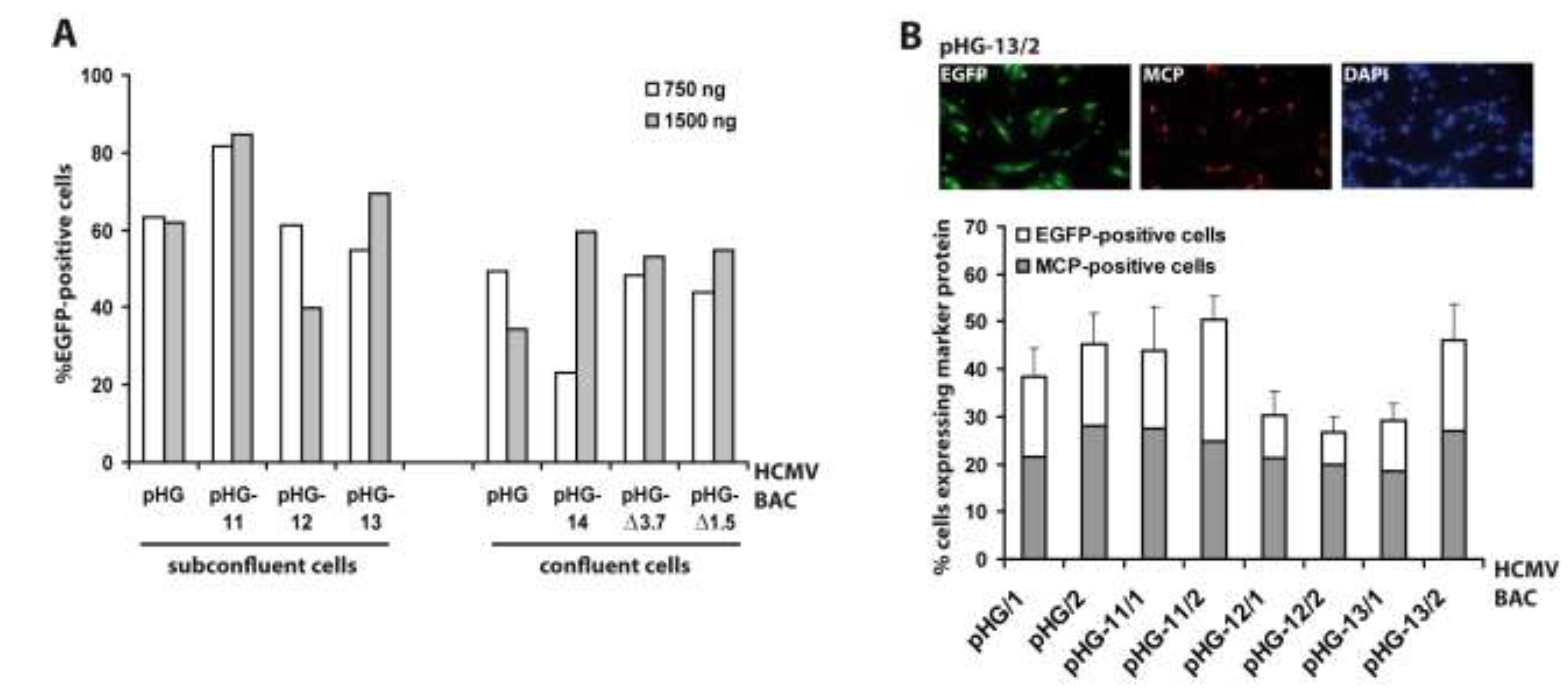
2.2. Viral Gene Expression in Adenofected Cells
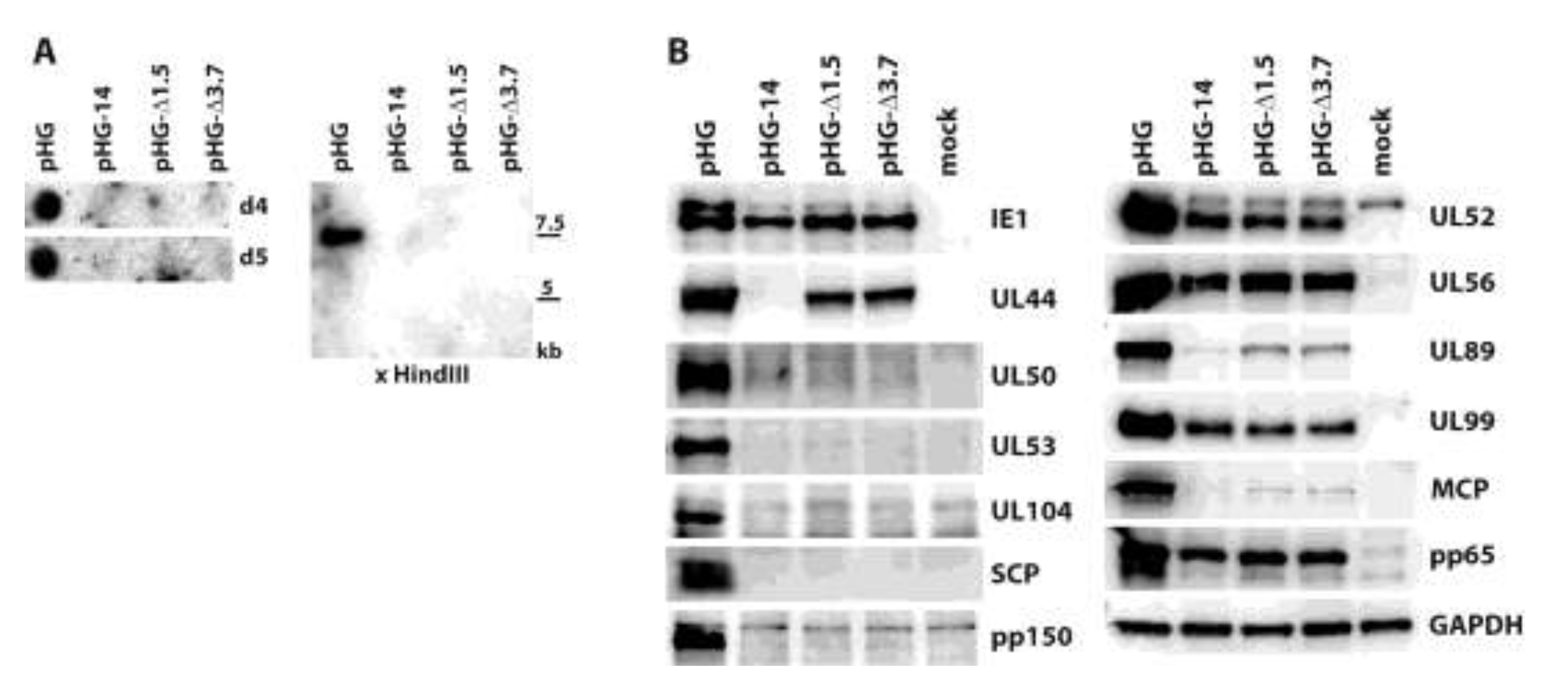
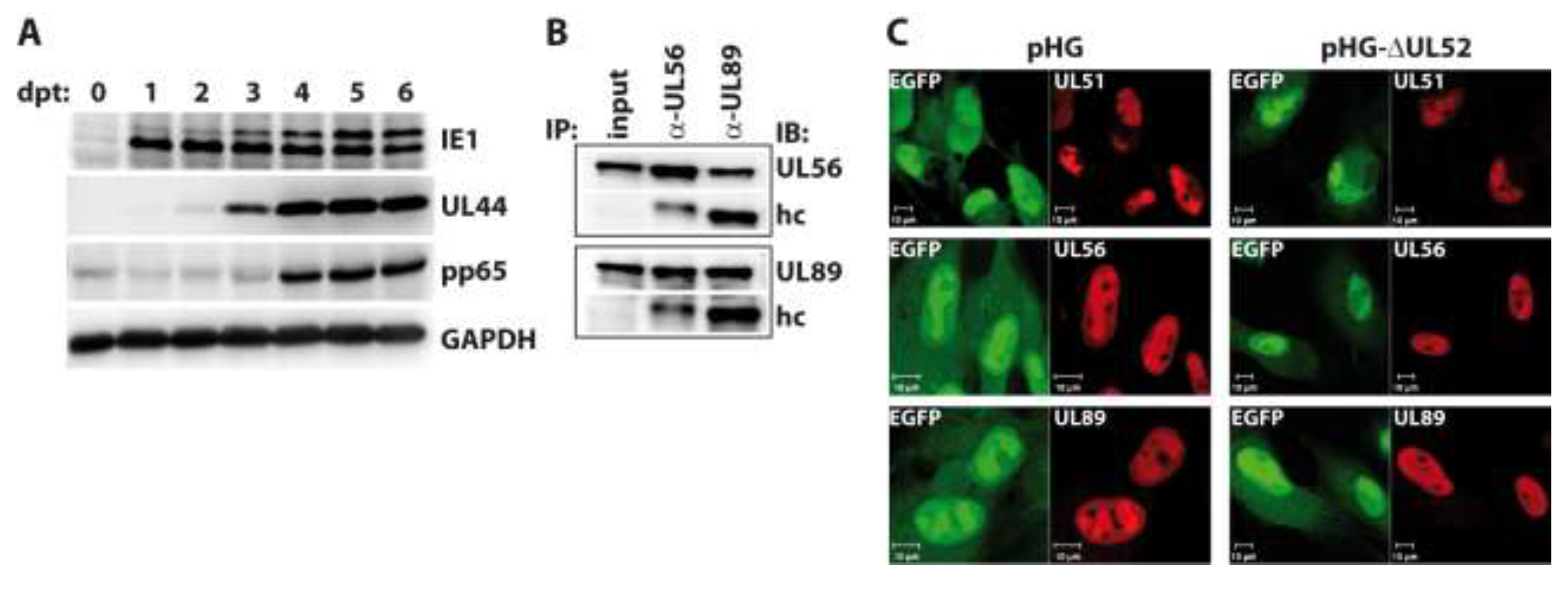
2.3. Impact of UL52 on HCMV Terminase Proteins
3. Experimental Section
3.1. Cells
3.2. HCMV Bacterial Artificial Chromosomes
| Primer name | Nucleotide sequence |
|---|---|
| UL77-ko.for | 5'-acgatgccatcacgggacccgccgccgccccgtctgacgtggaaaagtgccacctgcagat-3' |
| UL77-ko.rev | 5'-ccgaggacgttcgccctttatgcagcgagcgacacgtggtgcaggaacacttaacggctga-3' |
| UL104-ko.for | 5'-gctgtcgtcgtaggcggcggccacgatctcgccgaaggagagaaaagtgccacctgcagat-3' |
| UL104-ko.rev | 5'-gtgctgggcggcctccgcgacattttatatcagtacgccgcaggaacacttaacggctga-3' |
| UL44-ko.for | 5'-gggaccgcgccgtgcgcgcgttcccaggcacgcggcccgcgaaaagtgccacctgcagat-3' |
| UL44-ko.rev | 5'-tcgctcgctcgcgcccgctccttagtcgagacttgcacgctcaggaacacttaacggctga-3' |
| UL93-ko.for | 5'-agtcgaactataccgggcgttggacgcttatcgggcgtagtgactgattcgcgccgtgcgcaa-ggatgacgacgataag-3' |
| UL93-ko.rev | 5'-agtacagcgtagatctcgtcgcgcacggcgcgaatcagtcactacgcccgataagcgtccagcc-agtgttacaaccaatt-3' |
3.3. Preparation of Adenovirus Stocks and Transfection by Adenofection
3.4. Flow Cytometry
3.5. Viral DNA Replication Assay
3.6. Protein Biochemistry
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Griffiths, P.D. Burden of disease associated with human cytomegalovirus and prospects for elimination by universal immunisation. Lancet Infect. Dis. 2012, 12, 790–798. [Google Scholar] [CrossRef]
- Ljungman, P.; Hakki, M.; Boeckh, M. Cytomegalovirus in hematopoietic stem cell transplant recipients. Infect. Dis. Clin. N. Am. 2010, 24, 319–337. [Google Scholar] [CrossRef]
- Nigro, G.; Adler, S.P. Cytomegalovirus infections during pregnancy. Curr. Opin. Obstet. Gynecol. 2011, 23, 123–128. [Google Scholar] [CrossRef]
- Biron, K.K. Antiviral drugs for cytomegalovirus diseases. Antivir. Res. 2006, 71, 154–163. [Google Scholar] [CrossRef]
- Le Page, A.K.; Jager, M.M.; Iwasenko, J.M.; Scott, G.M.; Alain, S.; Rawlinson, W.D. Clinical aspects of cytomegalovirus antiviral resistance in solid organ transplant recipients. Clin. Infect. Dis. 2013, 56, 1018–1029. [Google Scholar] [CrossRef]
- Stern-Ginossar, N.; Weisburd, B.; Michalski, A.; Le, V.T.; Hein, M.Y.; Huang, S.X.; Ma, M.; Shen, B.; Qian, S.B.; Hengel, H.; et al. Decoding human cytomegalovirus. Science 2012, 338, 1088–1093. [Google Scholar] [CrossRef]
- Ruzsics, Z.; Borst, E.M.; Bosse, J.B.; Brune, W.; Messerle, M. Manipulating cytomegalovirus genomes by BAC mutagenesis: Strategies and applications. In Cytomegaloviruses: From Molecular Pathogenesis to Intervention; Reddehase, M.J., Ed.; Caister Academic Press: Hethersett, Norwich, UK, 2013; pp. 37–57. [Google Scholar]
- Davison, A.J.; Dolan, A.; Akter, P.; Addison, C.; Dargan, D.J.; Alcendor, D.J.; McGeoch, D.J.; Hayward, G.S. The human cytomegalovirus genome revisited: Comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 2003, 84, 17–28. [Google Scholar] [CrossRef]
- Dolan, A.; Cunningham, C.; Hector, R.D.; Hassan-Walker, A.F.; Lee, L.; Addison, C.; Dargan, D.J.; McGeoch, D.J.; Gatherer, D.; Emery, V.C.; et al. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 2004, 85, 1301–1312. [Google Scholar] [CrossRef]
- Yu, D.; Silva, M.C.; Shenk, T. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 2003, 100, 12396–12401. [Google Scholar] [CrossRef]
- Dunn, W.; Chou, C.; Li, H.; Hai, R.; Patterson, D.; Stolc, V.; Zhu, H.; Liu, F. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 2003, 100, 14223–14228. [Google Scholar]
- Mocarski, E.S.; Kemble, G.W.; Lyle, J.M.; Greaves, R.F. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA 1996, 93, 11321–11326. [Google Scholar]
- Silva, M.C.; Yu, Q.C.; Enquist, L.; Shenk, T. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 2003, 77, 10594–10605. [Google Scholar] [CrossRef]
- Borst, E.M.; Wagner, K.; Binz, A.; Sodeik, B.; Messerle, M. The essential human cytomegalovirus gene UL52 is required for cleavage-packaging of the viral genome. J. Virol. 2008, 82, 2065–2078. [Google Scholar] [CrossRef]
- Sanders, R.L.; Clark, C.L.; Morello, C.S.; Spector, D.H. Development of cell lines that provide tightly controlled temporal translation of the human cytomegalovirus IE2 proteins for complementation and functional analyses of growth-impaired and nonviable IE2 mutant viruses. J. Virol. 2008, 82, 7059–7077. [Google Scholar] [CrossRef]
- Mohr, H.; Mohr, C.A.; Schneider, M.R.; Scrivano, L.; Adler, B.; Kraner-Schreiber, S.; Schnieke, A.; Dahlhoff, M.; Wolf, E.; Koszinowski, U.H.; et al. Cytomegalovirus replicon-based regulation of gene expression in vitro and in vivo. PLoS Pathog. 2012, 8, e1002728. [Google Scholar] [CrossRef] [Green Version]
- McVoy, M.A.; Mocarski, E.S. Tetracycline-mediated regulation of gene expression within the human cytomegalovirus genome. Virology 1999, 258, 295–303. [Google Scholar] [CrossRef]
- Rupp, B.; Ruzsics, Z.; Sacher, T.; Koszinowski, U.H. Conditional cytomegalovirus replication in vitro and in vivo. J. Virol. 2005, 79, 486–494. [Google Scholar] [CrossRef]
- Glass, M.; Busche, A.; Wagner, K.; Messerle, M.; Borst, E.M. Conditional and reversible disruption of essential herpesvirus proteins. Nat. Methods 2009, 6, 577–579. [Google Scholar] [CrossRef]
- Qian, Z.; Leung-Pineda, V.; Xuan, B.; Piwnica-Worms, H.; Yu, D. Human cytomegalovirus protein pUL117 targets the mini-chromosome maintenance complex and suppresses cellular DNA synthesis. PLoS Pathog. 2010, 6, e1000814. [Google Scholar] [CrossRef]
- Perng, Y.C.; Qian, Z.; Fehr, A.R.; Xuan, B.; Yu, D. The human cytomegalovirus gene UL79 is required for the accumulation of late viral transcripts. J. Virol. 2011, 85, 4841–4852. [Google Scholar] [CrossRef]
- Tandon, R.; Mocarski, E.S. Cytomegalovirus pUL96 is critical for the stability of pp150-associated nucleocapsids. J. Virol. 2011, 85, 7129–7141. [Google Scholar] [CrossRef]
- Nitzsche, A.; Steinhausser, C.; Mucke, K.; Paulus, C.; Nevels, M. Histone H3 lysine 4 methylation marks postreplicative human cytomegalovirus chromatin. J. Virol. 2012, 86, 9817–9827. [Google Scholar] [CrossRef]
- Cotten, M.; Wagner, E.; Zatloukal, K.; Phillips, S.; Curiel, D.T.; Birnstiel, M.L. High-efficiency receptor-mediated delivery of small and large (48 kilobase gene constructs using the endosome-disruption activity of defective or chemically inactivated adenovirus particles. Proc. Natl. Acad. Sci. USA 1992, 89, 6094–6098. [Google Scholar]
- Cotten, M. Adenovirus-augmented, receptor-mediated gene delivery and some solutions to the common toxicity problems. Curr. Top. Microbiol. Immunol. 1995, 199, 283–295. [Google Scholar]
- Baker, A.; Cotten, M. Delivery of bacterial artificial chromosomes into mammalian cells with psoralen-inactivated adenovirus carrier. Nucleic Acids Res. 1997, 25, 1950–1956. [Google Scholar] [CrossRef]
- Baker, A.; Saltik, M.; Lehrmann, H.; Killisch, I.; Mautner, V.; Lamm, G.; Christofori, G.; Cotten, M. Polyethylenimine (PEI) is a simple, inexpensive and effective reagent for condensing and linking plasmid DNA to adenovirus for gene delivery. Gene Ther. 1997, 4, 773–782. [Google Scholar]
- Borst, E.M.; Messerle, M. Analysis of human cytomegalovirus oriLyt sequence requirements in the context of the viral genome. J. Virol. 2005, 79, 3615–3626. [Google Scholar] [CrossRef]
- Borst, E. Department of Virology, Hannover Medical School, Hannover, Germany. Unpublished observation. 2013. [Google Scholar]
- Cotten, M.; Baker, A.; Saltik, M.; Wagner, E.; Buschle, M. Lipopolysaccharide is a frequent contaminant of plasmid DNA preparations and can be toxic to primary human cells in the presence of adenovirus. Gene Ther. 1994, 1, 239–246. [Google Scholar]
- Cotten, M.; Saltik, M. Intracellular delivery of lipopolysaccharide during DNA transfection activates a lipid A-dependent cell death response that can be prevented by polymyxin B. Hum. Gene Ther. 1997, 8, 555–561. [Google Scholar] [CrossRef]
- Borst, E.; Elbasani, E. Department of Virology, Hannover Medical School, Hannover, Germany. Unpublished data. 2013. [Google Scholar]
- Borst, E.M.; Messerle, M. Development of a cytomegalovirus vector for somatic gene therapy. Bone Marrow Transplant. 2000, 25, S80–S82. [Google Scholar] [CrossRef]
- Kalejta, R.F. Functions of human cytomegalovirus tegument proteins prior to immediate early gene expression. Curr. Top. Microbiol. Immunol. 2008, 325, 101–115. [Google Scholar]
- Bishop, C.L.; Ramalho, M.; Nadkarni, N.; May, K.W.; Higgins, C.F.; Krauzewicz, N. Role for centromeric heterochromatin and PML nuclear bodies in the cellular response to foreign DNA. Mol. Cell Biol. 2006, 26, 2583–2594. [Google Scholar] [CrossRef]
- Baldick, C.J., Jr.; Marchini, A.; Patterson, C.E.; Shenk, T. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 1997, 71, 4400–4408. [Google Scholar]
- Pari, G.S.; Kacica, M.A.; Anders, D.G. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J. Virol. 1993, 67, 2575–2582. [Google Scholar]
- Silva, L.A.; Loregian, A.; Pari, G.S.; Strang, B.L.; Coen, D.M. The carboxy-terminal segment of the human cytomegalovirus DNA polymerase accessory subunit UL44 is crucial for viral replication. J. Virol. 2010, 84, 11563–11568. [Google Scholar] [CrossRef]
- Nevels, M.; Brune, W.; Shenk, T. SUMOylation of the human cytomegalovirus 72-kilodalton IE1 protein facilitates expression of the 86-kilodalton IE2 protein and promotes viral replication. J. Virol. 2004, 78, 7803–7812. [Google Scholar] [CrossRef]
- Borst, E.M.; Kleine-Albers, J.; Gabaev, I.; Babic, M.; Wagner, K.; Binz, A.; Degenhardt, I.; Kalesse, M.; Jonjic, S.; Bauerfeind, R.; et al. The human cytomegalovirus UL51 protein is essential for viral genome cleavage-packaging and interacts with the terminase subunits pUL56 and pUL89. J. Virol. 2013, 87, 1720–1732. [Google Scholar] [CrossRef]
- Wang, J.B.; Zhu, Y.; McVoy, M.A.; Parris, D.S. Changes in subcellular localization reveal interactions between human cytomegalovirus terminase subunits. Virol. J. 2012, 9, 315. [Google Scholar] [CrossRef]
- Borst, E.M.; Ständker, L.; Wagner, K.; Schulz, T.F.; Forssmann, W.G.; Messerle, M. A peptide inhibitor of cytomegalovirus infection from human hemofiltrate. Antimicrob. Agents Chemother. 2013, 57, 4751–4760. [Google Scholar] [CrossRef]
- Borst, E.M.; Benkartek, C.; Messerle, M. Use of bacterial artificial chromosomes in generating targeted mutations in human and mouse cytomegaloviruses. Current Protocols in Immunology, John Wiley & Sons: New York, NY, USA, 2007; Chapter 10, Unit 10.32. 10.32.1–10.32.30. [Google Scholar] [CrossRef]
- Tischer, B.K.; Smith, G.A.; Osterrieder, N. En passant mutagenesis: A two step markerless red recombination system. Methods Mol. Biol. 2010, 634, 421–430. [Google Scholar] [CrossRef]
- Bridge, E.; Ketner, G. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 1989, 63, 631–638. [Google Scholar]
- Weinberg, D.H.; Ketner, G. A cell line that supports the growth of a defective early region 4 deletion mutant of human adenovirus type 2. Proc. Natl. Acad. Sci. USA 1983, 80, 5383–5386. [Google Scholar] [CrossRef]
- Kaluza Cytometry Software, version 1.2; Beckman Coulter: Brea, CA, USA, 2012.
- Cotten, M. Axxima Pharmaceuticals AG, Munich, Germany. Personal communication, 2001. [Google Scholar]
- Murrell, I.; Tomasec, P.; Wilkie, G.S.; Dargan, D.J.; Davison, A.J.; Stanton, R.J. Impact of sequence variation in the UL128 locus on production of human cytomegalovirus in fibroblast and epithelial cells. J. Virol. 2013, 87, 10489–10500. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Elbasani, E.; Gabaev, I.; Steinbrück, L.; Messerle, M.; Borst, E.M. Analysis of Essential Viral Gene Functions after Highly Efficient Adenofection of Cells with Cloned Human Cytomegalovirus Genomes. Viruses 2014, 6, 354-370. https://doi.org/10.3390/v6010354
Elbasani E, Gabaev I, Steinbrück L, Messerle M, Borst EM. Analysis of Essential Viral Gene Functions after Highly Efficient Adenofection of Cells with Cloned Human Cytomegalovirus Genomes. Viruses. 2014; 6(1):354-370. https://doi.org/10.3390/v6010354
Chicago/Turabian StyleElbasani, Endrit, Ildar Gabaev, Lars Steinbrück, Martin Messerle, and Eva Maria Borst. 2014. "Analysis of Essential Viral Gene Functions after Highly Efficient Adenofection of Cells with Cloned Human Cytomegalovirus Genomes" Viruses 6, no. 1: 354-370. https://doi.org/10.3390/v6010354




