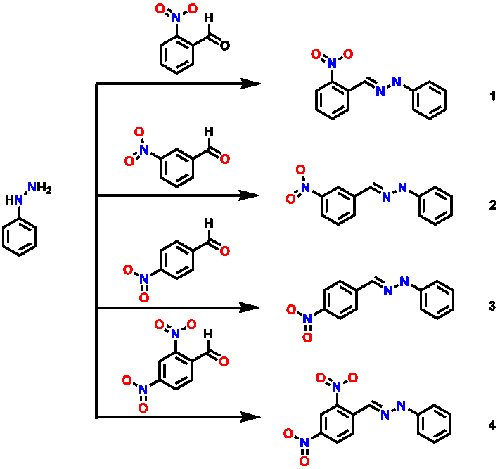
Potential Amoebicidal Activity of Hydrazone Derivatives: Synthesis, Characterization, Electrochemical Behavior, Theoretical Study and Evaluation of the Biological Activity
Abstract
:1. Introduction
2. Results and Discussion

| Chemical Shift | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| a | 6.87 | 6.85 | 6.86 | 6.82 |
| b | 7.27 | 7.27 | 7.26 | 7.26 |
| c | 7.12 | 7.14 | 7.21 | 7.13 |
| d | 9.15 | 8.98 | 10.0 | 11.42 |
| e | 8.23 | 7.85 | 7.94 | 8.63 |
| f | -- | 8.98 | 7.88 | -- |
| g | 8.19 | -- | 8.22 | 8.09 |
| h | 7.44 | 8.04 | -- | -- |
| i | 7.64 | 7.58 | 8.22 | 8.41 |
| j | 7.91 | 8.05 | 7.88 | 8.35 |
2.1. Electrochemistry
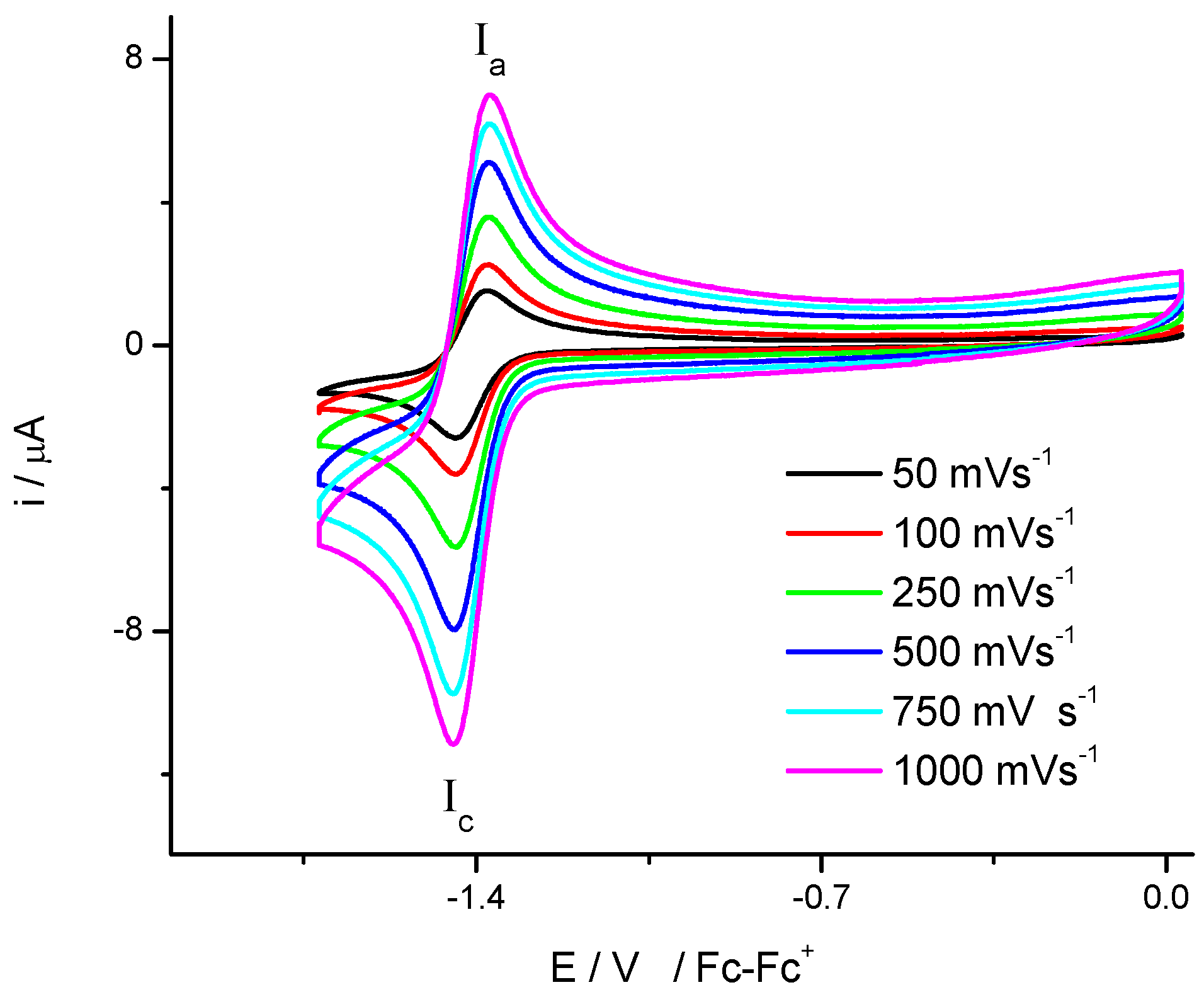
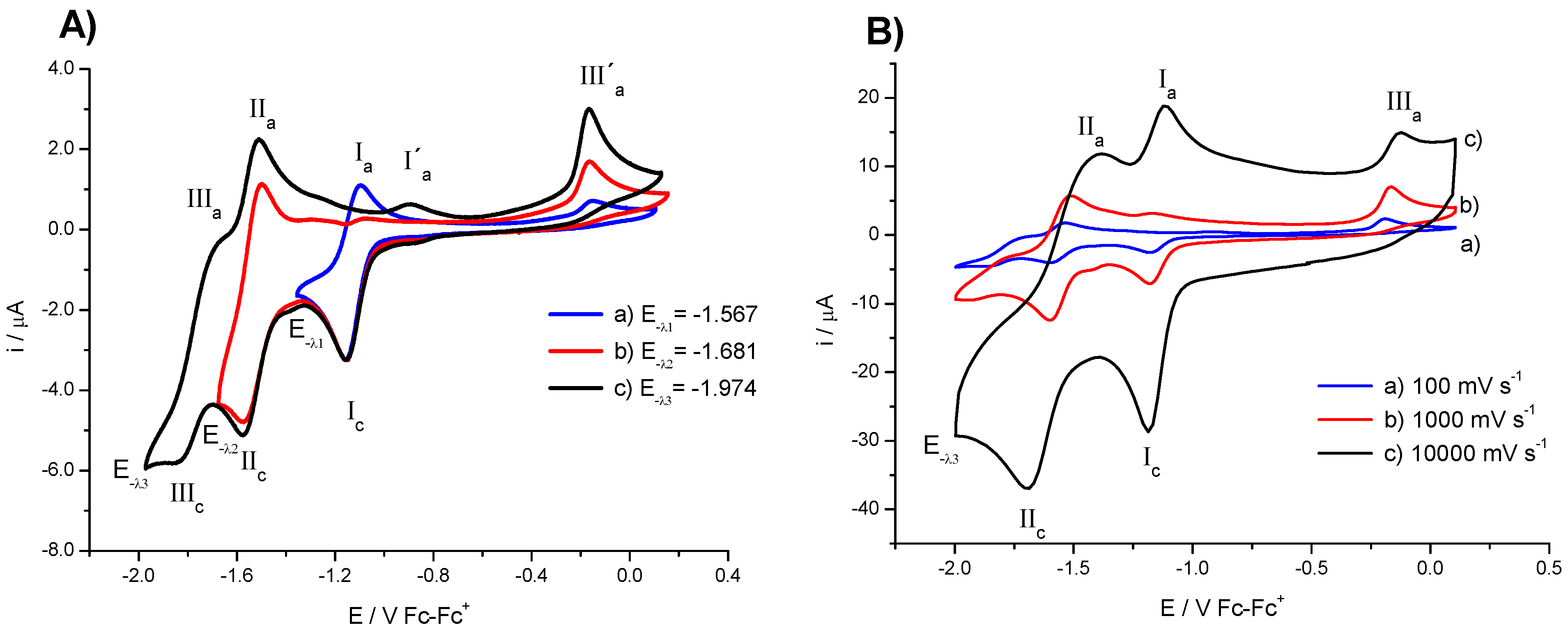
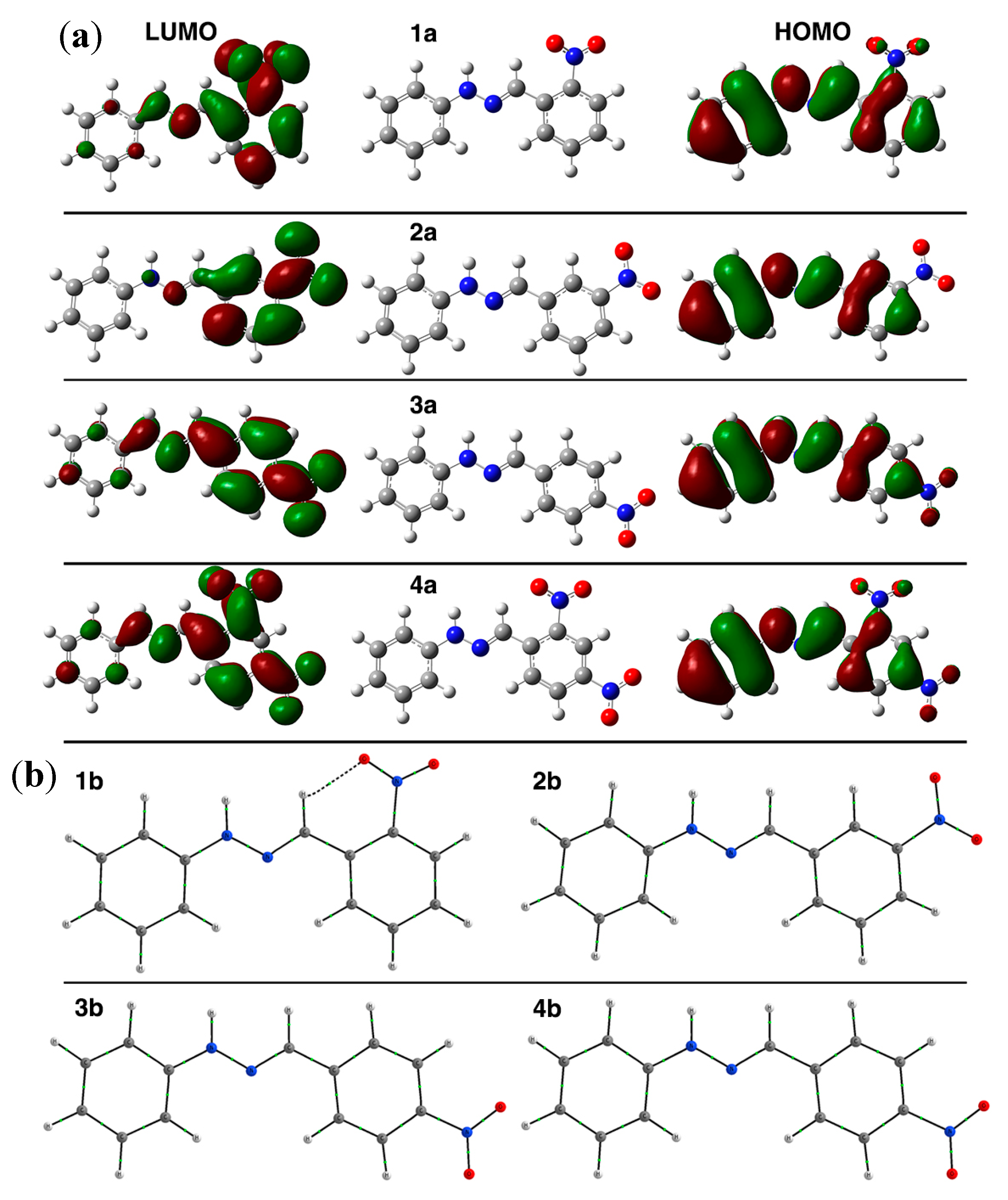
| Compound | E1/2 (I) (V) a | E1/2 (II) (V) a | Do (cm·s−1) | Total Energy d | HOMO d | LUMO d | Energy Gap (H-L) d | IC50 (µM) |
|---|---|---|---|---|---|---|---|---|
| 1 | −1.436 (−0.796) b | n.o. c | 0.94 × 10−5 | −816.045 | −0.21008 | −0.08477 | −0.12531 | 75 |
| 2 | −1.415 (−0.775) b | n.o. c | 1.08 × 10−5 | −816.050 | −0.21551 | −0.09123 | −0.12428 | 0.84 |
| 3 | −1.411 (−0.771) b | n.o. c | 0.91 × 10−5 | −816.051 | −0.22004 | −0.09413 | −0.12591 | 7 |
| 4 | −1.148 (−0.508) b | −1.539 (−0.899) | 0.81 × 10−5 | −1020.45 | −0.22572 | −0.11294 | −0.11278 | 23 |
| Metronidazole | −0.486 b | 6.3 |
2.2. Computational Methods
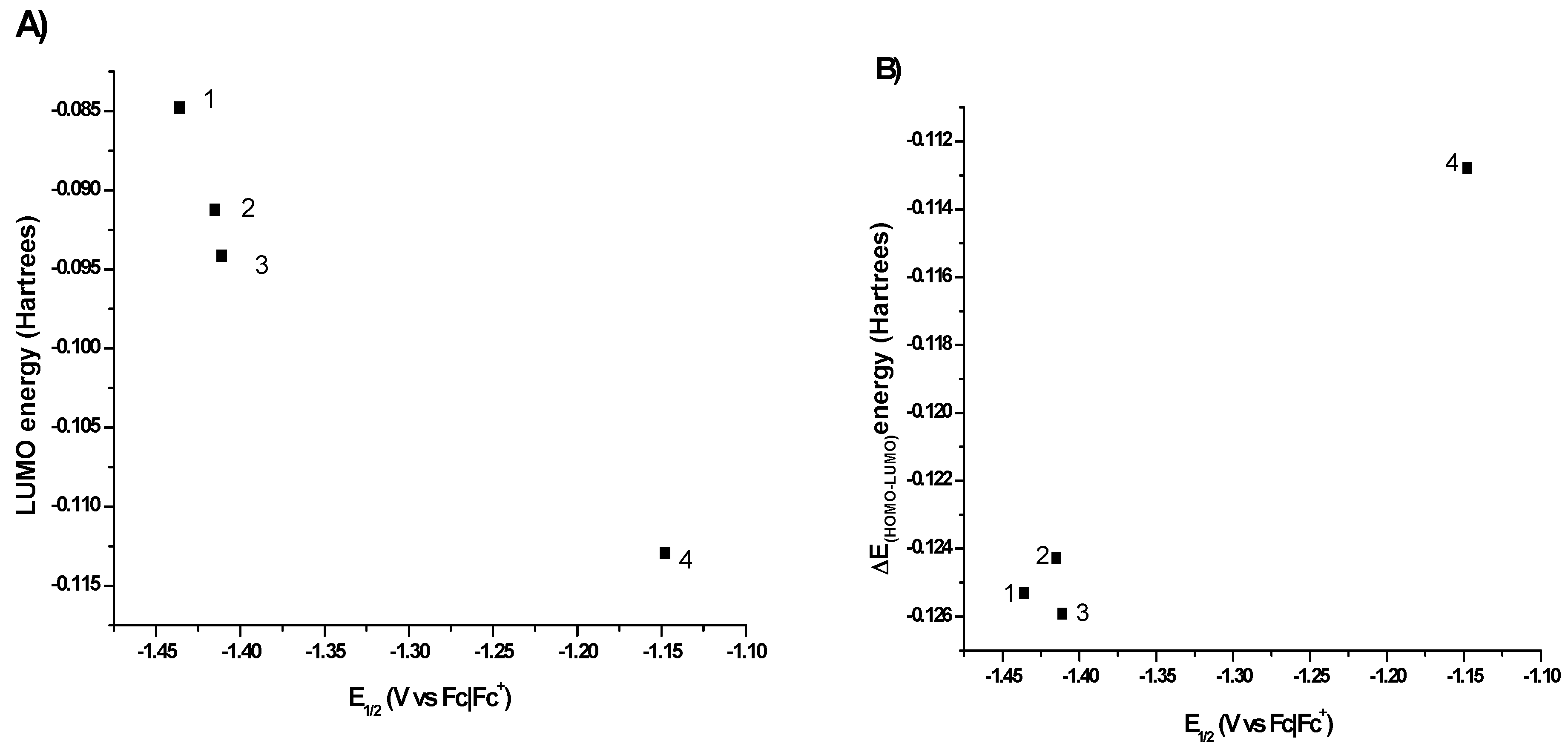

2.3. X-ray Diffraction Studies
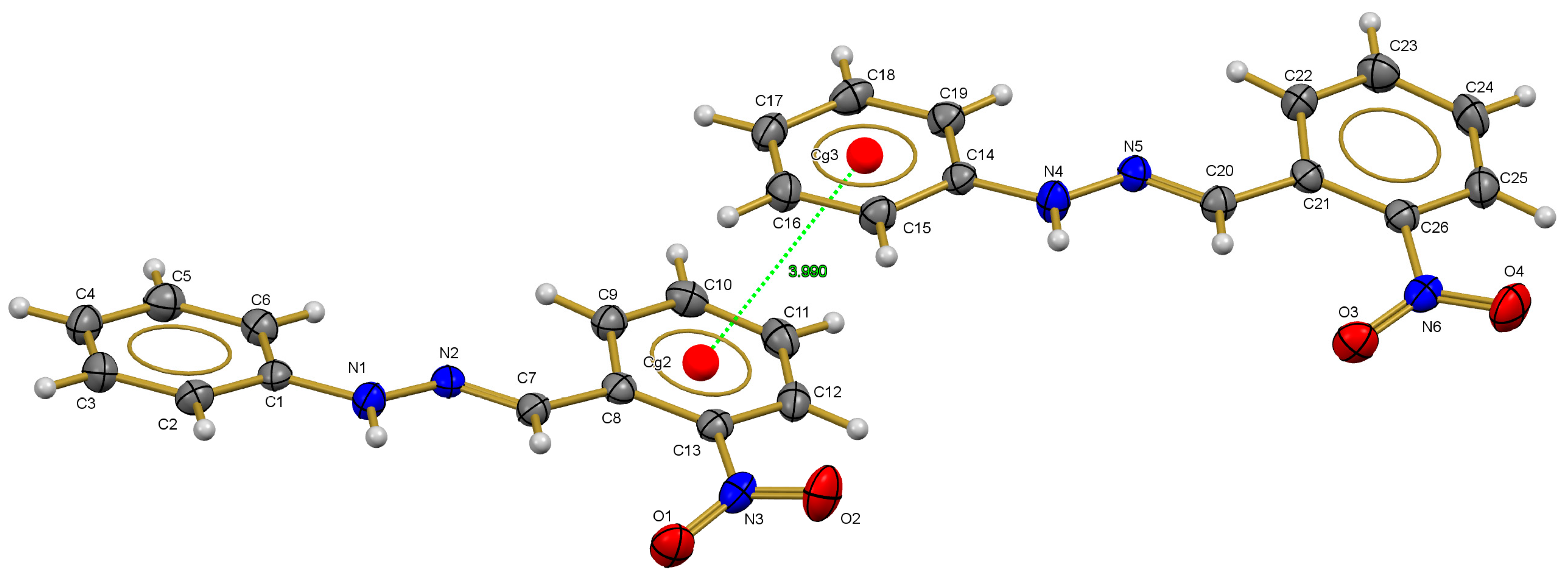
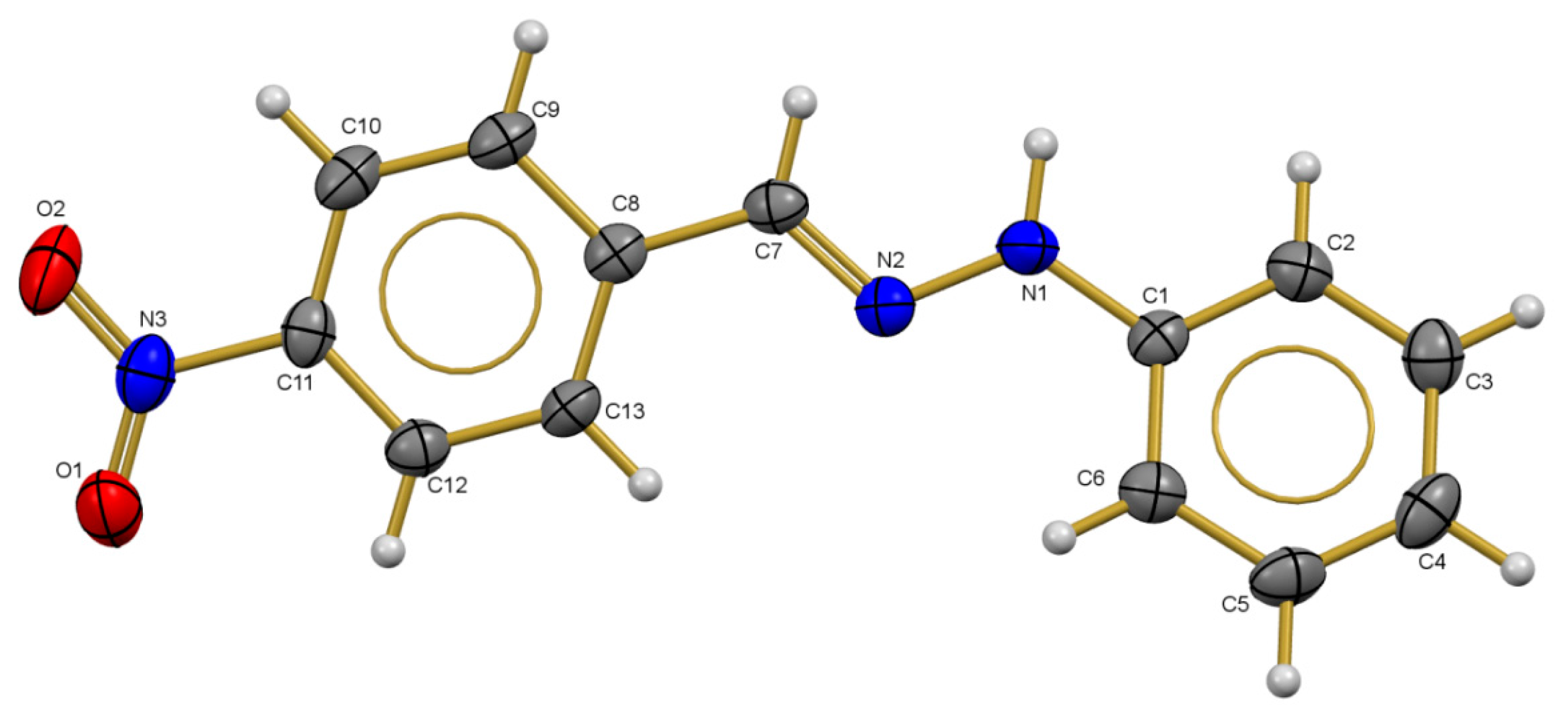
2.4. Amoebicidal Activity
| Compound a | #Carom-N | N-N | N=C | C-*Carom | *Carom-NO2 | N-O | Ref. |
|---|---|---|---|---|---|---|---|
| 1 | 1.407 (1.393) | 1.341 (1.327) | 1.286 (1.288) | 1.450 (1.460) | 1.489 (1.469) | 1.219 (1.220), 1.216 (1.224) | This work |
| 3 | 1.381 (1.394) | 1.354 (1.327) | 1.282 (1.286) | 1.455 (1.451) | 1.464 (1.463) | 1.242 (1.221), 1.226 (1.222) | This work |
| (E)-1-(2-Nitrobenzylidene)-2,2-diphenylhydrazine | 1.471 | 1.287 | 1.359 | 1.441 | 1.467 | 1.226, 1.227 | [36] |
| (E)-1-(4-Nitrobenzylidene)-2,2-diphenylhydrazine | 1.437 | 1.363 | 1.285 | 1.456 | 1.464 | 1.216, 1.218 | [37] |
| (E)-1-(2,4-Dinitrobenzylidene)-2,2-diphenylhydrazine | 1.434 | 1.353 | 1.289 | 1.459 | 1.468 | 1.219, 1.212 | [38] |
| (E)-1-(Benzylidene)-2,2-diphenylhydrazine | 1.441 | 1.369 | 1.279 | 1.469 | ------ | ------ | [39] |
| (E)-Benzaldehyde phenylhydrazone | 1.436 | 1.330 | 1.312 | 1.416 | ------ | ------ | [40] |
| (E)-2-Nitrobenzaldehyde-4-nitrophenylhydrazone | 1.369 | 1.355 | 1.283 | 1.467 | 1.471 | 1.227, 1.225 | [41] |
| (E)-2-Nitrobenzaldehyde-4-nitrophenylhydrazone | 1.371 | 1.351 | 1.275 | 1.464 | 1.471 | 1.215, 1.212 | [42] |
| (E)-Benzaldehyde-2,4-dinitrophenylhydrazone | 1.350 | 1.374 | 1.275 | 1.461 | ------ | ------ | [43] |

3. Experimental Section
3.1. Synthesis and Characterization
3.2. Electrochemistry
3.3. X-ray Crystallography. Experimental Part
| Identification Code | 1 | 3 |
|---|---|---|
| Empirical formula | C13 H11 N3 O2 | C13 H11 N3 O2 |
| Formula weight | 241.25 | 241.25 |
| Temperature | 130(2) K | 298(2) K |
| Wavelength | 0.71073 Å | 0.71073 Å |
| Crystal system | Orthorhombic | Orthorhombic |
| Space group | P b c a | P 21 21 21 |
| Unit cell dimensions | a = 19.0466(8) Å | a = 6.0462(11) Å |
| b = 12.0301(6) Å | b = 11.3924(17) Å | |
| c = 19.8489(7) Å | c = 17.004(3) Å | |
| Volume | 4548.0(3) Å3 | 1171.3(3) Å3 |
| Z | 16 | 4 |
| Density (calculated) | 1.409 Mg/m3 | 1.368 Mg/m3 |
| Absorption coefficient | 0.099 mm−1 | 0.096 mm−1 |
| F(000) | 2016 | 504 |
| Crystal size | 0.57 × 0.35 × 0.17 mm3 | 0.40 × 0.29 × 0.22 mm3 |
| Theta range for data collection | 3.539 to 29.501° | 3.576 to 29.499° |
| Index ranges | −26 ≤ h ≤ 24, −9 ≤ k ≤ 15, −27 ≤ l ≤ 18 | −8 ≤ h ≤ 7, −10 ≤ k ≤ 15, −15 ≤ l ≤ 22 |
| Reflections collected | 16711 | 3932 |
| Independent reflections | 5478 [R(int) = 0.0300] | 2571 [R(int) = 0.0226] |
| Completeness to theta = 25.242° | 99.8% | 99.8% |
| Refinement method | Full-matrix least-squares on F2 | Full-matrix least-squares on F2 |
| Data/restraints/parameters | 5478/0/331 | 2571/0/166 |
| Goodness-of-fit on F2 | 1.017 | 1.094 |
| Final R indices [I > 2 sigma(I)] | R1 = 0.0444, wR2 = 0.0925 | R1 = 0.0389, wR2 = 0.0868 |
| R indices (all data) | R1 = 0.0736, wR2 = 0.1055 | R1 = 0.0448, wR2 = 0.0929 |
| Largest diff. peak and hole | 0.232 and −0.249 e.Å−3 | 0.149 and −0.231 e.Å−3 |
3.4. Amoebicidal Activity
3.4.1. Parasite culture
3.4.2. Parasite Viability
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Brandt, H.; Pérez-Tamayo, R. Pathology of human amebiasis. Hum. Pathol. 1970, 1, 351–385. [Google Scholar] [CrossRef]
- Stanley, S.L., Jr. Amoebiasis. Lancet 2003, 361, 1025–1034. [Google Scholar] [CrossRef]
- Fotedar, R.; Stark, D.; Beebe, N.; Marriot, D.; Ellis, J.; Harkness, J. Laboratory Diagnostic Techniques for Entamoeba Species. Clin. Microbiol. Rev. 2007, 20, 511–532. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Available online: http://www.who.int/ith/diseases/amoebiasis/en/ (accessed on 2 February 2015).
- Johnson, P.J. Metronidazole and drug resistance. Parasitol. Today 1993, 9, 183–186. [Google Scholar] [CrossRef]
- Kim, D.; Park, J.; Yoon, B.; Baek, M.J.; Kim, J.E.; Kim, S.Y. Metronidazole-induced encephalopathy. J. Neurol. Sci. 2004, 224, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Kuriyama, A.; Jackson, J.L.; Doi, A.; Kamiya, T. Metronidazole-induced central nervous system toxicity: A systematic review. Clin. Neuropharmacol. 2011, 34, 241–247. [Google Scholar] [CrossRef] [PubMed]
- National Toxicology Program. Metronidazole. Rep. Carcinog. 2011, 12, 269–270. [Google Scholar]
- Goldman, P.; Koch, R.L.; Yeung, T.C.; Chrystal, E.J.; Beaulieu, B.B., Jr.; McLafferty, M.A.; Sudlow, G. Comparing the reduction of nitroimidazoles in bacteria and mammalian tissues and relating it to biological activity. Biochem. Pharmacol. 1986, 35, 43–51. [Google Scholar] [CrossRef]
- Singh, S.; Bharti, N.; Mohapatra, P.P. Chemistry and Biology of Synthetic and Naturally Occurring Antiamoebic Agents. Chem. Rev. 2009, 109, 1900–1947. [Google Scholar] [CrossRef] [PubMed]
- Wani, M.Y.; Bath, A.R.; Azam, A.; Athar, F. Nitroimidazolyl hydrazones are better amoebicides than their cyclized 1,3,4-oxadiazoline analogues: In vitro studies and Lipophilic efficiency analysis. Eur. J. Med. Chem. 2013, 64, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Saddiqui, S.M.; Salahuddin, A.; Azam, A. Synthesis, characterization and antiamoebic activity of some hydrazone and azole derivatives bearing pyridyl moiety as a promising heterocyclic scaffold. Eur. J. Med. Chem. 2012, 49, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Toledano-Magaña, Y.; Meléndrez-Luévano, R.; Navarro-Olivarria, M.; García-Ramos, J.C.; Flores-Alamo, M.; Ortiz-Frade, L.; Ruiz-Azuara, L.; Cabrera-Vivas, B.M. Synthesis, characterization and evaluation of the substituent effect on the amoebicide activity of new hydrazone derivatives. Med. Chem. Commun. 2014, 5, 989–996. [Google Scholar] [CrossRef]
- Navakovski de Oliveira, K.; Costa, P.; Santin, J.R.; Mazzambani, L.; Bürger, C.; Mora, C.; Nunes, R.J.; de Souza, M.M. Synthesis and antidepressant-like activity evaluation of sulphonamides and sulphonyl-hydrazones. Bioorg. Med. Chem. 2011, 19, 4295–4306. [Google Scholar] [CrossRef] [PubMed]
- Can, O.D.; Altintop, M.D.; Ozkay, U.D.; Uçel, U.I.; Doğruer, B.; Kaplancikli, Z.A. Synthesis of thiadiazole derivatives bearing hydrazone moieties and evaluation of their pharmacological effects on anxiety, depression, and nociception parameters in mice. Arch. Pharm. Res. 2012, 35, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Lacerda, R.B.; da Silva, L.L.; de Lima, C.K.F.; Miguez, E.; Miranda, A.L.P.; Laufer, S.A.; Barreiro, E.J.; Fraga, C.A.M. Discovery of Novel Orally Active Anti-Inflammatory N-Phenylpyrazolyl-N-Glycinyl-Hydrazone Derivatives That Inhibit TNF-α Production. PLoS ONE 2012, 7, e46925. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Narasimhan, B. Hydrazides/hydrazones as antimicrobial and anticancer agents in the new millennium. Mini Rev. Med. Chem. 2013, 13, 971–987. [Google Scholar] [CrossRef] [PubMed]
- Pieczonka, A.M.; Strzelczyk, A.; Sadowska, B.; Mlostoń, G.; Stączek, P. Synthesis and evaluation of antimicrobial activity of hydrazones derived from 3-oxido-1H-imidazole-4-carbohydrazides. Eur. J. Med. Chem. 2013, 64, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Dolla, N.K.; Casadei, G.; Bremner, J.B.; Lewis, K.; Kelso, M.J. Diarylacylhydrazones: Clostridium-selective antibacterials with activity against stationary-phase cells. Bioorg. Med. Chem. 2014, 24, 595–600. [Google Scholar] [CrossRef] [PubMed]
- More, U.A.; Joshi, S.D.; Aminabhavi, T.M.; Gadad, A.K.; Nadagouda, M.N.; Kulkarni, V.H. Design, synthesis, molecular docking and 3D-QSAR studies of potent inhibitors of enoyl-acyl carrier protein reductase as potential antimycobacterial agents. Eur. J. Med. Chem. 2014, 71, 199–216. [Google Scholar] [CrossRef] [PubMed]
- Coimbra, E.S.; Antinarelli, L.M.R.; da Silva, A.D.; Bispo, M.L.F.; Kaiser, C.R.; de Souza, M.V.N. 7-Chloro-4-quinolinyl Hydrazones: A Promising and Potent Class of Antileishmanial Compounds. Chem. Biol. Drug Des. 2013, 81, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Moreira, D.R.; Lima Leite, A.C.; Cardoso, M.V.; Srivastava, R.M.; Hernandes, M.Z.; Rabello, M.M.; da Cruz, L.F.; Ferreira, R.S.; de Simone, C.A.; Meira, C.S.; et al. Structural Design, Synthesis and Structure-Activity Relationships of Thiazolidinones with Enhanced Anti-Trypanosomacruzi Activity. ChemMedChem 2014, 9, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Flores-Alamo, M.; Cabrera-Vivas, B.M.; Meléndrez-Luévano, R.; Hernández, P.J.M.; Ruiz-Azuara, L. 1-{(E)-[5-(2-Nitro-phen-yl)furan-2-yl]methyl-idene}-2,2-diphenyl-hydrazine. Acta Crystallogr. E. 2013, 69. [Google Scholar] [CrossRef] [PubMed]
- Bard, A.J.; Faulkner, L.R. Electrochemical Methods, Fundamentals and Applications, 2nd ed.; John Wiley and Sons: New York, NY, USA, 2001. [Google Scholar]
- Amatore, C.; Capobianco, G.; Farnia, G.; Sandona, G.; Savéant, J.M.; Severin, M.G.; Vianello, E. Kinetics and mechanism of self-protonation reactions in organic electrochemical processes. J. Am. Chem. Soc. 1985, 107, 1815–1824. [Google Scholar] [CrossRef]
- Bader, R.F.W. Atoms in Molecules: A Quantum Theory; Oxford University Press: Oxford, UK, 1990. [Google Scholar]
- Liu, X.H.; Pan, L.; Tan, C.X.; Weng, J.Q.; Wang, B.L.; Li, Z.M. Synthesis, Crystal structure, bioactivity and DFT calculation of new oximeester derivatives containing cyclopropane moiety. Pestic. Biochem. Physiol. 2011, 101, 143–147. [Google Scholar] [CrossRef]
- Liu, X.H.; Zhao, W.G.; Wang, B.L.; Li, Z.M. Synthesis, bioactivity and DFT structure activity relationship study of a novel 1,2,3-thiadiazole derivatives. Res. Chem. Intermed. 2012, 38, 1999–2008. [Google Scholar] [CrossRef]
- Lundgren, R.J.; Stradiotto, M. Palladium-Catalyzed Cross-Coupling of Aryl Chlorides and Tosylates with Hydrazine. Angew. Chem. Int. Edit. 2010, 49, 8686–8690. [Google Scholar] [CrossRef] [PubMed]
- Grzesiek, S.; Cordier, F.; Jaravine, V.; Barfield, M. Insights into biomolecular hydrogen bonds from hydrogenbond scalar couplings. Prog. Nucl. Magn. Reson. Spectrosc. 2004, 45, 275–300. [Google Scholar] [CrossRef]
- Allen, F.H.; Kennard, O.; Watson, D.G.; Brammer, L.; Orpen, A.G.; Taylor, R. Tables of bond lengths determined by X-ray and Neutron diffraction. Part 1. Bond length in Organic Compounds. J. Chem. Soc. Perkin Trans. 2 1987, 12, S1–S19. [Google Scholar] [CrossRef]
- Tekwani, B.L.; Mehlotra, R.K. Molecular basis of defense against oxidative stress in Entamoeba histolytica and Giardia lamblia. Microbes Infect. 1999, 1, 385–394. [Google Scholar] [CrossRef]
- Samuelson, J. Why metronidazole is active against both bacteria and parasites. Antimicrob. Agents Chemother. 1999, 43, 1533–1541. [Google Scholar] [PubMed]
- Samarawickrema, N.A.; Brown, D.M.; Upcroft, J.A.; Thammapalerd, N.; Upcroft, P. Involvement of superoxide dismutase and pyruvate:ferredoxin oxidoreductase in mechanisms of metronidazole resistance in Entamoeba histolytica. J. Antimicrob. Chemother. 1997, 40, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Wassmann, C.; Helberg, A.; Tannich, E.; Bruchhaus, I. Metronidazole Resistance in the Protozoan Parasite Entamoeba histolytica Is Associated with Increased Expression of Iron-containing Superoxide Dismutase and Peroxiredoxin and Decreased Expression of Ferredoxin 1 and Flavin Reductase. J. Biol. Chem. 1999, 274, 26051–26056. [Google Scholar] [CrossRef] [PubMed]
- Flores-Alamo, M.; Meléndrez-Luevano, R.; Ortiz-Márquez, J.A.; Sansinenea Royano, E.; Cabrera-Vivas, B.M. (E)-1-(2-nitrobenzylidene)-2,2-diphenylhydrazine. Acta Crystallogr. E 2014, 70, o909–o910. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, A.; Meléndrez-Luevano, R.; Cabrera-Vivas, B.M.; Acoltzi-X, C.; Flores-Alamo, M. (E)-1-(4-nitrobenzylidene)-2,2-diphenylhydrazine. Acta Crystallogr. E 2012, 68, o3238. [Google Scholar] [CrossRef] [PubMed]
- Meléndrez-Luevano, R.; Cabrera-Vivas, B.M.; Flores-Alamo, M.; Ramírez, J.C.; Conde-Sánchez, P. (E)-1-(2,4-dinitrobenzylidene)-2,2-diphenylhydrazine. Acta Crystallogr. E 2013, 69, o1039. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, A.; Meléndrez-Luevano, R.; Cabrera-Vivas, B.M.; Lozano-Marquez, C.D.; Carranza, V. (E)-1-Benzylidene-2,2-diphenylhydrazine. Acta Crystallogr. E 2012, 68, o434. [Google Scholar] [CrossRef] [PubMed]
- Vickery, B.; Willey, G.R.; Drew, M.G.B. (E)-Benzaldehyde phenylhydrazone. Acta Crystallogr. 1985, 41, 1072–1075. [Google Scholar] [CrossRef]
- Wardell, J.L.; Skakle, J.M.S.; Low, J.N.; Glidewell, C. Hydrogen bonded sheets in (E)-2-nitrobenzaldehyde 4-nitrophenylhydrazone and a hydrogen-bonded framework structure in (E)-4-nitrobenzaldehyde-4-nitrophenylhydrazone. Acta Crystallogr. C 2005, 61, o10–o14. [Google Scholar] [CrossRef] [PubMed]
- Shan, S.; Wang, X.J.; Hu, W.X.; Xu, D.J. 2-Nitrobenzaldehyde-4-nitrophenylhydrazone. Acta Crystallogr. E 2004, 60, o1954–o1956. [Google Scholar] [CrossRef]
- Shan, S.; Xu, D.J.; Hung, C.H.; Wu, J.Y.; Chiang, M.J. Benzaldehyde-2,4-dinitrophenylhydrazone. Acta Crystallogr. C 2003, 59, o135–o136. [Google Scholar] [CrossRef] [PubMed]
- Gritzner, G.; Küta, J. Recommendations on reporting electrode potentials in nonaqueous solvents. Pure Appl. Chem. 1984, 4, 461–466. [Google Scholar] [CrossRef]
- CrysAlisPro—Data Collection and Processing Software for Agilent X-ray Diffractometers; Agilent Technologies: Yarnton, UK, 2012. Available online: http://www.chem.agilent.com/Library/usermanuals/Public/CrysAlis_Pro_User_Manual.pdf (accessed on 3 May 2015).
- Clark, R.C.; Reid, J.S. The analytical calculation of absorption in multifaceted crystals. Acta Crystallogr. A 1995, 51, 887–897. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. A 2008, 64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, L.J. WinGX suite for small-molecule single-crystal crystallography. J. Appl. Crystallogr. 1999, 32, 837–838. [Google Scholar] [CrossRef]
- Diamond, L.S. Axenic cultivation of Entamoeba histolytica. Science 1961, 134, 336–337. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds 1–4 are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toledano-Magaña, Y.; García-Ramos, J.C.; Navarro-Olivarria, M.; Flores-Alamo, M.; Manzanera-Estrada, M.; Ortiz-Frade, L.; Galindo-Murillo, R.; Ruiz-Azuara, L.; Meléndrez-Luevano, R.M.; Cabrera-Vivas, B.M. Potential Amoebicidal Activity of Hydrazone Derivatives: Synthesis, Characterization, Electrochemical Behavior, Theoretical Study and Evaluation of the Biological Activity. Molecules 2015, 20, 9929-9948. https://doi.org/10.3390/molecules20069929
Toledano-Magaña Y, García-Ramos JC, Navarro-Olivarria M, Flores-Alamo M, Manzanera-Estrada M, Ortiz-Frade L, Galindo-Murillo R, Ruiz-Azuara L, Meléndrez-Luevano RM, Cabrera-Vivas BM. Potential Amoebicidal Activity of Hydrazone Derivatives: Synthesis, Characterization, Electrochemical Behavior, Theoretical Study and Evaluation of the Biological Activity. Molecules. 2015; 20(6):9929-9948. https://doi.org/10.3390/molecules20069929
Chicago/Turabian StyleToledano-Magaña, Yanis, Juan Carlos García-Ramos, Marisol Navarro-Olivarria, Marcos Flores-Alamo, Mayra Manzanera-Estrada, Luis Ortiz-Frade, Rodrigo Galindo-Murillo, Lena Ruiz-Azuara, Ruth Ma. Meléndrez-Luevano, and Blanca M. Cabrera-Vivas. 2015. "Potential Amoebicidal Activity of Hydrazone Derivatives: Synthesis, Characterization, Electrochemical Behavior, Theoretical Study and Evaluation of the Biological Activity" Molecules 20, no. 6: 9929-9948. https://doi.org/10.3390/molecules20069929