pH-Dependent Deformations of the Energy Landscape of Avidin-like Proteins Investigated by Single Molecule Force Spectroscopy
Abstract
:1. Introduction
2. Results and Discussion
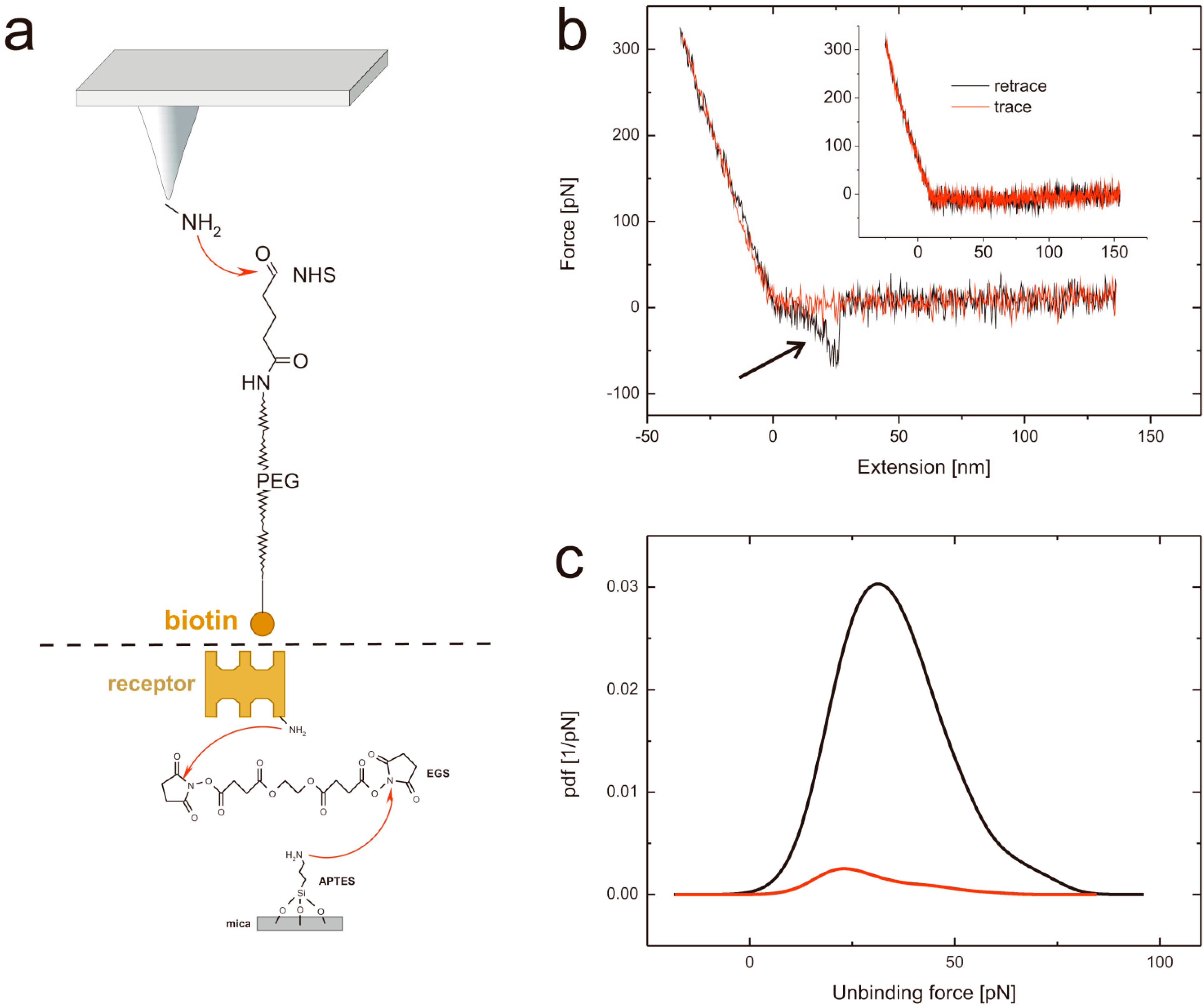
2.1. Tip and Support Functionalization
2.2. Force Distance Cycles, Data Evaluation and Specificity Proof Experiments
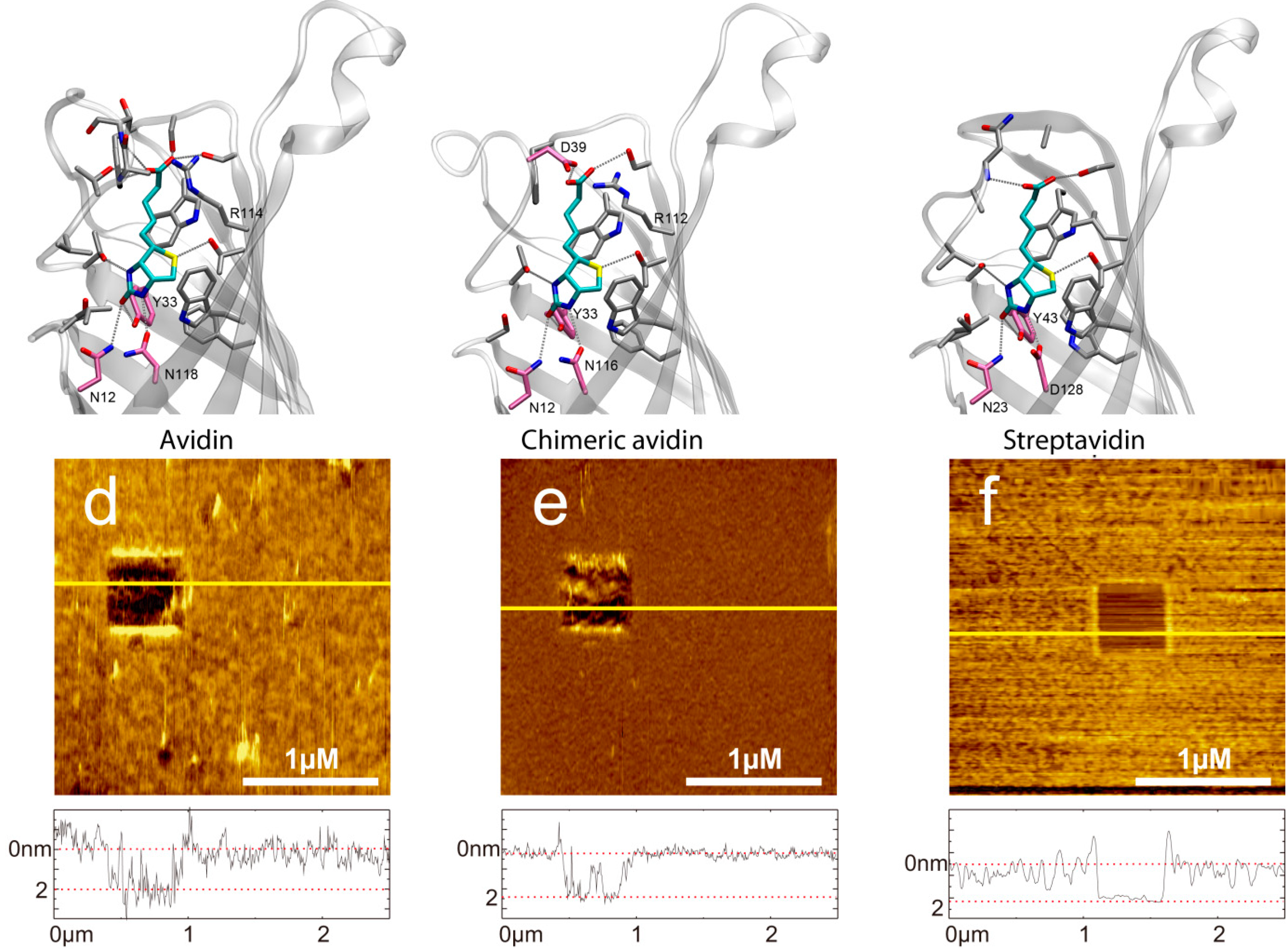
2.3. Influence of the pH Value on the Binding Probability

| Avidinkoff [s-1] | xβ [Å] | Chimeric avidinkoff [s-1] | xβ [Å] | |
|---|---|---|---|---|
| pH 1 | 2.08 ± 0.08 | 4.18 ± 0.04 | 0.76 ± 0,05 | 7.31 ± 0.08 |
| pH 2 | 3.00 ± 0.18 | 3.59 ± 0.07 | 2.10 ± 0.12 | 4.01 ± 0.06 |
| pH 3 | 3.93 ± 0.19 | 4.70 ± 0.07 | 2.13 ± 0. 18 | 5.12 ± 0.10 |
| pH 7 | 2.37 ± 0.14 | 4.49 ± 0.07 | 2.74 ± 0.18 | 4.29 ± 0.08 |
| pH 11 | 0.88 ± 0.07 | 4.52 ± 0.08 | 2.22 ± 0.13 | 3.89 ± 0.06 |
2.4. pH Induced Changes of the Bond Energy Landscape
3. Experimental Section
3.1. Materials
3.1.1. Chemicals
3.1.2. Cantilevers
3.1.3. Buffer
3.2. Methods
3.2.1. AFM Tip Functionalization for SMFS Experiments
3.2.2. Sample Preparation
3.3. Experimental Approach
3.3.1. Single Molecule Force Spectroscopy
3.3.2. AFM Imaging
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Määttä, J.A.; Eisenberg‐Domovich, Y.; Nordlund, H.R.; Hayouka, R.; Kulomaa, M.S.; Livnah, O.; Hytönen, V.P. Chimeric avidin shows stability against harsh chemical conditions—biochemical analysis and 3D structure. Biotechnol. Bioeng. 2011, 108, 481–490. [Google Scholar]
- Hytönen, V.P.; Määttä, J.A.; Nyholm, T.K.; Livnah, O.; Eisenberg-Domovich, Y.; Hyre, D.; Nordlund, H.R.; Hörhä, J.; Niskanen, E.A.; Paldanius, T.; et al. Design and construction of highly stable, protease-resistant chimeric avidins. J. Biol. Chem. 2005, 280, 10228–10233. [Google Scholar] [CrossRef]
- Chivers, C.E.; Crozat, E.; Chu, C.; Moy, V.T.; Sherratt, D.J.; Howarth, M. A streptavidin variant with slower biotin dissociation and increased mechanostability. Nat. Methods 2010, 7, 391–393. [Google Scholar] [CrossRef]
- Wilchek, M.; Bayer, E.A.; Livnah, O. Essentials of biorecognition: The avidin–biotin system as a model for protein–protein and protein–ligand interaction. Immunol. Lett. 2006, 103, 27–32. [Google Scholar] [CrossRef]
- Wilchek, M.; Bayer, E.A. Avidin-Biotin Technology. Methods Enzymol. 1990, 184. [Google Scholar]
- Laitinen, O.H.; Nordlund, H.R.; Hytönen, V.P.; Kulomaa, M.S. Brave new avidins in biotechnology. Trends Biotechnol. 2007, 25, 269–277. [Google Scholar]
- Green, N. Avidin. 4. Stability at extremes of pH and dissociation into sub-units by guanidine hydrochloride. Biochem. J. 1963, 89, 609–620. [Google Scholar]
- Wei, R.-D.; Wright, L.D. Heat Stability of Avidin and Avidin-Biotin Complex and Influence of Ionic Strength on Affinity of Avidin for Biotin. Exp. Biol. Med. 1964, 117, 341–344. [Google Scholar] [CrossRef]
- Evans, E. Energy landscapes of biomolecular adhesion and receptor anchoring at interfaces explored with dynamic force spectroscopy. Faraday Discuss 1998, 1–16. [Google Scholar]
- Merkel, R.; Nassoy, P.; Leung, A.; Ritchie, K.; Evans, E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature 1999, 397, 50–53. [Google Scholar]
- Israelachvili, J.; Min, Y.; Akbulut, M.; Alig, A.; Carver, G.; Greene, W.; Kristiansen, K.; Meyer, E.; Pesika, N.; Rosenberg, K.; et al. Recent advances in the surface forces apparatus (SFA) technique. Rep. Prog. Phys. 2010, 73, 036601. [Google Scholar] [CrossRef]
- Wong, J.; Chilkoti, A.; Moy, V.T. Direct force measurements of the streptavidin–biotin interaction. Biomol. Eng. 1999, 16, 45–55. [Google Scholar] [CrossRef]
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491. [Google Scholar] [CrossRef]
- Pincet, F.; Husson, J. The solution to the streptavidin-biotin paradox: The influence of history on the strength of single molecular bonds. Biophys. J. 2005, 89, 4374–4381. [Google Scholar] [CrossRef]
- Florin, E.L.; Moy, V.T.; Gaub, H.E. Adhesion forces between individual ligand-receptor pairs. Science 1994, 264, 415–7. [Google Scholar]
- Moy, V.T.; Florin, E.L.; Gaub, H.E. Intermolecular forces and energies between ligands and receptors. Science 1994, 266, 257–257. [Google Scholar]
- Lee, G.U.; Kidwell, D.A.; Colton, R.J. Sensing Discrete Streptavidin-Biotin Interactions with Atomic Force Microscopy. Langmuir 1994, 10, 354–357. [Google Scholar] [CrossRef]
- De Paris, R.; Strunz, T.; Oroszlan, K.; Güntherodt, H.-J.; Hegner, M. Force spectroscopy and dynamics of the biotin-avidin bond studied by scanning force microscopy. Single Mol. 2000, 1, 285–290. [Google Scholar] [CrossRef]
- Yuan, C.; Chen, A.; Kolb, P.; Moy, V.T. Energy landscape of streptavidin-biotin complexes measured by atomic force microscopy. Biochemistry 2000, 39, 10219–10223. [Google Scholar]
- Chilkoti, A.; Boland, T.; Ratner, B.D.; Stayton, P.S. The relationship between ligand-binding thermodynamics and protein-ligand interaction forces measured by atomic force microscopy. Biophysi. J. 1995, 69, 2125–2130. [Google Scholar] [CrossRef]
- Rangl, M.; Leitner, M.; Riihimäki, T.; Lehtonen, S.; Hytönen, V.P.; Gruber, H.J.; Kulomaa, M.; Hinterdorfer, P.; Ebner, A. Investigating the binding behaviour of two avidin-based testosterone binders using molecular recognition force spectroscopy. J. Mol. Recognit. 2014, 27, 92–97. [Google Scholar] [CrossRef]
- Rico, F.; Moy, V.T. Energy landscape roughness of the streptavidin–biotin interaction. J. Mol. Recognit. 2007, 20, 495–501. [Google Scholar]
- Taninaka, A.; Takeuchi, O.; Shigekawa, H. Hidden variety of biotin–streptavidin/avidin local interactions revealed by site-selective dynamic force spectroscopy. Phys. Chem. Chem. Phys. 2010, 12, 12578–12583. [Google Scholar] [CrossRef]
- Guo, S.; Ray, C.; Kirkpatrick, A.; Lad, N.; Akhremitchev, B.B. Effects of multiple-bond ruptures on kinetic parameters extracted from force spectroscopy measurements: Revisiting biotin-streptavidin interactions. Biophys. J. 2008, 95, 3964–3976. [Google Scholar]
- Teulon, J.M.; Delcuze, Y.; Odorico, M.; Chen, S.W.W.; Parot, P.; Pellequer, J.L. Single and multiple bonds in (strept) avidin–biotin interactions. J. Mol. Recognit. 2011, 24, 490–502. [Google Scholar] [CrossRef]
- Takeuchi, O.; Miyakoshi, T.; Taninaka, A.; Tanaka, K.; Cho, D.; Fujita, M.; Yasuda, S.; Jarvis, S.P.; Shigekawa, H. Dynamic-force spectroscopy measurement with precise force control using atomic-force microscopy probe. J. Appl. Phys. 2006, 100, 074315. [Google Scholar] [CrossRef]
- Friddle, R.W.; Noy, A.; de Yoreo, J.J. Interpreting the widespread nonlinear force spectra of intermolecular bonds. Proc. Natl. Acad. Sci. USA 2012, 109, 13573–13578. [Google Scholar]
- Hyre, D.E.; Amon, L.M.; Penzotti, J.E.; Le Trong, I.; Stenkamp, R.E.; Lybrand, T.P.; Stayton, P.S. Early mechanistic events in biotin dissociation from streptavidin. Nat. Struct. Mol. Biol. 2002, 9, 582–585. [Google Scholar]
- Zhou, J.; Zhang, L.; Leng, Y.; Tsao, H.K.; Sheng, Y.J.; Jiang, S. Unbinding of the streptavidin-biotin complex by atomic force microscopy: A hybrid simulation study. J. Chem. Phys. 2006, 125, 104905. [Google Scholar]
- Izrailev, S.; Stepaniants, S.; Balsera, M.; Oono, Y.; Schulten, K. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys. J. 1997, 72, 1568–1581. [Google Scholar] [CrossRef]
- Ebner, A.; Hinterdorfer, P.; Gruber, H.J. Comparison of different aminofunctionalization strategies for attachment of single antibodies to AFM cantilevers. Ultramicroscopy 2007, 107, 922–927. [Google Scholar]
- Riener, C.K.; Stroh, C.M.; Ebner, A.; Klampfl, C.; Gall, A.A.; Romanin, C.; Lyubchenko, Y.L.; Hinterdorfer, P.; Gruber, H.J. Simple test system for single molecule recognition force microscopy. Anal. Chim. Acta 2003, 479, 59–75. [Google Scholar] [CrossRef]
- Baumgartner, W.; Hinterdorfer, P.; Schindler, H. Data analysis of interaction forces measured with the atomic force microscope. Ultramicroscopy 2000, 82, 85–95. [Google Scholar] [CrossRef]
- Livnah, O.; Bayer, E.A.; Wilchek, M.; Sussman, J.L. Three-dimensional structures of avidin and the avidin-biotin complex. Proc. Natl. Acad. Sci. USA 1993, 90, 5076–5080. [Google Scholar]
- Repo, S.; Paldanius, T.A.; Hytönen, V.P.; Nyholm, T.K.; Halling, K.K.; Huuskonen, J.; Pentikäinen, O.T.; Rissanen, K.; Slotte, J.P.; Airenne, T.T. Binding properties of HABA-type azo derivatives to avidin and avidin-related protein 4. Chem. Biol. 2006, 13, 1029–1039. [Google Scholar]
- Bell, G.I. Models for the specific adhesion of cells to cells. Science 1978, 200, 618–627. [Google Scholar]
- Strunz, T.; Oroszlan, K.; Schumakovitch, I.; Guntherodt, H.; Hegner, M. Model energy landscapes and the force-induced dissociation of ligand-receptor bonds. Biophys. J. 2000, 79, 1206–1212. [Google Scholar] [CrossRef]
- Noy, A.; Friddle, R.W. Practical single molecule force spectroscopy: How to determine fundamental thermodynamic parameters of intermolecular bonds with an atomic force microscope. Methods 2013, 60, 142–150. [Google Scholar] [CrossRef]
- De Odrowąż Piramowicz, M.; Czuba, P.; Targosz, M.; Burda, K.; Szymoński, M. Dynamic force measurements of avidin–biotin and streptavdin–biotin interactions using AFM. Acta Biochim. Pol. 2006, 53, 93–100. [Google Scholar]
- Hutter, J.L.; Bechhoefer, J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993, 64, 1868–1873. [Google Scholar]
- Kamruzzahan, A.S.M.; Kienberger, F.; Stroh, C.M.; Berg, J.; Huss, R.; Ebner, A.; Zhu, R.; Rankl, C.; Gruber, H.J.; Hinterdorfer, P. Imaging morphological details and pathological differences of red blood cells using tapping-mode AFM. Biol. Chem. 2004, 385, 955–960. [Google Scholar]
- Wildling, L.; Rankl, C.; Haselgrübler, T.; Gruber, H.J.; Holy, M.; Newman, A.H.; Zou, M.-F.; Zhu, R.; Freissmuth, M.; Sitte, H.H. Probing binding pocket of serotonin transporter by single molecular force spectroscopy on living cells. J. Biol. Chem. 2012, 287, 105–113. [Google Scholar] [CrossRef]
- Sample Availability: The NHS-PEG-Biotin linker is commercially available, you can get it also via our institute from Prof. Hermann Gruber (http://www.jku.at/biophysics/content/e54633/e54706/e201881). Also the Avidin and Streptavidin are commercially available and can be brought from Sigma-Aldrich. Chimericavidin is not purchasable, we got this protein mutant from our partner in Finland, Vesa Hytönen and Markuu Kulomaa from the University of Tampere.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Köhler, M.; Karner, A.; Leitner, M.; Hytönen, V.P.; Kulomaa, M.; Hinterdorfer, P.; Ebner, A. pH-Dependent Deformations of the Energy Landscape of Avidin-like Proteins Investigated by Single Molecule Force Spectroscopy. Molecules 2014, 19, 12531-12546. https://doi.org/10.3390/molecules190812531
Köhler M, Karner A, Leitner M, Hytönen VP, Kulomaa M, Hinterdorfer P, Ebner A. pH-Dependent Deformations of the Energy Landscape of Avidin-like Proteins Investigated by Single Molecule Force Spectroscopy. Molecules. 2014; 19(8):12531-12546. https://doi.org/10.3390/molecules190812531
Chicago/Turabian StyleKöhler, Melanie, Andreas Karner, Michael Leitner, Vesa P. Hytönen, Markku Kulomaa, Peter Hinterdorfer, and Andreas Ebner. 2014. "pH-Dependent Deformations of the Energy Landscape of Avidin-like Proteins Investigated by Single Molecule Force Spectroscopy" Molecules 19, no. 8: 12531-12546. https://doi.org/10.3390/molecules190812531






