1
State Key Laboratory of Emerging Infectious Diseases, The University of Hong Kong, Pokfulam, Hong Kong, China
2
Research Centre of Infection and Immunology, The University of Hong Kong, Pokfulam, Hong Kong, China
3
Carol Yu Centre for Infection, The University of Hong Kong, Pokfulam, Hong Kong, China
4
Department of Microbiology, The University of Hong Kong, Pokfulam, Hong Kong, China
5
Department of Medicine, The University of Hong Kong, Pokfulam, Hong Kong, China
6
Department of Pathology, The University of Hong Kong, Pokfulam, Hong Kong, China
†
These authors contributed equally to this work.
Int. J. Mol. Sci. 2016, 17(3), 307; https://doi.org/10.3390/ijms17030307 - 27 Feb 2016
Cited by 18 | Viewed by 7875
Abstract
To identify potential biomarkers for improving diagnosis of melioidosis, we compared plasma metabolome profiles of melioidosis patients compared to patients with other bacteremia and controls without active infection, using ultra-high-performance liquid chromatography-electrospray ionization-quadruple time-of-flight mass spectrometry. Principal component analysis (PCA) showed that the
[...] Read more.
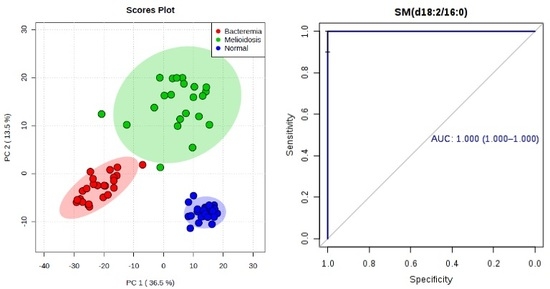
To identify potential biomarkers for improving diagnosis of melioidosis, we compared plasma metabolome profiles of melioidosis patients compared to patients with other bacteremia and controls without active infection, using ultra-high-performance liquid chromatography-electrospray ionization-quadruple time-of-flight mass spectrometry. Principal component analysis (PCA) showed that the metabolomic profiles of melioidosis patients are distinguishable from bacteremia patients and controls. Using multivariate and univariate analysis, 12 significant metabolites from four lipid classes, acylcarnitine (n = 6), lysophosphatidylethanolamine (LysoPE) (n = 3), sphingomyelins (SM) (n = 2) and phosphatidylcholine (PC) (n = 1), with significantly higher levels in melioidosis patients than bacteremia patients and controls, were identified. Ten of the 12 metabolites showed area-under-receiver operating characteristic curve (AUC) >0.80 when compared both between melioidosis and bacteremia patients, and between melioidosis patients and controls. SM(d18:2/16:0) possessed the largest AUC when compared, both between melioidosis and bacteremia patients (AUC 0.998, sensitivity 100% and specificity 91.7%), and between melioidosis patients and controls (AUC 1.000, sensitivity 96.7% and specificity 100%). Our results indicate that metabolome profiling might serve as a promising approach for diagnosis of melioidosis using patient plasma, with SM(d18:2/16:0) representing a potential biomarker. Since the 12 metabolites were related to various pathways for energy and lipid metabolism, further studies may reveal their possible role in the pathogenesis and host response in melioidosis.
Full article
(This article belongs to the Special Issue Microbial Genomics and Metabolomics)
▼
Show Figures