Curcumin Reverse Methicillin Resistance in Staphylococcus aureus
Abstract
:1. Introduction
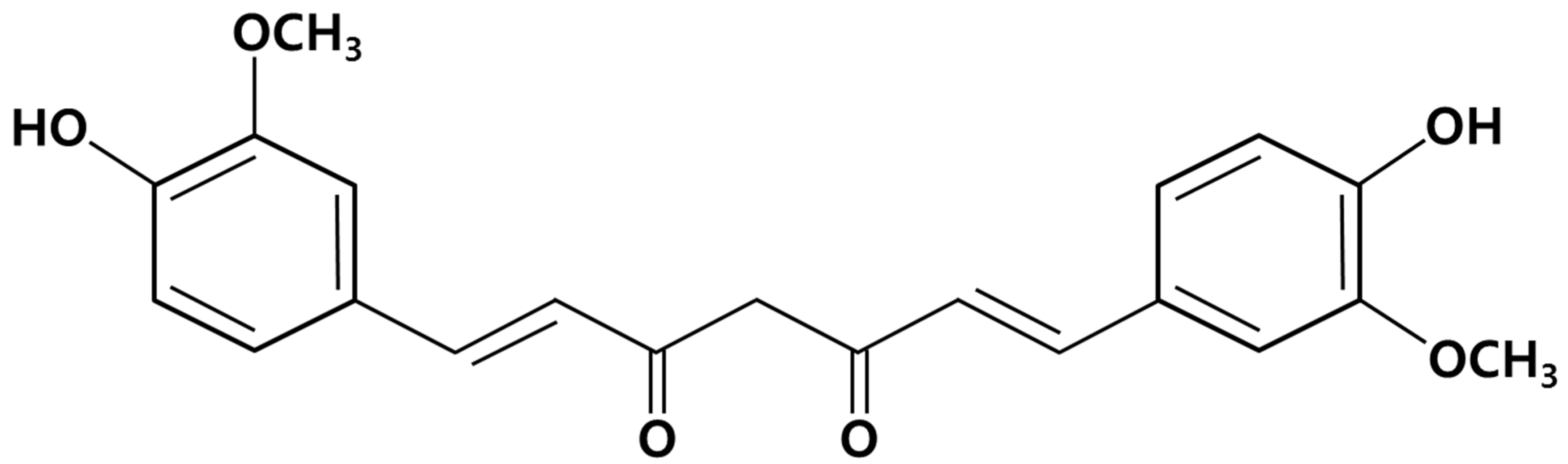
2. Results and Discussion
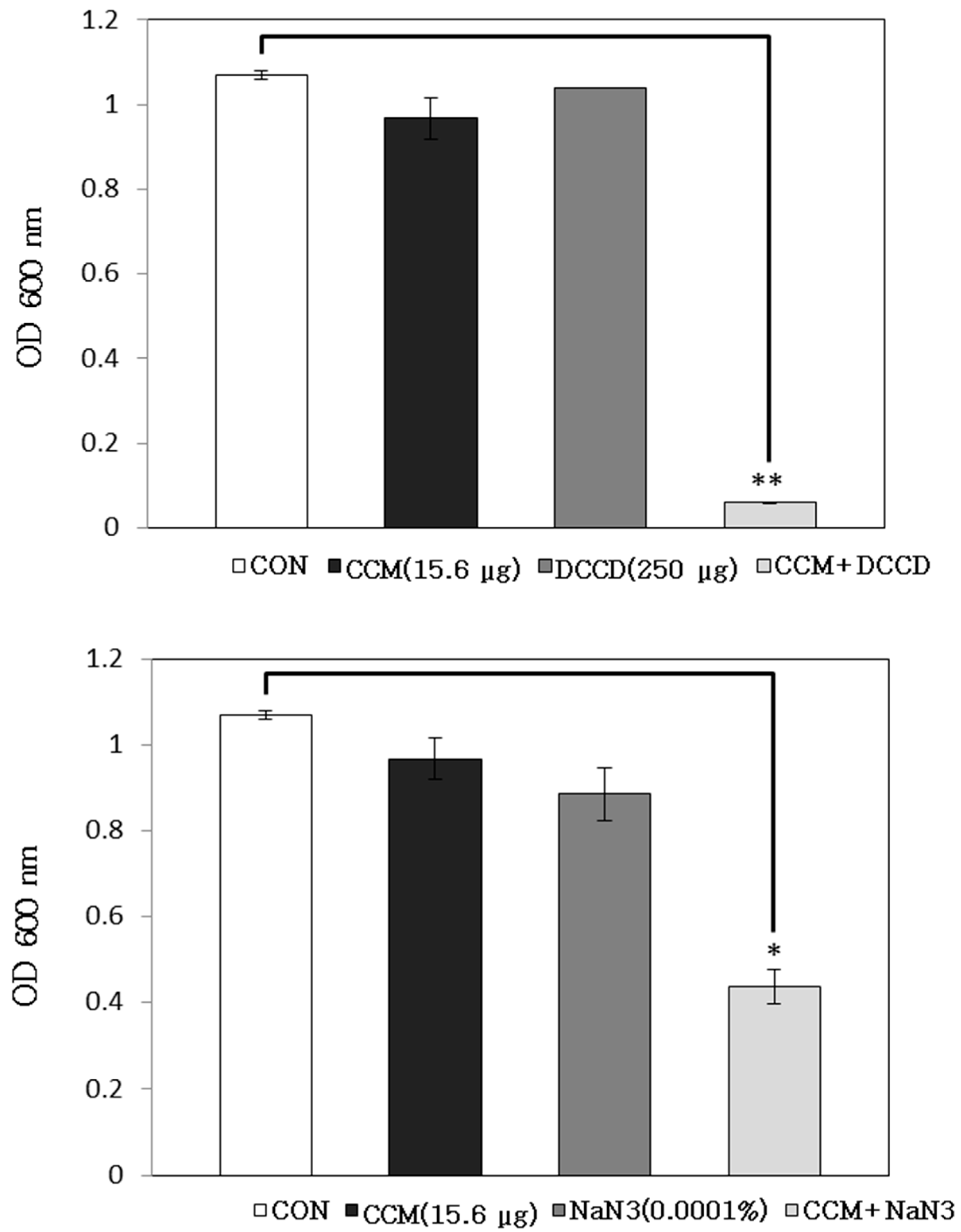
2.1. Antibacterial Activity of Curcumin with Detergents or ATPase Inhibitors

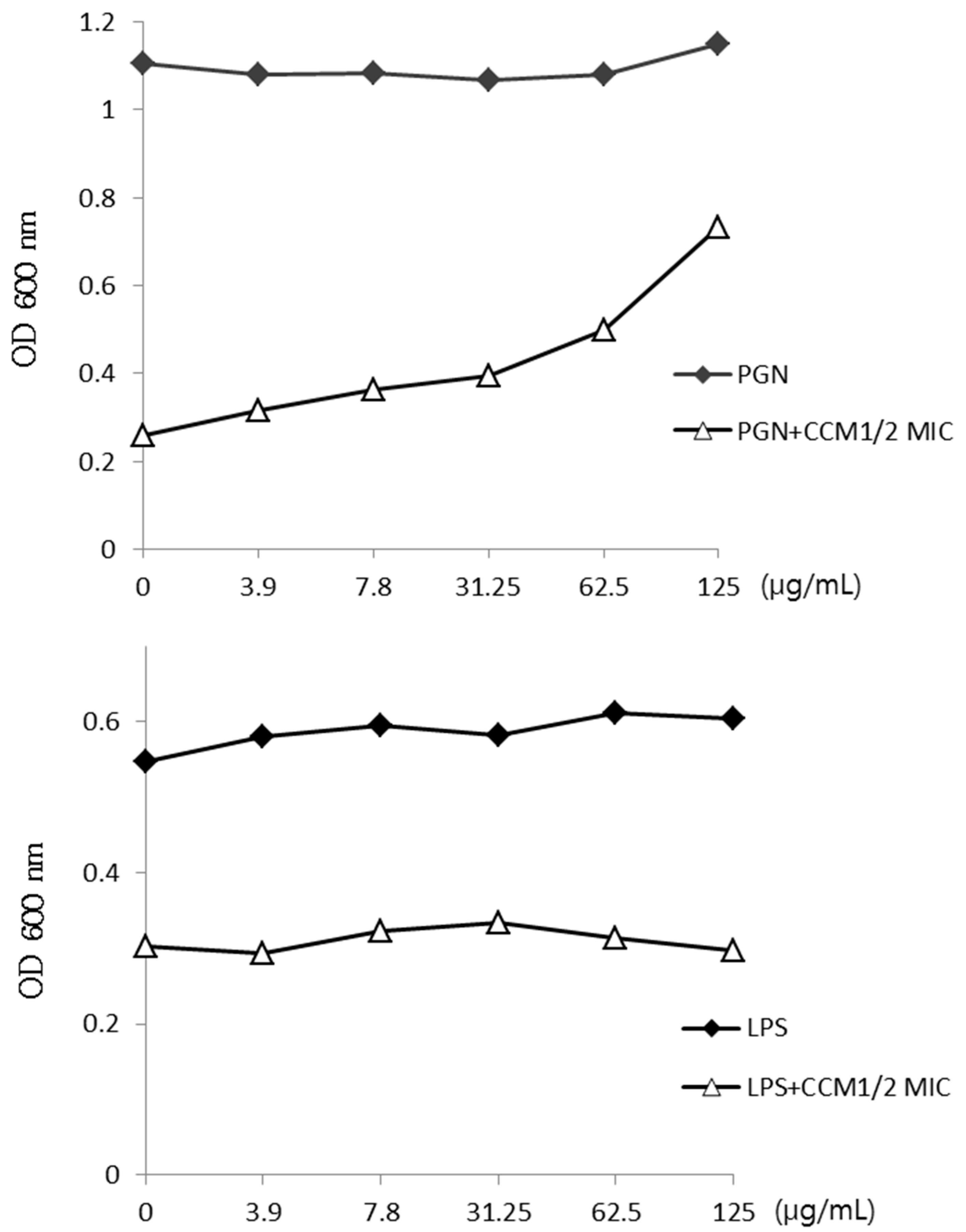
2.2. Activity of Curcumin by Binding of Peptidoglycan
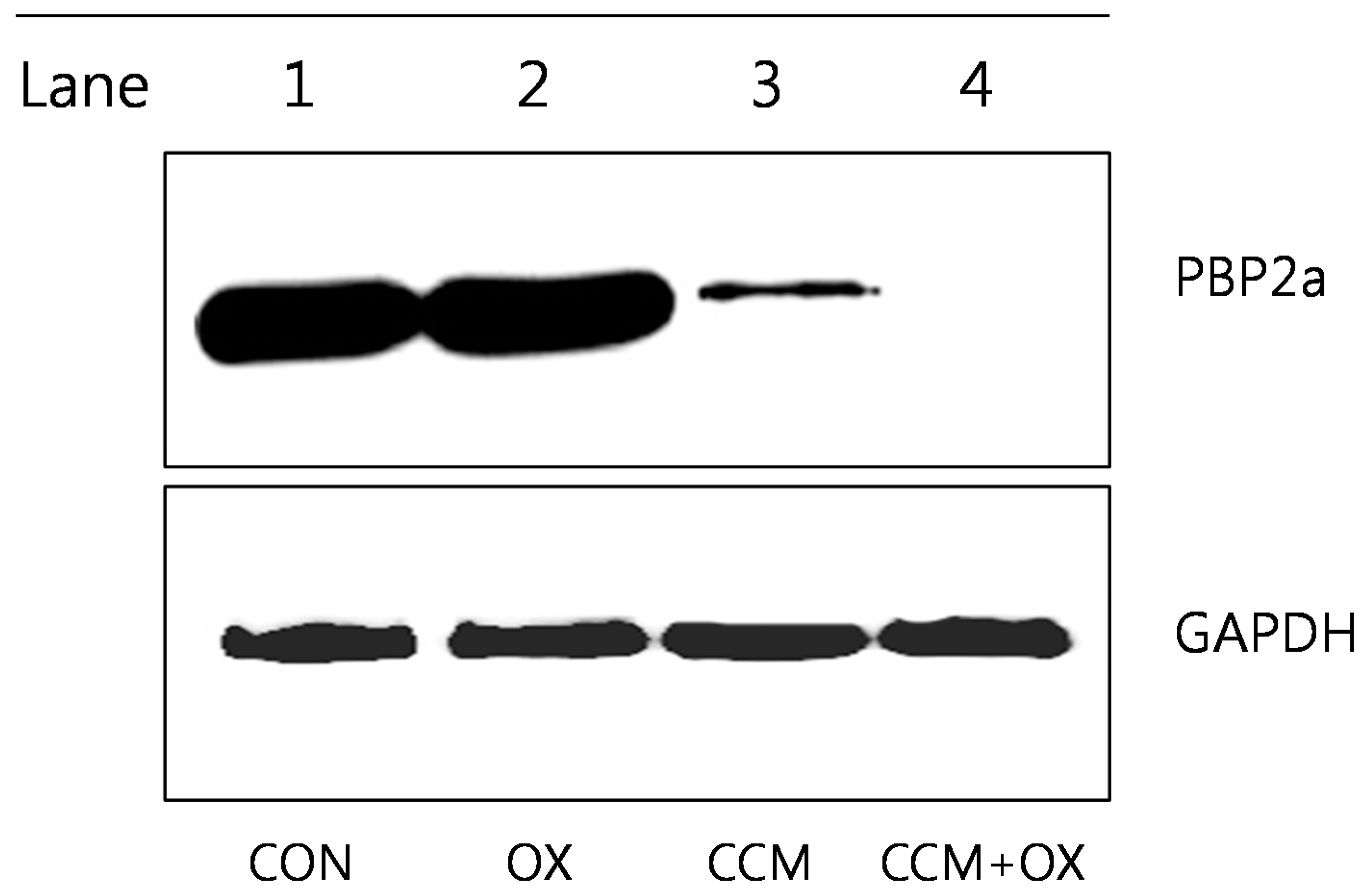
2.3. Curcumin Reduces PBP2a Levels in MRSA
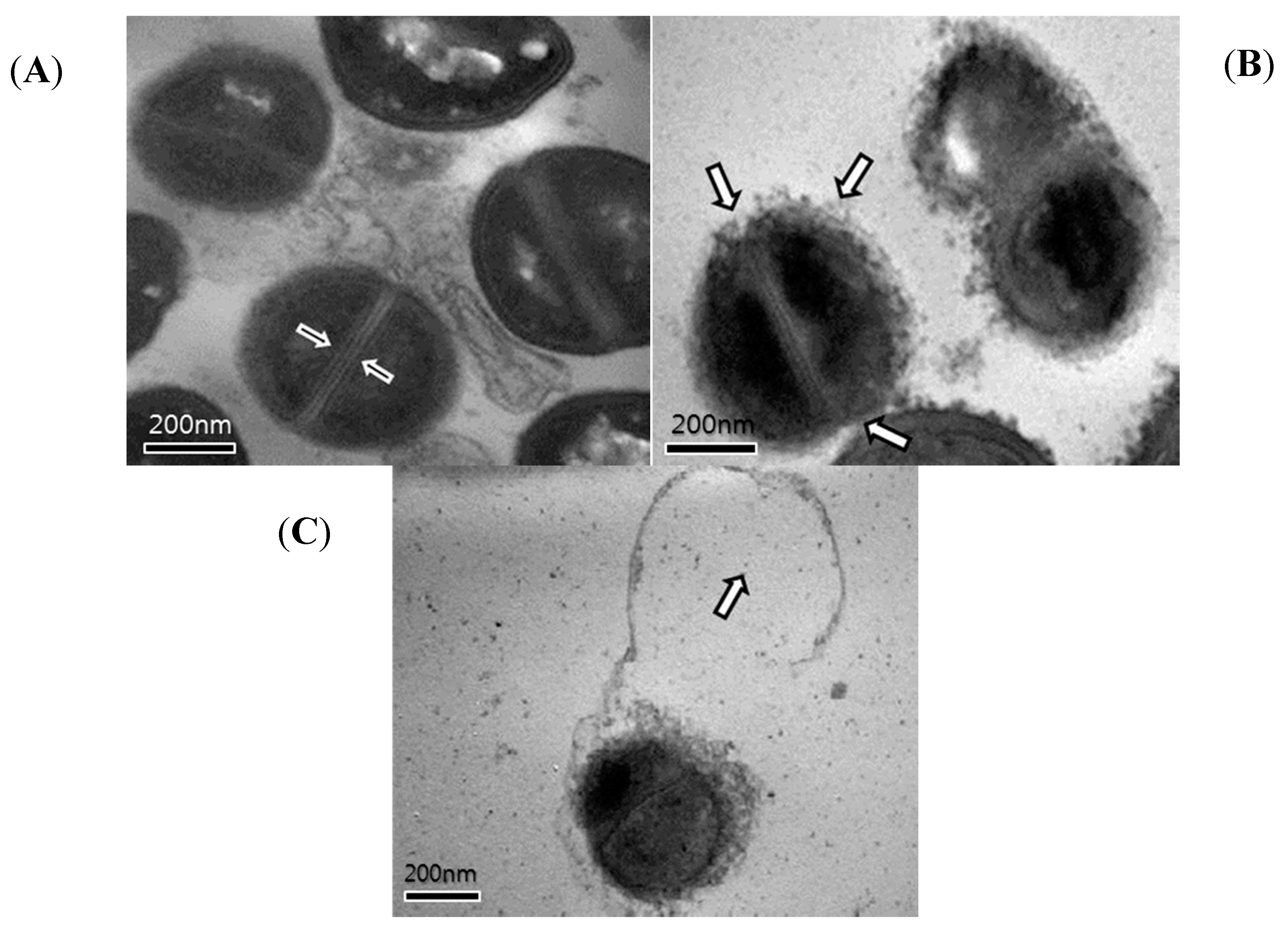
2.4. Transmission Electron Microscopy (TEM)
3. Experimental Section
3.1. Isolation and Purification of Curcumin
3.2. Bacterial Strains and Growth Conditions
3.3. Antimicrobial Resistance Testing
| S. aureus | Class | mecA Gene | β-Lactamase Activity | Antibiotic Resistance |
|---|---|---|---|---|
| ATCC 33591 | MRSA | + | + | AM, OX |
| ATCC 25923 | MSSA | − | − | − |
3.4. Reagents
3.5. Antibacterial Activity with Detergent or ATPase Inhibitors
3.6. Effect of Exogenous Peptidoglycan on Curcumin Activity
3.7. Western Blotting
3.8. Transmission Electron Microscopy (TEM)
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Stapleton, P.D.; Shah, S.; Ehlert, K.; Hara, Y.; Taylor, P.W. The beta-lactam-resistance modifier (−)-epicatechin gallate alters the architecture of the cell wall of Staphylococcus aureus. Microbiology 2007, 153, 2093–2103. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, K.; Hanaki, H.; Ino, T.; Yabuta, K.; Oguri, T.; Tenover, F.C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 1997, 40, 135–136. [Google Scholar] [CrossRef] [PubMed]
- Bachi, B.B.; Rohrer, S. Factors influencing methicillin resistance in Staphylococci. Arch. Microbiol. 2002, 178, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Barbour, E.K.; Al Sharif, M.; Sagherian, V.K.; Habre, A.N.; Talhouk, R.S.; Talhouk, S.N. Screening of select indigenous plants of Lebanon for antimicrobial activity. J. Etnopharmacol. 2004, 93, 1–7. [Google Scholar] [CrossRef]
- Mun, S.H.; Joung, D.K.; Kim, Y.S.; Kang, O.H. Synergistic antibacterial effect of curcumin against methicillin-resistant Staphylococcus aureus. Phytomedicine 2013, 20, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Yutani, R.; Morita, S.Y.; Teraoka, R.; Kitagawa, S. Distribution of polyphenols and a surfactant component in skin during Aerosol OT microemulsion-enhanced intradermal delivery. Chem. Pharm. Bull. 2012, 60, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Hegge, A.B.; Nielsen, T.T.; Larsen, K.L.; Bruzell, E.; Tønnesen, H.H. Impact of curcumin supersaturation in antibacterial photodynamic therapy—Effect of cyclodextrin type and amount: Studies on curcumin and curcuminoides XLV. J. Pharm. Sci. 2012, 101, 1524–1537. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Zhu, G.; Wang, Y.; Cai, L.; Cai, Y.; Hu, J.; Li, Y.; Yan, Y.; Wang, Z.; Li, X.; et al. Attenuation of high-glucose-induced inflammatory response by a novel curcumin derivative B06 contributes to its protection from diabetic pathogenic changes in rat kidney and heart. J. Nutr. Biochem. 2013, 24, 146–155. [Google Scholar]
- Anand, K.; Sarkar, A.; Kumar, A.; Ambasta, R.K.; Kumar, P. Combinatorial antitumor effect of naringenin and curcumin elicit angioinhibitory activities in vivo. Nutr. Cancer 2012, 64, 714–724. [Google Scholar] [PubMed]
- Fang, J.Y.; Hung, C.F.; Chiu, H.C.; Wang, J.J.; Chan, T.F. Efficacy and irritancy of enhancers on the in vitro and in vivo percutaneous absorption of curcumin. J. Pharm. Pharmacol. 2003, 55, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Egan, M.E.; Pearson, M.; Weiner, S.A.; Rajendran, V.; Rubin, D.; Glöckner-Pagel, J.; Canny, S.; Du, K.; Lukacs, G.L.; Caplan, M.J.; et al. Curcumin, a major constituent of turmeric, corrects cystic fibrosis defects. Science 2004, 304, 600–602. [Google Scholar]
- Wang, Y.; Lu, Z.; Wu, H.; Lv, F. Study on the antibiotic activity of microcapsule curcumin against foodborne pathogens. Int. J. Food Microbiol. 2009, 136, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Irvin, R.T.; MacAlister, T.J.; Costerton, J.W. Tris (hydroxymethyl) aminomethane buffer modification of Escherichia coli outer membrane permeability. J. Bacteriol. 1981, 145, 1397–1403. [Google Scholar] [PubMed]
- Raychaudhuri, D.; Chatterjee, A.N. Use of resistant mutants to study the interaction of Triton X-100 with Staphylococcus aureus. J. Bacteriol. 1985, 16, 1337–1349. [Google Scholar]
- Komatsuzawa, H.; Suzuki, J.; Sugai, M.; Miyake, Y.; Suginaka, H. The effect of Triton X-100 on the in vitro susceptibility of methicillin-resistant Staphylococcus aureus to oxacillin. J. Antimicrob. Chemother. 1994, 34, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Van der Heide, T.; Poolman, B. ABC transporters: One, two or four extra cytoplasmic substrate-binding sites? EMBO Rep. 2002, 3, 938–943. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.J.; Seu, Y.B.; Lee, D.G. Candicidal action of resveratrol isolated from grapes on human pathogenic yeast C. albicans. J. Microbiol. Biotechnol. 2007, 17, 1324–1329. [Google Scholar]
- Nester, E.W.; Roberts, C.E.; Pearsall, N.N.; Anderson, D.G.; Nester, M.T. Functional Anatomy of Prokaryotes and Eukaryotes. In Microbiology, 2nd ed.; Kane, K.T., Ronald, E., Terrance, S., Eds.; WCB/McGraw-Hill: New York, NY, USA, 1998; pp. 58–59. [Google Scholar]
- Ghuysen, J.M. Molecular structures of penicillin-binding proteins and beta-lactamases. Trends Microbiol. 1994, 2, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Goffin, C.; Ghuysen, J.M. Multimodular penicillin-binding proteins: An enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 1998, 62, 1079–1093. [Google Scholar]
- Otto, C.C.; Cunningham, T.M.; Hansen, M.R.; Haydel, S.E. Effects of antibacterial mineral leachates on the cellular ultrastructure, morphology, and membrane integrity of Escherichia coli and methicillin-resistant Staphylococcus aureus. Ann. Clin. Microbiol. Antimicrob. 2010, 9. [Google Scholar] [CrossRef]
- Al-Habib, A.; Al-Saleh, E.; Safer, A.M.; Afzal, M. Bactericidal effect of grape seed extract on methicillin-resistant Staphylococcus aureus (MRSA). J. Toxicol. Sci. 2010, 35, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Morse, S.A. Cell structure. In Medical Microbiology; Jawetz, E., Melnick, J.L., Adelberg, E.A., Brooks, G.F., Butel, J.S., Ornston, L.N., Eds.; Appleton & Lange: Norwalk, CT, USA, 1995; pp. 7–34. [Google Scholar]
- Song, E.K.; Cho, H.; Kim, J.S.; Kim, N.Y.; An, N.H.; Kim, J.A.; Lee, S.H.; Kim, Y.C. Diarylheptanoids with free radical scavenging and hepatoprotective activity in vitro from Curcuma longa. Planta Med. 2000, 67, 876–877. [Google Scholar] [CrossRef]
- Cordwell, S.J.; Larsen, M.R.; Cole, R.T.; Walsh, B.J. Comparative proteomics of Staphylococcus aureus and the response of methicillin-resistant and methicillin-sensitive strains to Triton X-100. Microbiology 2002, 148, 2765–2781. [Google Scholar]
- Shibata, H.; Saito, H.; Yomota, C.; Kawanishi, T.; Okuda, H. Alterations in the Detergent-Induced Membrane Permeability and Solubilization of Saturated Phosphatidylcholine/Cholesterol Liposomes: Effects of Poly(ethylene glycol)-Conjugated Lipid. Chem. Pharm. Bull. 2012, 60, 1105–1111. [Google Scholar] [CrossRef]
- Linnett, P.E.; Beechey, R.B. Inhibitors of the ATP synthetase system. Methods Enzymol. 1979, 55, 472–518. [Google Scholar] [PubMed]
- Jung, H.J.; Lee, D.G. Synergistic antibacterial effect between silybin and N,N′-dicyclohexylcarbodiimide in clinical Pseudomonas aeruginosa isolates. J. Microbiol. 2008, 46, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.H.; Hu, Z.Q.; Okubo, S.; Hara, Y.; Shimamura, T. Mechanism of synergy between epigallocatechin gallate and beta-lactams against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2001, 45, 1737–1742. [Google Scholar] [CrossRef] [PubMed]
- Klitgaard, J.K.; Skov, M.N.; Kallipolitis, B.H.; Kolmos, H.J. Reversal of methicillin resistance in Staphylococcus aureus by thioridazine. J. Antimicrob. Chemother. 2008, 62, 1215–1221. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.W.; Park, E.W. Ultrastructure of spined conidia and hyphae of the rice false smut fungus Ustilaginoidea virens. Micron 2007, 38, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Not available.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mun, S.-H.; Kim, S.-B.; Kong, R.; Choi, J.-G.; Kim, Y.-C.; Shin, D.-W.; Kang, O.-H.; Kwon, D.-Y. Curcumin Reverse Methicillin Resistance in Staphylococcus aureus. Molecules 2014, 19, 18283-18295. https://doi.org/10.3390/molecules191118283
Mun S-H, Kim S-B, Kong R, Choi J-G, Kim Y-C, Shin D-W, Kang O-H, Kwon D-Y. Curcumin Reverse Methicillin Resistance in Staphylococcus aureus. Molecules. 2014; 19(11):18283-18295. https://doi.org/10.3390/molecules191118283
Chicago/Turabian StyleMun, Su-Hyun, Sung-Bae Kim, Ryong Kong, Jang-Gi Choi, Youn-Chul Kim, Dong-Won Shin, Ok-Hwa Kang, and Dong-Yeul Kwon. 2014. "Curcumin Reverse Methicillin Resistance in Staphylococcus aureus" Molecules 19, no. 11: 18283-18295. https://doi.org/10.3390/molecules191118283




