The Evaluation of Magnetic Polymethacrylate-based Microspheres Used for Solid Phase DNA Micro-Extraction
Abstract
:1. Introduction
2. Materials
2.1. Chemicals and Equipment
| Microsphere | Fe content (%) | -COOH (mM g −1) | Diameter (μm) | PDI * |
|---|---|---|---|---|
| P(HEMA-co-GMA) (A1) | 5.94 | 0.00 | 2.86 | 1.04 |
| P(HEMA-co-GMA) (A2) | 5.94 | 0.41 | 2.86 | 1.04 |
| PGMA (B1) | 5.95 | 0.00 | 0.74 | 1.07 |
| PGMA (B2) | 5.95 | 0.42 | 0.74 | 1.07 |
| PGMA (C) | 2.1 | 2.61 | 1.16 | 1.05 |
| PGMA (D) | 10.02 | 0.76 | 2.23 | 1.81 |
2.2. Methods
2.2.1. Imaging of Magnetic Particles
2.2.2. Quantitative PCR
3. Results and Discussion
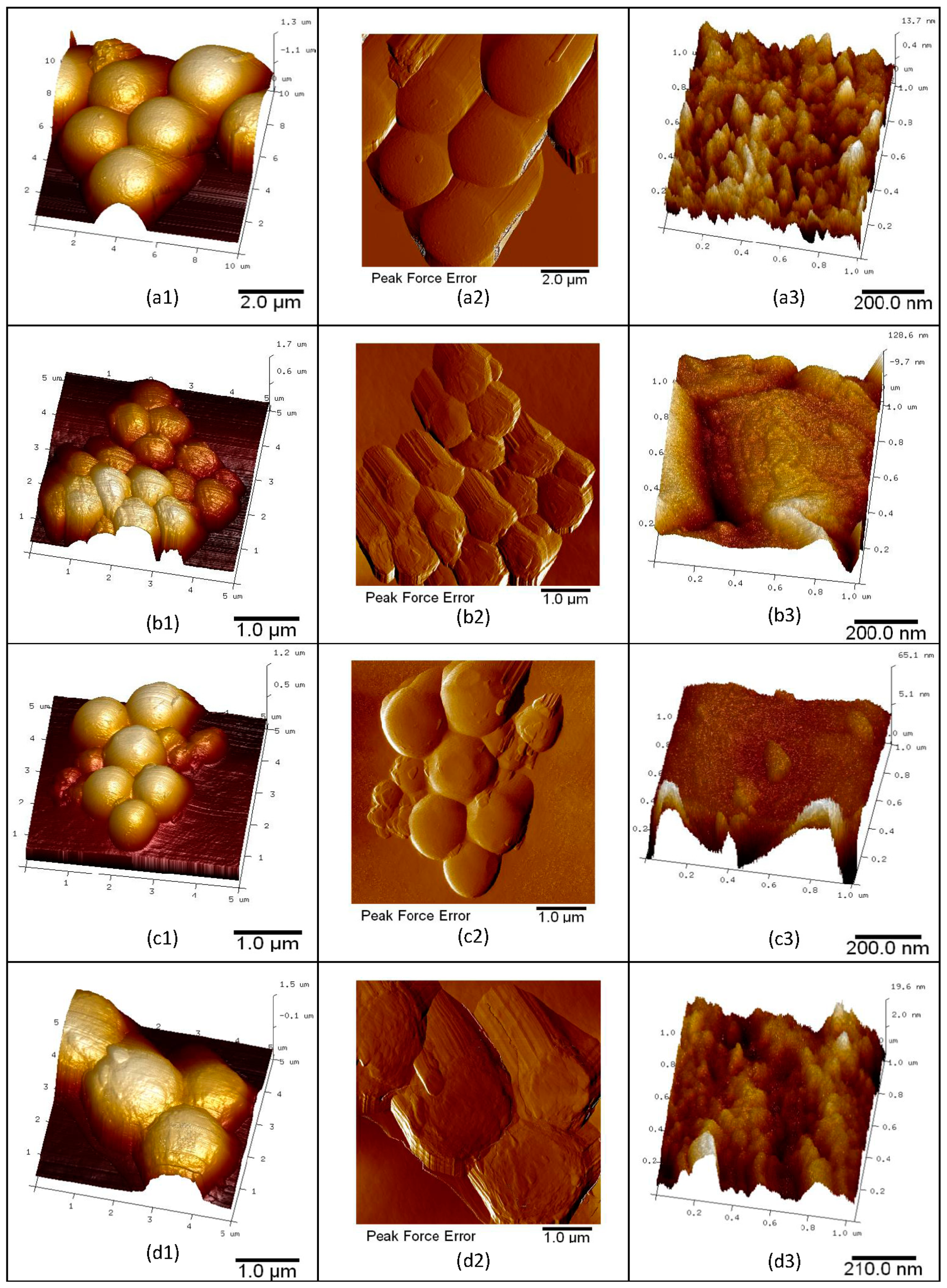
3.1. Determination of Microsphere Surface Morphology
3.2. Microsphere Interference in Quantitative PCR
| Microsphere | Microsphere concentration (mg mL−1) | Cq values | ||||
|---|---|---|---|---|---|---|
| microsphere volume (µL 25 µL−1 PCR mixture) | ||||||
| 0.0 | 0.5 | 1.0 | 2.5 | 5.0 | ||
| P(HEMA-co-GMA) (A1) | 20 | 12.8 ± 0.6 | 14.1 ± 0.3 | 15.3 ± 0.4 | 17.2 ± 0.6 | 27.5 ± 5.9 |
| 2 | 11.8 ± 0.0 | 11.9 ± 0.0 | 12.4 ± 0.0 | 12.7 ± 0.0 | 13.1 ± 0.0 | |
| P(HEMA-co-GMA) (A2) * | 20 | 12.9 ± 0.2 | 15.1 ± 0.4 | 15.7 ± 2.1 | 12.1 ± 1.2 | 10.4 ± 0.7 |
| 2 | 12.6 ± 0.1 | 13.2 ± 0.2 | 13.5 ± 0.3 | 14.8 ± 0.1 | 15.1 ± 0.3 | |
| PGMA (B1) | 20 | 13.0 ± 0.1 | 13.6 ± 0.2 | 13.7 ± 0.2 | 14.3 ± 0.3 | 14.7 ± 0.2 |
| 2 | 12.6 ± 0.0 | 12.9 ± 0.3 | 12.7 ± 0.0 | 13.2 ± 0.0 | 13.5 ± 0.0 | |
| PGMA (B2) * | 20 | 12.9 ± 0.3 | 14.9 ± 0.9 | 15.8 ± 1.1 | 22.8 ± 4.8 | 16.0 ± 1.2 |
| 2 | 12.8 ± 0.2 | 13.4 ± 0.0 | 14.4 ± 0.0 | 15.9 ± 0.4 | 15.8 ± 2.4 | |
| PGMA (C) * | 20 | 12.6 ± 0.6 | 13.6 ± 1.1 | n | n | n |
| 2 | 12.2 ± 0.2 | 12.3 ± 0.4 | 12.7 ± 0.7 | 13.1 ± 1.0 | 15.6 ± 3.4 | |
| PGMA (D) * | 20 | 12.8 ± 0.4 | 14.9 ± 0.7 | 15.4 ± 0.6 | 23.1 ± 5.5 | n |
| 2 | 12.3 ± 0.1 | 12.7 ± 0.3 | 13.2 ± 0.0 | 13.6 ± 0.0 | 14.0 ± 0.1 | |
4. Conclusion
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Chua, T.; Bhagwat, A.A. A rapid and simple DNA extraction procedure to detect Salmonella spp. and Listeria monocytogenes from fresh produce using real-time PCR. Food Anal. Methods 2009, 2, 96–101. [Google Scholar] [CrossRef]
- Martinez, N.; Martin, M.C.; Herrero, A.; Fernandez, M.; Alvarez, M.A.; Ladeto, V. qPCR as a powerful tool for microbial food spoilage quantification: Significance for food quality. Trends Food Sci. Technol. 2011, 22, 367–376. [Google Scholar] [CrossRef] [Green Version]
- Skelin, A.; Mrkonjić Fuka, M.; Čanžek Majhenić, A.; Redžepović, S.; Samaržija, D.; Bogović Matijasić, B. Phenotypic and genotypic characterization of indigenous Lactobacillus community from traditional Istrian ewe's cheese. Food Technol. Biotechnol. 2012, 50, 362–370. [Google Scholar]
- Boyer, M.; Combrisson, J. Analytical opportunities of quantitative polymerase chain reaction in dairy microbiology. Int. Dairy J. 2013, 30, 45–52. [Google Scholar] [CrossRef]
- Kuramae, E.E.; Yergeau, E.; Wong, L.C.; Pijl, A.S.; van Veen, J.A.; Kowalchuk, G.A. Soil characteristics more strongly influence soil bacterial communities than land-use type. FEMS Microbiol. Ecol. 2012, 79, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Shuga, J.; Zeng, Y.; Novak, R.; Mathies, R.A.; Hainaut, P.; Smith, M.T. Selected technologies for measuring acquired genetic damage in humans. Environ. Mol. Mutagen. 2010, 51, 851–870. [Google Scholar] [CrossRef] [PubMed]
- Kitpipit, T.; Chotigeat, W.; Linacre, A.; Thanakiatkrai, P. Forensic animal DNA analysis using economical two-step direct PCR. Forensic Sci. Med. Pathol. 2014, 10, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors–occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
- Kenk, M.; Panter, S.; Engler-Blum, G.; Bergemann, J. Sensitive DNA-based allergen detection depends on food matrix and DNA isolation method. Eur. Food Res. Technol. 2012, 234, 351–359. [Google Scholar] [CrossRef]
- Alaeddini, R. Forensic implications of PCR inhibition—A review. Forensic Sci. Int. Gen. 2012, 6, 297–305. [Google Scholar] [CrossRef]
- Opel, K.L.; Chung, D.; McCord, B.R. A Study of PCR Inhibition Mechanisms Using Real Time PCR. J. Forensic Sci. 2010, 55, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Deng, C.; Liu, Y.; Zhao, X.; Tang, Y.; Liu, R.; Xia, Q.; Yan, W.; Ge, G. Optimization of influencing factors of nucleic acid adsorption onto silica-coated magnetic particles: Application to viral nucleic acid extraction from serum. J. Chromatogr. A 2014, 1325, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Rödiger, S.; Liebsch, C.; Schmidt, C.; Lehmann, W.; Resch-Genger, U.; Schedler, U.; Schierack, P. Nucleic acid detection based on the use of microbeads: A review. Microchim. Acta 2014, 181, 1151–1168. [Google Scholar] [CrossRef]
- Španová, A.; Rittich, B.; Horák, D.; Lenfeld, J.; Prodělalová, J.; Sučiková, J.; Štrumcová, S. Immunomagnetic separation and detection of Salmonella cells using newly designed carriers. J. Chromatogr. A 2003, 1009, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Trachtová, Š.; Španová, A.; Tóth, J.; Prettl, Z.; Horák, D.; Gyenis, J.; Rittich, B. Solid-phase DNA isolation from food matrices using hydrophilic magnetic microspheres. Food Bioprod. 2014, in press. [Google Scholar]
- Zieliński, B.; Skumiel, A.; Józefczak, A.; Vandeweert, E. Determination of magnetic particle size using ultrasonic, magnetic and atomic force microscopy methods. Mol. Quant. Acoust. 2005, 26, 309–316. [Google Scholar]
- Moskalenko, A.V.; Yarova, P.L.; Gordeev, S.N.; Smirnov, S.V. Single protein molecule mapping with magnetic atomic force microscopy. Biophys. J. 2010, 98, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Müller, D.J.; Dufrêne, Y.F. Atomic force microscopy: A nanoscopic window on the cell surface. Trends Cell Biol. 2011, 21, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Španová, A.; Rittich, B.; Beneš, M.J.; Horák, D. Ferrite supports for isolation of DNA from complex samples and polymerase chain reaction amplification. J. Chromatogr. A 2005, 1080, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Trachtová, Š.; Kaman, O.; Španová, A.; Veverka, P.; Pollert, E.; Rittich, B. Silica-coated La0.75Sr0.25MnO3 nanoparticles for magnetically driven DNA isolation. J. Sep. Sci. 2011, 34, 3077–3082. [Google Scholar] [CrossRef] [PubMed]
- King, C.E.; Debruyne, R.; Kuch, M.; Schwarz, M.C.; Poinar, H.N. A quantitative approach to detect and overcome PCR inhibition in ancient DNA extracts. BioTechniques 2009, 47, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Haugland, R.A.; Siefring, S.; Lavender, J.; Varma, M. Influences of sample interference and interference controls on quantification of enterococci fecal indicator bacteria in surface water samples by the qPCR method. Water Res. 2012, 46, 5989–6001. [Google Scholar] [CrossRef] [PubMed]
- Horák, D.; Semenyuk, N.; Lednický, F. Effect of the reaction parameters on the particle size in the dispersion polymerization of 2-hydroxyethyl and glycidyl methacrylate in the presence of a ferrofluid. J. Polym. Sci. Pol. Chem. 2003, 41, 1848–1863. [Google Scholar] [CrossRef]
- Horák, D.; Rittich, B.; Španová, A. Carboxyl-functionalized magnetic microparticle carrier for isolation and identification of DNA in dairy products. J. Magn. Magn. Mater. 2007, 311, 249–254. [Google Scholar] [CrossRef]
- Haarman, M.; Knol, J. Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. Appl. Environ. Microbiol. 2006, 72, 2359–2365. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, R. Quantification on the LightCycler. In Rapid Cycle Real-time PCR, Methods and Applications; Meuer, S., Wittwer, C., Nakagawara, K., Eds.; Springer Press: Heidelberg, Germany, 2001; pp. 21–34. [Google Scholar]
- Heid, C.A.; Stevens, J.; Livak, K.J.; Williams, P.M. Real time quantitative PCR. Genome Res. 1996, 6, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Stahlberg, A.P.; Aman, P.; Ridell, B.; Mostad, P.; Kubista, M. Quantitative real-time PCR method for detection of B-lymphocyte monoclonality by comparison of kappa and lambda immunoglobulin light chain expression. Clin. Chem. 2003, 49, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Tarnok, A. SYTO dyes and histoproteins—Myriad of applications. Cytom. Part A 2008, 73A, 477–479. [Google Scholar] [CrossRef]
- Monis, P.T.; Giglio, S.; Saint, C.P. Comparison of SYTO9 and SYBR Green I for real-time polymerase chain reaction and investigation of the effect of dye concentration on amplification and DNA melting curve analysis. Anal. Biochem. 2005, 340, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Koubková, J.; Müller, P.; Hlídková, H.; Plichta, Z.; Proks, V.; Vojtěšek, B.; Horák, D. Magnetic poly(glycidyl methacrylate) microspheres for protein capture. New Biotechnol. 2014, 31, 482–491. [Google Scholar] [CrossRef]
- Horák, D.; Hlídková, H.; Trachtová, Š.; Šlouf, M.; Rittich, B.; Španová, A. Evaluation of poly(ethylene glycol)-coated monodispersed magnetic poly(2-hydroxyethyl methacrylate) and poly(glycidyl methacrylate) microspheres by PCR. Eur. Polym. J. 2015. [Google Scholar] [CrossRef]
- Monroe, C.; Grier, C.; Kemp, B.M. Evaluating the efficacy of various thermo-stable polymerases against co-extracted PCR inhibitors in ancient DNA samples. Forensic Sci. Int. 2013, 228, 142–153. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trachtová, Š.; Zapletalová, H.; Španová, A.; Horák, D.; Kolářová, H.; Rittich, B. The Evaluation of Magnetic Polymethacrylate-based Microspheres Used for Solid Phase DNA Micro-Extraction. Chromatography 2015, 2, 156-166. https://doi.org/10.3390/chromatography2020156
Trachtová Š, Zapletalová H, Španová A, Horák D, Kolářová H, Rittich B. The Evaluation of Magnetic Polymethacrylate-based Microspheres Used for Solid Phase DNA Micro-Extraction. Chromatography. 2015; 2(2):156-166. https://doi.org/10.3390/chromatography2020156
Chicago/Turabian StyleTrachtová, Štěpánka, Hana Zapletalová, Alena Španová, Daniel Horák, Hana Kolářová, and Bohuslav Rittich. 2015. "The Evaluation of Magnetic Polymethacrylate-based Microspheres Used for Solid Phase DNA Micro-Extraction" Chromatography 2, no. 2: 156-166. https://doi.org/10.3390/chromatography2020156
APA StyleTrachtová, Š., Zapletalová, H., Španová, A., Horák, D., Kolářová, H., & Rittich, B. (2015). The Evaluation of Magnetic Polymethacrylate-based Microspheres Used for Solid Phase DNA Micro-Extraction. Chromatography, 2(2), 156-166. https://doi.org/10.3390/chromatography2020156





