A Novel Three-Dimensional Wound Healing Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells
2.2. 2D Scratch Wound Assay
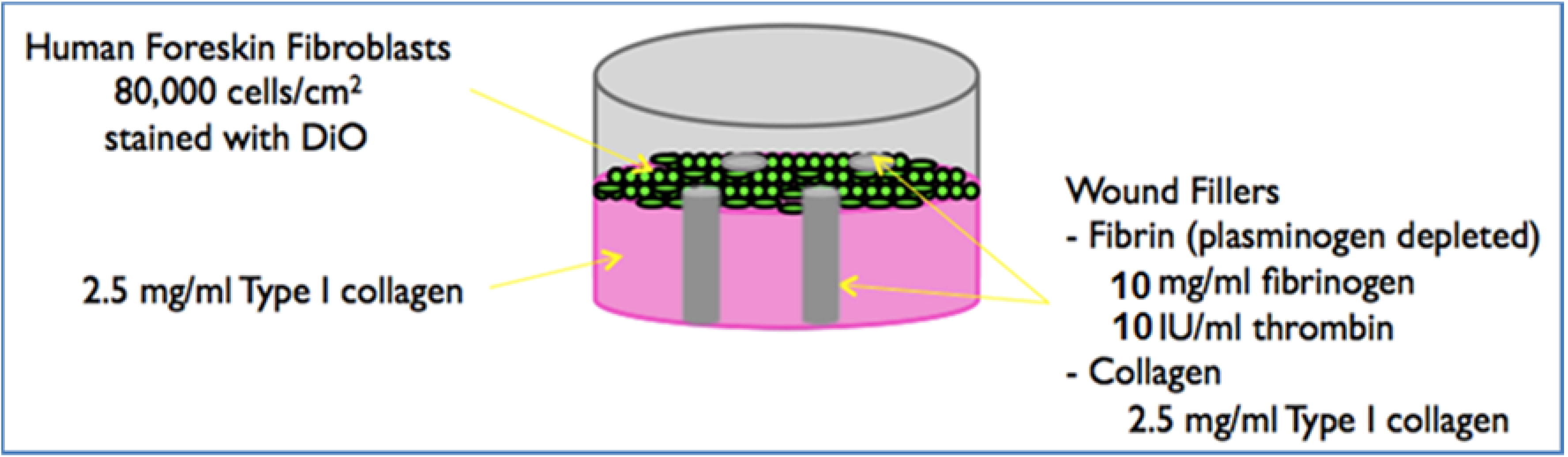
2.3. Preparation of the 3D Model
Biopsy Punch Fabrication
2.4. Preparing Conditioned Media
2.5. Measuring Cell Migration
2.6. Statistical Analysis
3. Results
3.1. Cell Migration in 2D Scratch Assays
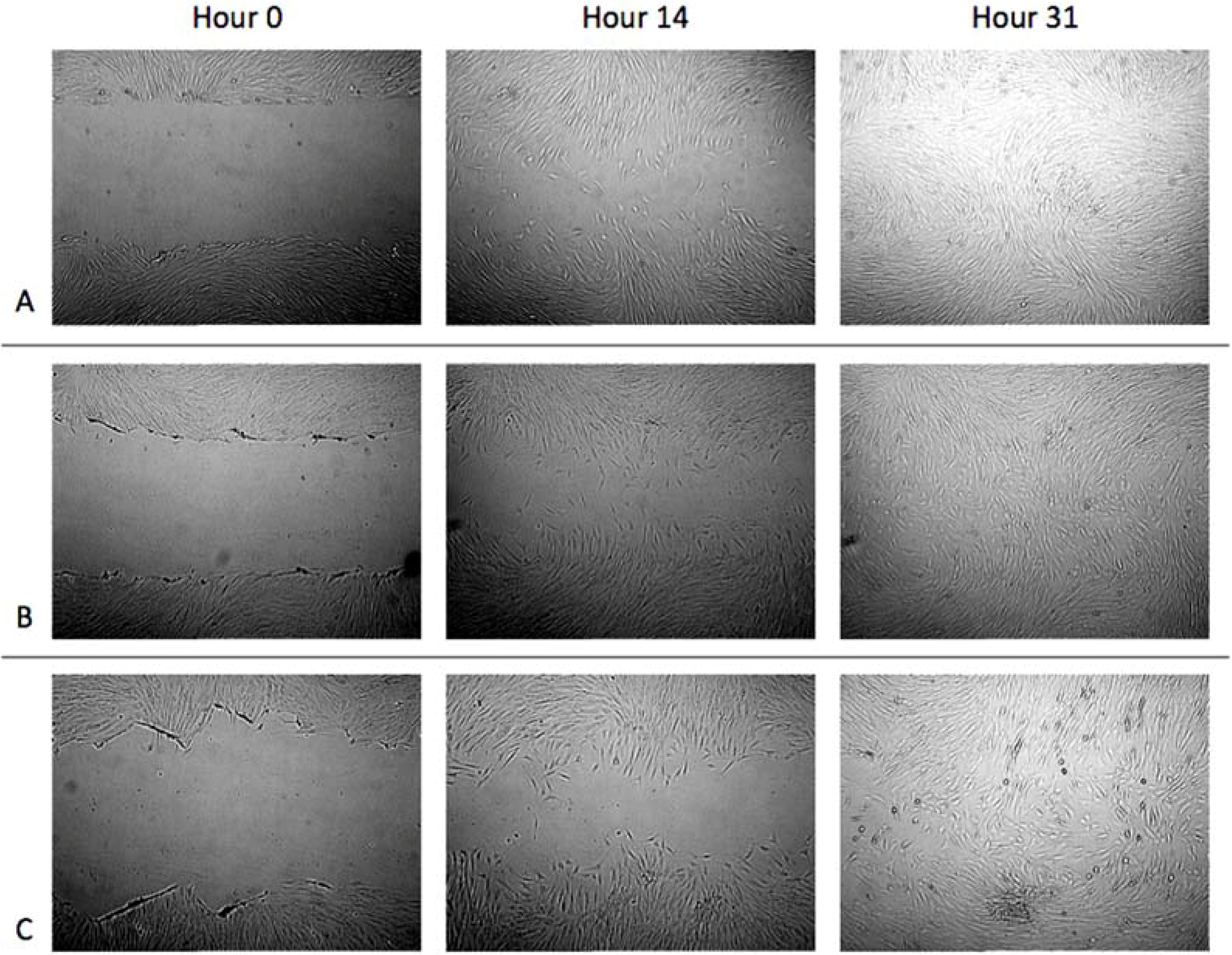

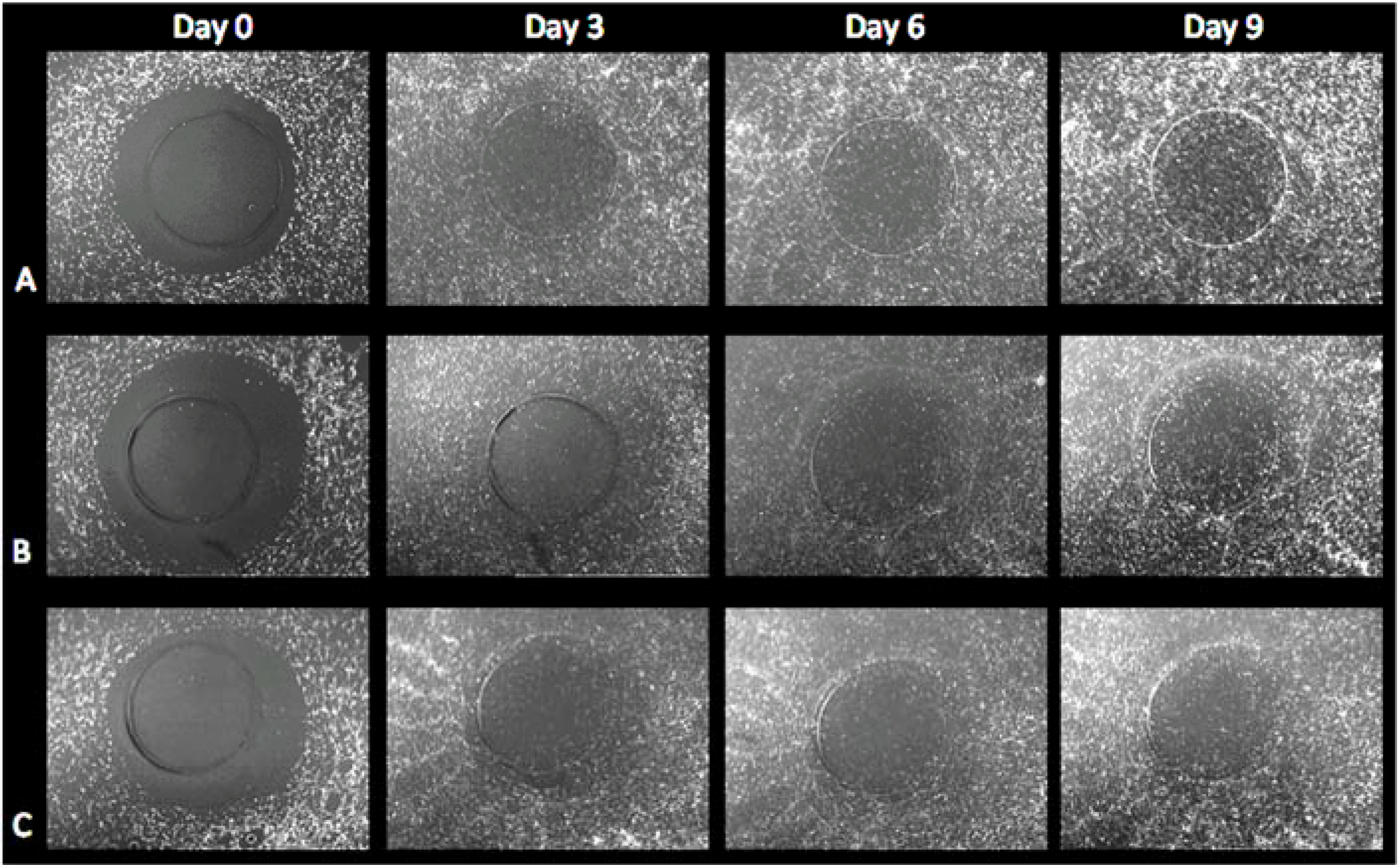
3.2. Cell Migration on 3D Models
3.2.1. HFF Migration from Collagen to Collagen
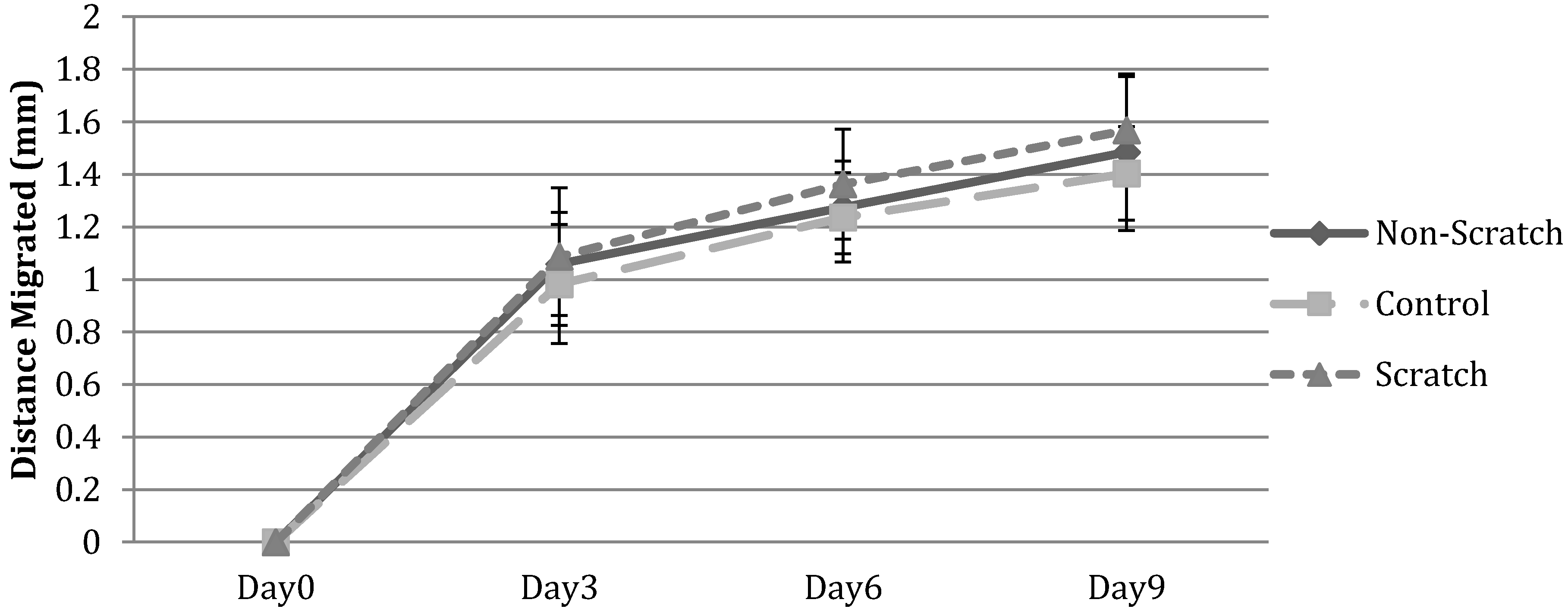
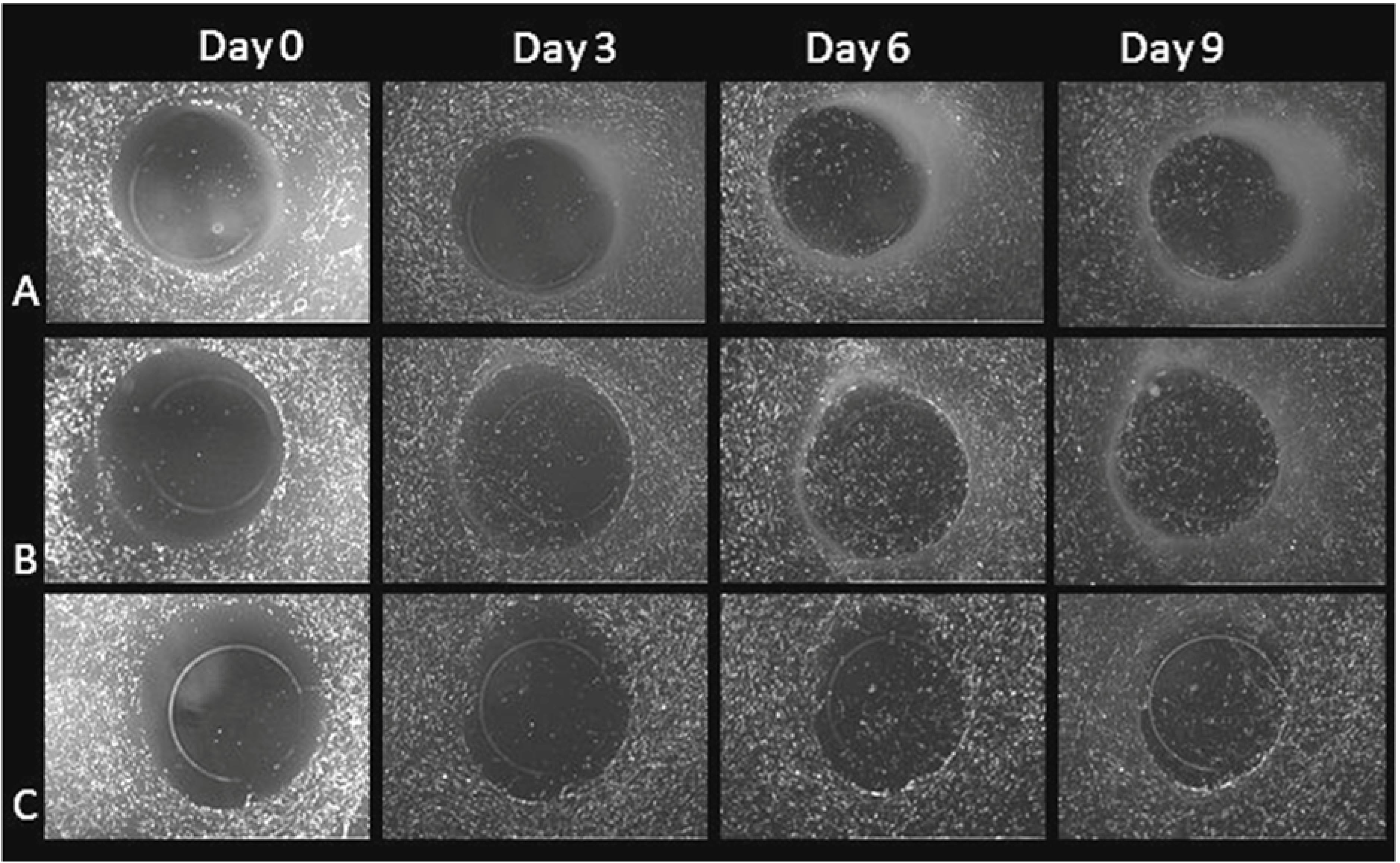
3.2.2. HFF Migration from Collagen to Fibrin
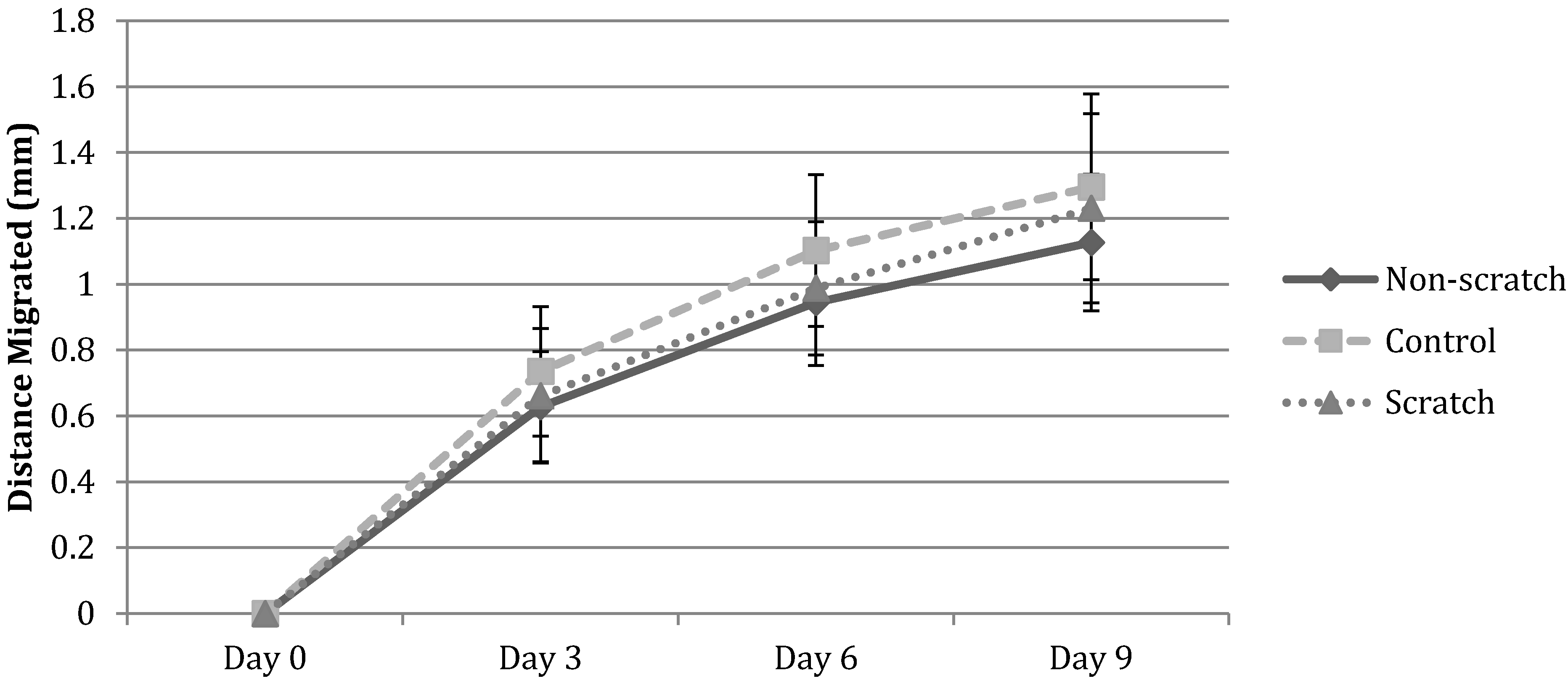
3.2.3. HFF Migration: Biopsy Punch vs. Mold Model

4. Discussion
5. Conclusion
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Brown, G.L.; Nanney, L.B.; Griffen, J.; Cramer, A.B.; Yancey, J.M.; Curtsinger, L.J., 3rd; Holtzin, L.; Schultz, G.S.; Jurkiewicz, M.J.; Lynch, J.B. Enhancement of wound healing by topical treatment with epidermal growth factor. N. Engl. J. Med. 1989, 321, 76–79. [Google Scholar] [CrossRef]
- Brown, L.F.; Berse, B.; Yeo, T.K.; Senger, D.R.; Dvorak, H.F.; van de Water, L. Expression of vascular permeability factor (vascular endothelial growth factor) by epidermal keratinocytes during wound healing. J. Exp. Med. 1992, 176, 1375–1379. [Google Scholar] [CrossRef]
- Carlson, M.W.; Dong, S.; Garlick, J.A.; Egles, C. Tissue-engineered models for the study of cutaneous wound-healing. In Studies in Mechanobiology, Tissue Engineering and Biomaterials; Gefen, A., Ed.; Springer: New York, NY, USA, 2009; Volume 1, pp. 263–280. [Google Scholar]
- Catelas, I.; Sese, N.; Wu, B.; Dunn, J.; Helgerson, S.; Tawil, B. Human mesenchymal stem cells proliferation and osteogenic differentiation in fibrin gels in vitro. Tissue Eng. 2006, 12, 2385–2396. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.; Hecht, V.; Duong, H.; Wu, B.; Tawil, B. Permeability of three-dimensional fibrin constructs corresponds to fibrinogen and thrombin concentrations. BioRes. 2012, 1, 34–40. [Google Scholar]
- Clark, R.A.F. Cuntaneous wound healing. N. Engl. J. Med. 1999, 341, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Cox, S.; Cole, M.; Tawil, N. Behavior of human dermal fibroblasts in 3 dimensional fibrin clots: Dependence on the fibrinogen and thrombin concentration. Tissue Eng. 2004, 10, 942–954. [Google Scholar] [CrossRef] [PubMed]
- Duong, H.; Wu, B.; Tawil, B. Modulation of 3-dimensional fibrin matrix stiffness by intrinsic fibrinogen-thrombin compositions and by extrinsic cellular activity. Tissue Eng. 2009, 15, 1865–1876. [Google Scholar] [CrossRef]
- Geer, D.; Swartz, D.; Andreadis, S. In vivo model of wound healing based on transplanted tissue-engineered skin. Tissue Eng. 2004, 10, 1006–1017. [Google Scholar] [CrossRef] [PubMed]
- Hebda, P.A.; Klingbeil, C.K.; Abraham, J.A.; Fiddes, J.C. Basic fibroblast growth factor stimulation of epidermal wound healing in pigs. J. Invest. Dermatol. 1990, 95, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Ho, W.; Tawil, B.; Dunn, J.C.Y.; Wu, B.M. The behavior of human mesenchymal stem cells in 3D fibrin clots: Dependence on fibrinogen concentration and clot structure. Tissue Eng. 2006, 12, 1587–1595. [Google Scholar] [CrossRef] [PubMed]
- Karamichos, D.; Lakshman, N.; Petroll, M. An experimental model for assessing fibroblast migration in 3-D collagen matrices. Cell Motil. Cytoskel. 2009, 66, 1–9. [Google Scholar] [CrossRef]
- Linsley, C.; Wu, B.; Tawil, B. The effect of fibrinogen, collagen type I and fibronectin on mesenchymal stem cells growth and differentiation into osteoblasts. Tissue Eng. 2013, 19, 1416–1423. [Google Scholar] [CrossRef]
- Lynch, S.E.; Robert, B.C.; Harry, N.A. Growth factors in wound healing. Single and synergistic effects on partial thickness porcine skin wounds. J. Clin. Invest. 1989, 84, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Mana, M.; Cole, M.; Cox, S.; Tawil, B. Human U937 monocyte behavior and protein expression on various formulations of three-dimensional fibrin clots. Wound Repair Regen. 2006, 14, 72–80. [Google Scholar] [CrossRef]
- Mooney, R.; Tawil, B.; Mahoney, M. Specific fibrinogen and thrombin concentrations promote neuronal rather than glial growth when primary neural cells are seeded within plasma-derived fibrin gels. Tissue Eng. 2010, 16, 1607–1619. [Google Scholar] [CrossRef]
- Puchtler, H.; Waldrop, F.S.; Valentine, L.S. Polarization microscopic studies of connective tissue stained with picrosirius red FBA. Beitr. Pathol. 1973, 150, 174–187. [Google Scholar] [CrossRef] [PubMed]
- Rich, L.; Whittaker, P. Collagen and picrosirius red staining: A polarized light assessment of fibrillar hue and sptail distribution. Braz. J. morphol. Sci. 2005, 22, 97–104. [Google Scholar]
- Sese, N.; Cole, M.; Tawil, B. Cross-talk between human dermal fibroblasts and keratinocytes co-cultured in 3 dimensional fibrin clots. Tissue Eng. 2011, 17, 429–437. [Google Scholar] [CrossRef]
- Tao, H.; Berno, A.; Cox, D.; Frazer, K. In vitro human keratinocyte migration rates are associated with SNPs in the KRT1 interval. PLOS ONE 2007, 2, e697. [Google Scholar] [CrossRef] [PubMed]
- Tawil, B.; Wu, B. Three-dimensional fibrin constructs in tissue engineering. Review. In An Introduction to Biomaterials, 2nd ed.; Hollinger, J.O., Ed.; CRC Taylor & Francis: New York, NY, USA, 2012; pp. 249–262. [Google Scholar]
- Xie, Y.; Rizzi, S.C.; Dawson, R.; Lynam, E.; Richards, S.; Leavesley, D.I.; Upton, Z. Development of a three-dimensional human skin equivalent wound model for investigating novel wound healing therapies. Tissue Eng. Part C 2010, 16, 1111–1123. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.J.; Yang, J.P.; Wu, B.M.; Tawil, B. A Novel Three-Dimensional Wound Healing Model. J. Dev. Biol. 2014, 2, 198-209. https://doi.org/10.3390/jdb2040198
Chen ZJ, Yang JP, Wu BM, Tawil B. A Novel Three-Dimensional Wound Healing Model. Journal of Developmental Biology. 2014; 2(4):198-209. https://doi.org/10.3390/jdb2040198
Chicago/Turabian StyleChen, Zhuo J., Jessica P. Yang, Benjamin M. Wu, and Bill Tawil. 2014. "A Novel Three-Dimensional Wound Healing Model" Journal of Developmental Biology 2, no. 4: 198-209. https://doi.org/10.3390/jdb2040198
APA StyleChen, Z. J., Yang, J. P., Wu, B. M., & Tawil, B. (2014). A Novel Three-Dimensional Wound Healing Model. Journal of Developmental Biology, 2(4), 198-209. https://doi.org/10.3390/jdb2040198



