Epitope Prediction Assays Combined with Validation Assays Strongly Narrows down Putative Cytotoxic T Lymphocyte Epitopes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Prediction Algorithms and Nucleotide Sequences
2.2. Synthetic Peptides
2.3. MHC Class I Stabilization Assay
2.4. Mice
2.5. Recombinant Semliki Forest Virus (rSFV) Particles Production and Immunizations
2.6. Degranulation and IFN-γ Staining
3. Results and Discussion
3.1. Selection and Characteristics of HCV Synthetic Long Peptides that May Contain CTL Epitopes
| Protein/Position | Sequence (CTL Epitopes Are Underlined) | MHC I Peptides Prediction | MHC I Stabilization Assay (Figure 1) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SYFPEITHI (>20 strong binding) | NetMHCpan 2.8 (<0.5 strong binding) | IEDB (<0.5 strong binding) | Peptide Concentration = 10 μM 1,2 | Selected for Short Peptides Synthesis (v) | ||||||
| H-2Db | H-2Kb | H-2Db | H-2Kb | H-2Db | H-2Kb | H-2Db | H-2Kb | |||
| NS372-87 | IQMYTNVDQDLVGWPA | 24 | 10 | 0.8 | 3 | 2.05 | 2.25 | + | - | |
| NS3165-180 | KAVDFIPVENLGTTMR | 30 | 8 | 2 | 32 | 2.5 | 18.5 | - | + | |
| NS3214-228 | VPAAYAAQGYKVLVL | 0 | 22 | 10 | 15 | 24 | 6.8 | ++ | + | |
| NS3323-340 | ATPPGSVTVSHPNIEEVA | 23 | 9 | 0.08 | 8 | 0.3 | 9.45 | + | - | v |
| NS3383-400 | ALGINAVAYYRGLDVSVI | 0 | 22 | 32 | 1.5 | 19.1 | 1.15 | ++ | - | |
| NS3507-524 | AETTVRLRAYMNTPGLPV | 22 | 11 | 0.1 | 0.25 | 0.7 | 0.7 | +++ | - | v |
| NS3525-542 | CQDHLEFWEGVFTGLTHI | 0 | 21 | 50 | 32 | 13.95 | 7.9 | ++ | - | |
| NS3547-563 | LSQTKQSGENFPYLVAY | 28 | 22 | 0.3 | 1.5 | 0.4 | 1.15 | ++ | - | |
| NS3601-618 | RLGAVQNEVTLTHPITKY | 29 | 12 | 0.08 | 32 | 0.2 | 12.95 | + | - | v |
| NS5A58-75 | HCGAEITGHVKNGTMRIV | 24 | 8 | 5 | 32 | 2.65 | 36.5 | - | - | |
| NS5A98-115 | CTPLPAPNYKFALWRVSA | 20 | 12 | 7 | 32 | 19 | 19 | - | - | |
| NS5A140-157 | CPCQIPSPEFFTELDGVR | 21 | 22 | 8 | 1.5 | 6.3 | 0.5 | - | - | |
| NS5A269-284 | ITRVESENKVVILDSF | 24 | 7 | 4 | 50 | 5.1 | 45.5 | - | - | |
| NS5B1-16 | SMSYSWTGALVTPCAA | 13 | 11 | 2 | 0.03 | 5.3 | 0.2 | + | + | v |
| NS5B46-63 | CQRQKKVTFDRLQVLDSH | 15 | 11 | 15 | 0.05 | 27 | 0.25 | - | - | v |
| NS5B152-169 | GGRKPARLIVFPDLGVRV | 15 | 22 | 32 | 15 | 22 | 12.35 | - | ++ | v |
| NS5B249-266 | ARVAIKSLTERLYVGGPL | 22 | 12 | 0.8 | 3 | 1.35 | 2.8 | - | - | |
| NS5B329-346 | VQEDAASLRAFTEAMTRY | 20 | 12 | 0.4 | 1.5 | 9.7 | 13.95 | ++ | + | |
| NS5B402-419 | HTPVNSWLGNIIMFAPTL | 21 | 8 | 7 | 8 | 12 | 2.7 | - | - | |
| NS5B423-439 | MILMTHFFSVLIARDQL | 13 | 21 | 8 | 0.17 | 9.1 | 0.3 | - | - | v |
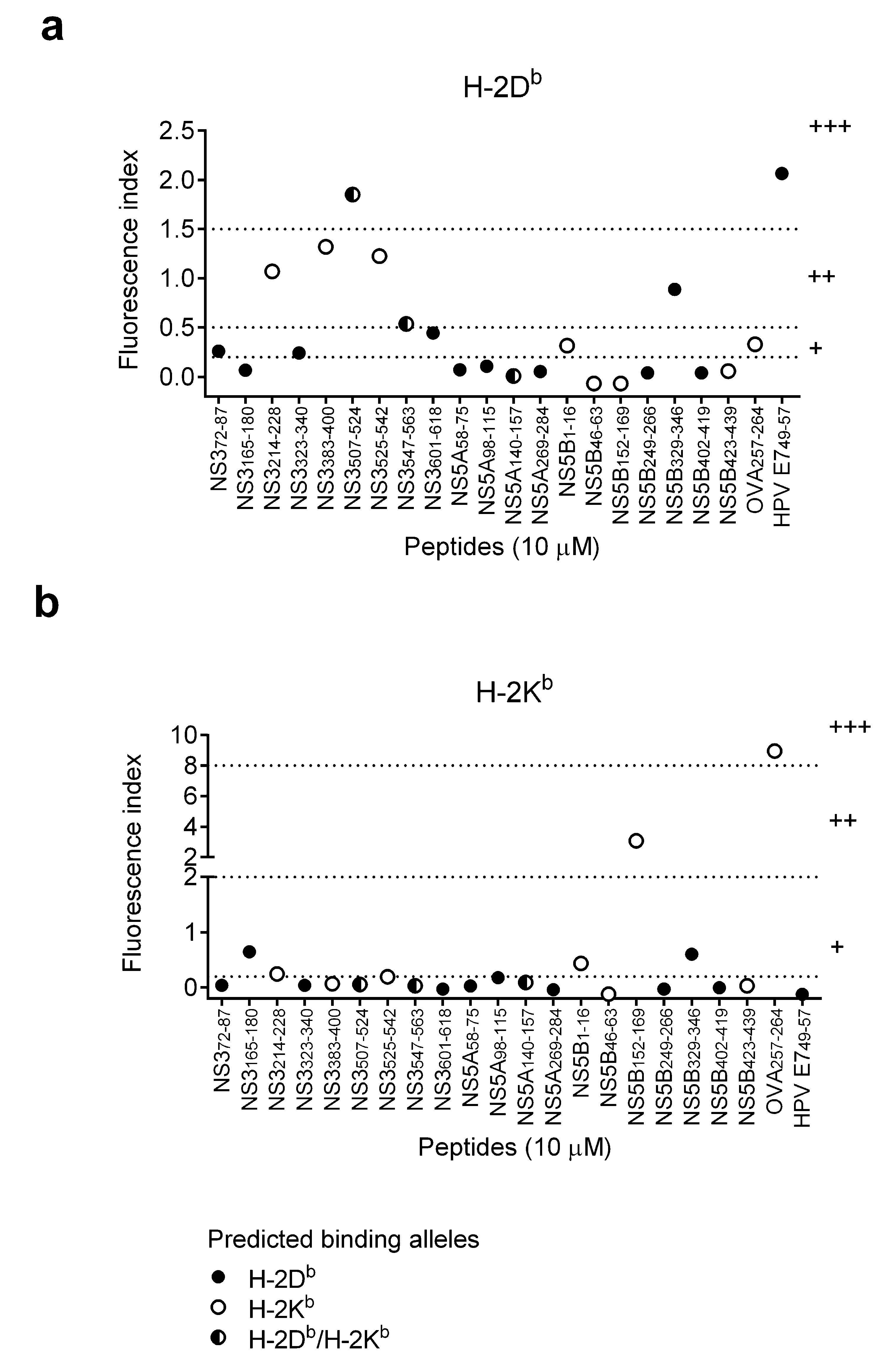
3.2. Binding Affinity of HCV Short Peptides to MHC Class I Molecules

3.3. The Presence of Proteasomal Degradation Sites at the Carboxyterminal Site of the Predicted CTL Epitopes
3.4. The Presence of MHC Class II Epitopes Flanking the Predicted CTL Epitopes
3.5. Induction of Peptide-Specific Effector CD8+ T Cells in Vivo
| a | MHC I Stabilization Assay 1 | MHC Class I Prediction | Proteasomal Cleavage | MHC Class II Prediction | |||||||
| SYFPEITHI (>20 strong binding) | NetMHCpan 2.8 (<0.5 strong binding) | IEDB (<0.5 strong binding) | MAPPP (Cleavage Probability 3) | PAProC I (Score 4) | Netchop (Cleavage Probability 3) | IEDB- H-2-IAb (<10 strong binding) | |||||
| Protein/Position | Sequence | H-2Db | H-2Db | H-2Db | H-2Db | 5' of CTL Epitope | Complete CTL Epitope | 3' of CTL Epitope | |||
| NS2139-147 | YVYNHLTPL | +++ | 13 | 0.15 | 0.7 | 1 | 0- | 0.95 | 31.64–58.25 | 1.82–3.90 | 27.51–64.83 |
| NS3331-339 | VSHPNIEEV | +++ | 23 | 0.08 | 0.3 | 0.7502 | 121+++ | 0.97 | 2.21–12.13 | 10.72–48.25 | 31.02–54.21 |
| NS3514-522 | RAYMNTPGL | +++ | 22 | 0.1 | 0.7 | 0 | 78++ | 0.95 | 5.17–73.56 | 5.97–22.88 | 16.57–87.59 |
| NS3603-611 | GAVQNEVTL | +++ | 29 | 0.08 | 0.2 | 0 | 178+++ | 0.88 | 9.35–55.70 | 18.51–32.13 | 16.70–51.34 |
| HPV E749-57 | RAHYNIVTF | +++ | 23 | 0.08 | 0.2 | 1 | N.D. | 0.91 | 33.38–53.38 | 56.31–79.26 | 69.56–86.81 |
| NS5B52-60 | VTFDRLQVL | +++ | 15 | 15 | 27 | 0.5976 | 0- | 0.96 | 10.39–86.63 | 71.97–79.66 | 28.42–85.75 |
| OVA257-264 | SIINFEKL | ++ | 0 | 4 | 0.2 | 1 | N.D. | 0.97 | 51.45–69.97 | 51.88–81.68 | 9.57–82.66 |
| NS4B38-46 | AVQTNWQKL | ++ | 23 | 1 | 1.9 | 0 | 0- | 0.72 | 12.81–32.62 | 23.98–74.51 | 21.59–58.54 |
| NS5B2-10 | MSYSWTGAL | ++ | 13 | 2 | 5.3 | 1 | 0- | 0.87 | N.D. | 0.93–0.95 | 2.62–56.47 |
| NS5B425-433 | LMTHFFSVL | ++ | 13 | 8 | 9.1 | 0.5009 | 40++ | 0.96 | 11.68–61.33 | 36.91–72.20 | 46.08–77.76 |
| NS3265-273 | ITYSTYGKF | ++ | 9 | 32 | 14.3 | 1 | 0- | 0.209 | 10.30–47.29 | 36.17–47.83 | 49.48–84.06 |
| NS5A280-287 | ILDSFDPL | ++ | 0 | 15 | 0.2 | 0.906 | 0- | 0.92 | 51.43–86.16 | 18.50–86.11 | 20.39–86.16 |
| NS5B157-165 | ARLIVFPDL | ++ | 15 | 32 | 22 | 0.6886 | 0- | 0.96 | 44.16–67.37 | 47.47–56.37 | 51.99–81.87 |
| b | MHC I Stabilization Assay 2 | MHC Class I Prediction | Proteasomal Cleavage | MHC Class II Prediction | |||||||
| SYFPEITHI (>20 strong binding) | NetMHCpan 2.8 (<0.5 strong binding) | IEDB (<0.5 strong binding) | MAPPP (Cleavage Probability 3) | PAProC I (Score 4) | Netchop (Cleavage Probability 3) | IEDB- H-2-IAb (<10 strong binding) | |||||
| Protein/Position | Sequence | H-2Kb | H-2Kb | H-2Kb | H-2Kb | 5' of CTL Epitope | Complete CTL Epitope | 3' of CTL Epitope | |||
| OVA257-264 | SIINFEKL | +++ | 25 | 1.5 | 0.35 | 1 | N.D. | 0.97 | 51.45–69.97 | 51.88–81.68 | 9.57–82.66 |
| NS5B2-10 | MSYSWTGAL | +++ | 11 | 0.03 | 0.2 | 1 | 0- | 0.87 | N.D. | 0.93–0.95 | 2.62–56.47 |
| NS3265-273 | ITYSTYGKF | +++ | 11 | 0.4 | 0.3 | 1 | 0- | 0.209 | 10.30–47.29 | 36.17–47.83 | 49.48–84.06 |
| NS5B157-165 | ARLIVFPDL | +++ | 22 | 15 | 12.35 | 0.6886 | 0- | 0.96 | 44.16–67.37 | 47.47–56.37 | 51.99–81.87 |
| NS2139-147 | YVYNHLTPL | +++ | 11 | 0.01 | 0.2 | 1 | 0- | 0.95 | 31.64–58.25 | 1.82–3.90 | 27.51–64.83 |
| NS5B52-60 | VTFDRLQVL | +++ | 11 | 0.05 | 0.25 | 0.5976 | 0- | 0.96 | 10.39–86.63 | 71.97–79.66 | 28.42–85.75 |
| NS3514-522 | RAYMNTPGL | ++ | 11 | 0.25 | 0.7 | 0 | 78++ | 0.95 | 5.17–73.56 | 5.97–22.88 | 16.57 – 87.59 |
| NS5B425-433 | LMTHFFSVL | ++ | 21 | 0.17 | 0.3 | 0.5009 | 40++ | 0.96 | 11.68–61.33 | 36.91–72.20 | 46.08–77.76 |
| NS5A280-287 | ILDSFDPL | ++ | 21 | 6 | 1.45 | 0.906 | 0- | 0.92 | 51.43–86.16 | 18.50–86.11 | 20.39–86.16 |
| NS4B38-46 | AVQTNWQKL | + | 11 | 15 | 8.2 | 0 | 0- | 0.72 | 12.81–32.62 | 23.98–74.51 | 21.59–58.54 |
| NS3331-339 | VSHPNIEEV | + | 9 | 8 | 9.45 | 0.7502 | 121+++ | 0.97 | 2.21–12.13 | 10.72–48.25 | 31.02–54.21 |
| NS3603-611 | GAVQNEVTL | - | 12 | 32 | 12.95 | 0 | 178+++ | 0.88 | 9.35–55.70 | 18.51–32.13 | 16.70–51.34 |
| HPV E749-57 | RAHYNIVTF | - | 8 | 5 | 11.2 | 1 | N.D. | 0.91 | 33.38–53.38 | 56.31–79.26 | 69.56–86.81 |

4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ogata, N.; Alter, H.J.; Miller, R.H.; Purcell, R.H. Nucleotide sequence and mutation rate of the h strain of Hepatitis C virus. Proc. Natl. Acad. Sci. USA 1991, 88, 3392–3396. [Google Scholar] [CrossRef] [PubMed]
- Erickson, A.L.; Kimura, Y.; Igarashi, S.; Eichelberger, J.; Houghton, M.; Sidney, J.; McKinney, D.; Sette, A.; Hughes, A.L.; Walker, C.M. The outcome of Hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity 2001, 15, 883–895. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, I.M.; McHutchison, J.G.; Dusheiko, G.; di Bisceglie, A.M.; Reddy, K.R.; Bzowej, N.H.; Marcellin, P.; Muir, A.J.; Ferenci, P.; Flisiak, R.; et al. Telaprevir for previously untreated chronic Hepatitis C virus infection. N. Engl. J. Med. 2011, 364, 2405–2416. [Google Scholar] [CrossRef] [PubMed]
- Poordad, F.; McCone, J., Jr.; Bacon, B.R.; Bruno, S.; Manns, M.P.; Sulkowski, M.S.; Jacobson, I.M.; Reddy, K.R.; Goodman, Z.D.; Boparai, N.; et al. Boceprevir for untreated chronic HCV genotype 1 infection. N. Engl. J. Med. 2011, 364, 1195–1206. [Google Scholar] [CrossRef] [PubMed]
- Rehermann, B. Hepatitis C virus versus innate and adaptive immune responses: A tale of coevolution and coexistence. J. Clin. Investig. 2009, 119, 1745–1754. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.H.; Cox, A.; Hoover, D.R.; Wang, X.H.; Mao, Q.; Ray, S.; Strathdee, S.A.; Vlahov, D.; Thomas, D.L. Protection against persistence of Hepatitis C. Lancet 2002, 359, 1478–1483. [Google Scholar] [CrossRef] [PubMed]
- Shoukry, N.H.; Grakoui, A.; Houghton, M.; Chien, D.Y.; Ghrayeb, J.; Reimann, K.A.; Walker, C.M. Memory CD8+ T cells are required for protection from persistent Hepatitis C virus infection. J. Exp. Med. 2003, 197, 1645–1655. [Google Scholar] [CrossRef] [PubMed]
- McKiernan, S.M.; Hagan, R.; Curry, M.; McDonald, G.S.; Kelly, A.; Nolan, N.; Walsh, A.; Hegarty, J.; Lawlor, E.; Kelleher, D. Distinct MHC class I and II alleles are associated with Hepatitis C viral clearance, originating from a single source. Hepatology 2004, 40, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Neumann-Haefelin, C.; McKiernan, S.; Ward, S.; Viazov, S.; Spangenberg, H.C.; Killinger, T.; Baumert, T.F.; Nazarova, N.; Sheridan, I.; Pybus, O.; et al. Dominant influence of an HLA-B27 restricted CD8+ T cell response in mediating HCV clearance and evolution. Hepatology 2006, 43, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Fitzmaurice, K.; Petrovic, D.; Ramamurthy, N.; Simmons, R.; Merani, S.; Gaudieri, S.; Sims, S.; Dempsey, E.; Freitas, E.; Lea, S.; et al. Molecular footprints reveal the impact of the protective HLA-A*03 allele in Hepatitis C virus infection. Gut 2011, 60, 1563–1571. [Google Scholar] [CrossRef] [PubMed]
- Ip, P.P.; Boerma, A.; Regts, J.; Meijerhof, T.; Wilschut, J.; Nijman, H.W.; Daemen, T. Alphavirus-based vaccines encoding nonstructural proteins of Hepatitis C virus induce robust and protective T-cell responses. Mol. Ther. 2014, 22, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Kolykhalov, A.A.; Agapov, E.V.; Blight, K.J.; Mihalik, K.; Feinstone, S.M.; Rice, C.M. Transmission of Hepatitis C by intrahepatic inoculation with transcribed RNA. Science 1997, 277, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Rammensee, H.; Bachmann, J.; Emmerich, N.P.; Bachor, O.A.; Stevanovic, S. Syfpeithi: Database for MHC ligands and peptide motifs. Immunogenetics 1999, 50, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Hoof, I.; Peters, B.; Sidney, J.; Pedersen, L.E.; Sette, A.; Lund, O.; Buus, S.; Nielsen, M. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 2009, 61, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.; Sidney, J.; Bourne, P.; Bui, H.H.; Buus, S.; Doh, G.; Fleri, W.; Kronenberg, M.; Kubo, R.; Lund, O.; et al. The immune epitope database and analysis resource: From vision to blueprint. PLOS Biol. 2005, 3, e91. [Google Scholar] [CrossRef] [PubMed]
- Shirai, M.; Akatsuka, T.; Pendleton, C.D.; Houghten, R.; Wychowski, C.; Mihalik, K.; Feinstone, S.; Berzofsky, J.A. Induction of cytotoxic T cells to a cross-reactive epitope in the Hepatitis C virus nonstructural RNA polymerase-like protein. J. Virol. 1992, 66, 4098–4106. [Google Scholar] [PubMed]
- Lauer, G.M.; Barnes, E.; Lucas, M.; Timm, J.; Ouchi, K.; Kim, A.Y.; Day, C.L.; Robbins, G.K.; Casson, D.R.; Reiser, M.; et al. High resolution analysis of cellular immune responses in resolved and persistent Hepatitis C virus infection. Gastroenterology 2004, 127, 924–936. [Google Scholar] [CrossRef] [PubMed]
- Christinck, E.R.; Luscher, M.A.; Barber, B.H.; Williams, D.B. Peptide binding to class I MHC on living cells and quantitation of complexes required for CTL lysis. Nature 1991, 352, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.C.; Jardetzky, T.S.; Garrett, T.P.; Lane, W.S.; Strominger, J.L.; Wiley, D.C. Different length peptides bind to HLA-AW68 similarly at their ends but bulge out in the middle. Nature 1992, 360, 364–366. [Google Scholar] [CrossRef] [PubMed]
- Crowe, S.R.; Miller, S.C.; Brown, D.M.; Adams, P.S.; Dutton, R.W.; Harmsen, A.G.; Lund, F.E.; Randall, T.D.; Swain, S.L.; Woodland, D.L. Uneven distribution of MHC class II epitopes within the influenza virus. Vaccine 2006, 24, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Dow, C.; Oseroff, C.; Peters, B.; Nance-Sotelo, C.; Sidney, J.; Buchmeier, M.; Sette, A.; Mothe, B.R. Lymphocytic choriomeningitis virus infection yields overlapping CD4+ and CD8+ T-cell responses. J. Virol. 2008, 82, 11734–11741. [Google Scholar] [CrossRef] [PubMed]
- Mothe, B.R.; Stewart, B.S.; Oseroff, C.; Bui, H.H.; Stogiera, S.; Garcia, Z.; Dow, C.; Rodriguez-Carreno, M.P.; Kotturi, M.; Pasquetto, V.; et al. Chronic lymphocytic choriomeningitis virus infection actively down-regulates CD4+ T cell responses directed against a broad range of epitopes. J. Immunol. 2007, 179, 1058–1067. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, R.; Celis, E. Multiple roles for CD4+ T cells in anti-tumor immune responses. Immunol. Rev. 2008, 222, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, M.K.; Moon, J.J. The role of naive T cell precursor frequency and recruitment in dictating immune response magnitude. J. Immunol. 2012, 188, 4135–4140. [Google Scholar] [CrossRef] [PubMed]
- Kotturi, M.F.; Scott, I.; Wolfe, T.; Peters, B.; Sidney, J.; Cheroutre, H.; von Herrath, M.G.; Buchmeier, M.J.; Grey, H.; Sette, A. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J. Immunol. 2008, 181, 2124–2133. [Google Scholar] [CrossRef] [PubMed]
- Ahlen, G.; Nystrom, J.; Pult, I.; Frelin, L.; Hultgren, C.; Sallberg, M. In vivo clearance of Hepatitis C virus nonstructural 3/4A-expressing hepatocytes by DNA vaccine-primed cytotoxic T lymphocytes. J. Infect. Dis. 2005, 192, 2112–2116. [Google Scholar] [CrossRef] [PubMed]
- Wolk, B.; Trautwein, C.; Buchele, B.; Kersting, N.; Blum, H.E.; Rammensee, H.G.; Cerny, A.; Stevanovic, S.; Moradpour, D.; Brass, V. Identification of naturally processed Hepatitis C virus-derived major histocompatibility complex class I ligands. PLOS ONE 2012, 7, e29286. [Google Scholar] [CrossRef] [PubMed]
- De Lucas, S.; Bartolome, J.; Rodriguez-Inigo, E.; Casqueiro, M.; Millan, A.; Ruiz-Moreno, M.; Oliva, H.; Carreno, V. Distribution of Hepatitis C virus infection in liver biopsies from children and adults with chronic Hepatitis C. J. Med. Virol. 2001, 64, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.M.; Diamond, D.L.; Chan, E.Y.; Gritsenko, M.A.; Qian, W.; Stastna, M.; Baas, T.; Camp, D.G., 2nd.; Carithers, R.L., Jr.; Smith, R.D.; et al. Proteome analysis of liver cells expressing a full-length Hepatitis C virus (HCV) replicon and biopsy specimens of posttransplantation liver from HCV-infected patients. J. Virol. 2005, 79, 7558–7569. [Google Scholar] [CrossRef] [PubMed]
- Rotzschke, O.; Falk, K.; Stevanovic, S.; Jung, G.; Walden, P.; Rammensee, H.G. Exact prediction of a natural T cell epitope. Eur. J. Immunol. 1991, 21, 2891–2894. [Google Scholar] [CrossRef] [PubMed]
- Sette, A.; Buus, S.; Appella, E.; Smith, J.A.; Chesnut, R.; Miles, C.; Colon, S.M.; Grey, H.M. Prediction of major histocompatibility complex binding regions of protein antigens by sequence pattern analysis. Proc. Natl. Acad. Sci. USA 1989, 86, 3296–3300. [Google Scholar] [CrossRef] [PubMed]
- Falk, K.; Rotzschke, O.; Stevanovic, S.; Jung, G.; Rammensee, H.G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 1991, 351, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Groettrup, M.; Kirk, C.J.; Basler, M. Proteasomes in immune cells: More than peptide producers? Nat. Rev. Immunol. 2010, 10, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Rock, K.L.; Goldberg, A.L. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 1999, 17, 739–779. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Norbury, C.C.; Cho, Y.; Yewdell, J.W.; Bennink, J.R. Immunoproteasomes shape immunodominance hierarchies of antiviral CD8+ T cells at the levels of T cell repertoire and presentation of viral antigens. J. Exp. Med. 2001, 193, 1319–1326. [Google Scholar] [CrossRef] [PubMed]
- Guillaume, B.; Chapiro, J.; Stroobant, V.; Colau, D.; van Holle, B.; Parvizi, G.; Bousquet-Dubouch, M.P.; Theate, I.; Parmentier, N.; van den Eynde, B.J. Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc. Natl. Acad. Sci. USA 2010, 107, 18599–18604. [Google Scholar] [CrossRef] [PubMed]
- Kesmir, C.; Nussbaum, A.K.; Schild, H.; Detours, V.; Brunak, S. Prediction of proteasome cleavage motifs by neural networks. Protein Eng. 2002, 15, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Piontkivska, H. Frequent associations between CTL and T-helper epitopes in HIV-1 genomes and implications for multi-epitope vaccine designs. BMC Microbiol. 2010. [Google Scholar] [CrossRef]
- Rice, J.; Ottensmeier, C.H.; Stevenson, F.K. DNA vaccines: Precision tools for activating effective immunity against cancer. Na. Rev. Cancer 2008, 8, 108–120. [Google Scholar] [CrossRef]
- Tsang, J.Y.; Chai, J.G.; Lechler, R. Antigen presentation by mouse CD4+ T cells involving acquired MCH class II: Peptide complexes: Another mechanism to limit clonal expansion? Blood 2003, 101, 2704–2710. [Google Scholar] [CrossRef] [PubMed]
- Umeshappa, C.S.; Huang, H.; Xie, Y.; Wei, Y.; Mulligan, S.J.; Deng, Y.; Xiang, J. CD4+ Th-APC with acquired peptide/MHC class I and II complexes stimulate type 1 helper CD4+ and central memory CD8+ T cell responses. J. Immunol. 2009, 182, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, S.A.; McKeithan, T.W.; Parker, D.C. Peptide-specific intercellular transfer of MHC class II to CD4+ T cells directly from the immunological synapse upon cellular dissociation. J. Immunol. 2005, 174, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Gromme, M.; Uytdehaag, F.G.; Janssen, H.; Calafat, J.; van Binnendijk, R.S.; Kenter, M.J.; Tulp, A.; Verwoerd, D.; Neefjes, J. Recycling MHC class I molecules and endosomal peptide loading. Proc. Natl. Acad. Sci. USA 1999, 96, 10326–10331. [Google Scholar] [CrossRef] [PubMed]
- Reimann, J.; Schirmbeck, R. Alternative pathways for processing exogenous and endogenous antigens that can generate peptides for MHC class I-restricted presentation. Immunol. Rev. 1999, 172, 131–152. [Google Scholar] [CrossRef] [PubMed]
- Lanzavecchia, A. From antigen presentation to T-cell activation. Res. Immunol. 1998, 149, 626. [Google Scholar] [CrossRef] [PubMed]
- Ossevoort, M.A.; de Bruijn, M.L.; van Veen, K.J.; Kast, W.M.; Melief, C.J. Peptide specificity of alloreactive CD4 positive T lymphocytes directed against a major histocompatibility complex class I disparity. Transplantation 1996, 62, 1485–1491. [Google Scholar] [CrossRef] [PubMed]
- Ip, P.P.; Boerma, A.; Walczak, M.; Oosterhuis, K.; Haanen, J.B.; Schumacher, T.N.; Nijman, H.W.; Daemen, T. Antigen design enhances the immunogenicity of Semliki Forest virus-based therapeutic human papillomavirus vaccines. Gene Ther. 2015. [Google Scholar] [CrossRef]
- Kedl, R.M.; Kappler, J.W.; Marrack, P. Epitope dominance, competition and T cell affinity maturation. Curr. Opin. Immunol. 2003, 15, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Henrickson, S.E.; Perro, M.; Loughhead, S.M.; Senman, B.; Stutte, S.; Quigley, M.; Alexe, G.; Iannacone, M.; Flynn, M.P.; Omid, S.; et al. Antigen availability determines CD8+ T cell-dendritic cell interaction kinetics and memory fate decisions. Immunity 2013, 39, 496–507. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ip, P.P.; Nijman, H.W.; Daemen, T. Epitope Prediction Assays Combined with Validation Assays Strongly Narrows down Putative Cytotoxic T Lymphocyte Epitopes. Vaccines 2015, 3, 203-220. https://doi.org/10.3390/vaccines3020203
Ip PP, Nijman HW, Daemen T. Epitope Prediction Assays Combined with Validation Assays Strongly Narrows down Putative Cytotoxic T Lymphocyte Epitopes. Vaccines. 2015; 3(2):203-220. https://doi.org/10.3390/vaccines3020203
Chicago/Turabian StyleIp, Peng Peng, Hans W. Nijman, and Toos Daemen. 2015. "Epitope Prediction Assays Combined with Validation Assays Strongly Narrows down Putative Cytotoxic T Lymphocyte Epitopes" Vaccines 3, no. 2: 203-220. https://doi.org/10.3390/vaccines3020203
APA StyleIp, P. P., Nijman, H. W., & Daemen, T. (2015). Epitope Prediction Assays Combined with Validation Assays Strongly Narrows down Putative Cytotoxic T Lymphocyte Epitopes. Vaccines, 3(2), 203-220. https://doi.org/10.3390/vaccines3020203





