Comparative Analysis of Cotton Small RNAs and Their Target Genes in Response to Salt Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Measurement of Na+ Contents and Proline Level
2.3. Total RNA Isolation andConstruction of Small RNA and Degradome Libraries
2.4. Transcriptome Sequencing andde novoAssembly
2.5. Analysis of Small RNA and Degradome Sequencing Data
2.6. qRT-PCR Analysis
2.7. Statistical Analysis
3. Results
3.1. Deep-Sequencing of sRNAs under Salt Stress
3.2. Small RNA Annotation and miRNA Identification
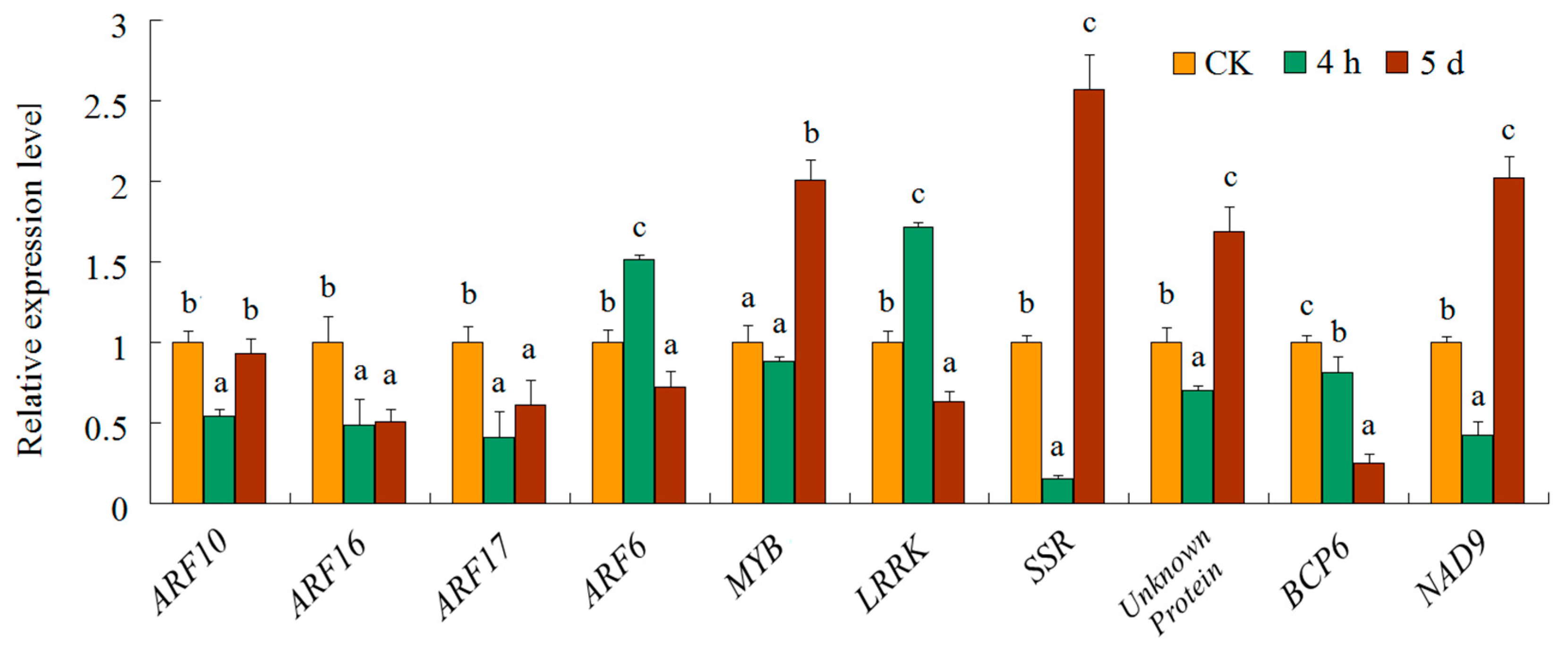
3.3. miRNA Expression Patterns under Different Salt Stress Treatment
3.4. Retrotransposons and Their Endogenous siRNAs
3.5. Transcriptome-Wide Identification of miRNA Targets in Cotton
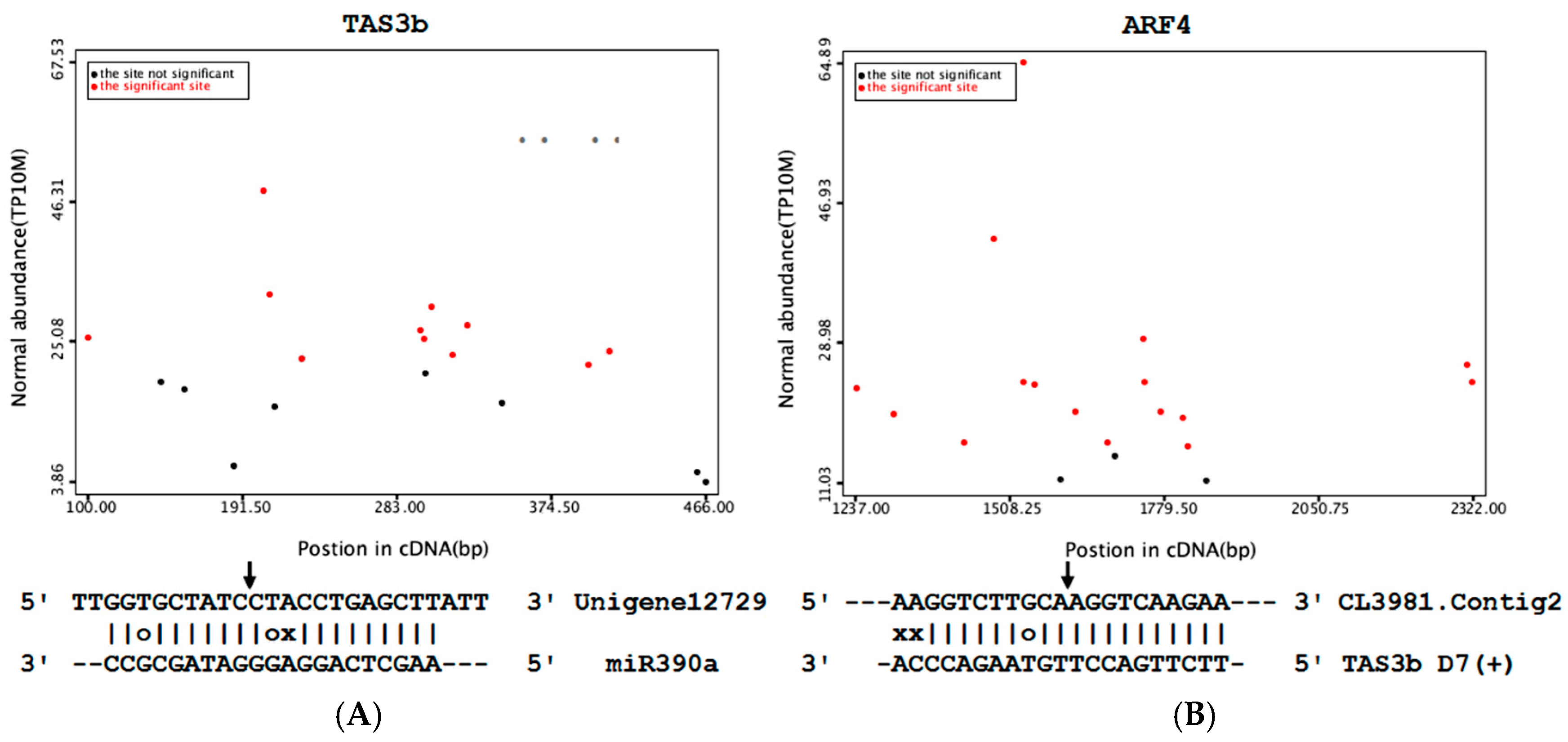
3.6. Cotton miR390/tasiRNA-ARF/ARF4 Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Xiong, L.; Schumaker, K.S.; Zhu, J.K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef] [PubMed]
- Shavrukov, Y. Salt stress or salt shock: Which genes are we studying? J. Exp. Bot. 2013, 64, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.F.; Song, J.; Fan, H.; Zhou, S.; Zhao, M. Growth response to ionic and osmotic stress of NaCl in salt-tolerant and salt-sensitive maize. J. Integr. Plant Biol. 2010, 52, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Duan, H.; Li, J.; Deng, X.W.; Yin, W.; Xia, X. Global identification of miRNAs and targets in Populus euphratica under salt stress. Plant Mol. Biol. 2013, 81, 525–539. [Google Scholar] [CrossRef] [PubMed]
- Busov, V.B.; Brunner, A.M.; Strauss, S.H. Genes for control of plant stature and form. New Phytol. 2008, 177, 589–607. [Google Scholar] [CrossRef] [PubMed]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pan, X.; Cannon, C.H.; Cobb, G.P.; Anderson, T.A. Conservation and divergence of plant microRNA genes. Plant J. 2006, 46, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J.; Snyder, J.A.; Bartel, D.P. Common functions for diverse small RNAs of land plants. Plant Cell 2007, 19, 1750–1769. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. microRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 2013, 64, 137–159. [Google Scholar] [CrossRef] [PubMed]
- Shukla, L.I.; Chinnusamy, V.; Sunkar, R. The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochim. Biophys. Acta 2008, 1779, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Nonogaki, H. MicroRNA gene regulation cascades during early stages of plant development. Plant Cell Physiol. 2010, 51, 1840–1846. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Hu, L.; Han, N.; Hu, J.; Yang, Y.; Xiang, T.; Zhang, X.; Wang, L. Overexpression of a miR393-resistant form of transport inhibitor response protein 1 (mTIR1) enhances salt tolerance by increased osmoregulation and Na+ exclusion in Arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Bai, X.; Yang, L.; Lv, D.; Pan, X.; Li, Y.; Cai, H.; Ji, W.; Chen, Q.; Zhu, Y. Osa-miR393: A salinity- and alkaline stress-related microRNA gene. Mol. Biol. Rep. 2010, 38, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Wang, Q.; Sun, R.; Zhang, B. Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J. Exp. Bot. 2015, 66, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, Q.; Zhang, B. Response of miRNAs and their targets to salt and drought stresses in cotton (Gossypium hirsutum L.). Gene 2013, 530, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Chinnusamy, V.; Zhu, J.; Zhu, J.K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007, 12, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Datt Pant, B.; Stitt, M.; Scheible, W.R. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Li, Y.; Yu, J.; Liu, Y.; Li, C.; Han, X.; Shen, F. Difference in miRNA expression profiles between two cotton cultivars with distinct salt sensitivity. Mol. Biol. Rep. 2012, 39, 4961–4970. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.; Zhang, L.; Wang, H.; Liu, Z.; Zhang, Z.; Zheng, Y. Differential expression of miRNAs in response to salt stress in maize roots. Ann. Bot. 2009, 103, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Leidi, E.O.; Saiz, J.F. Is salinity tolerance related to Na+ accumulation in upland cotton (Gossypium hirsutum) seedlings? Plant Soil 1997, 190, 67–75. [Google Scholar] [CrossRef]
- Carra, A.; Gambino, G.; Schubert, A. A cetyltrimethylammonium bromide-based method to extract low-molecular-weight RNA from polysaccharide-rich plant tissues. Anal. Biochem. 2007, 360, 318–320. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Li, Y.; Han, X.; Shen, F. Genome-wide profiling of miRNAs and other small non-coding RNAs in the Verticillium dahliae-inoculated cotton roots. PLoS ONE 2012, 7, e35765. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.; Landgraf, P.; Ludwig, J.; Rice, A.; Ojo, T.; Lin, C.; Holoch, D.; Lim, C.; Tuschl, T. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 2008, 44, 3–12. [Google Scholar] [CrossRef] [PubMed]
- German, M.A.; Pillay, M.; Jeong, D.H.; Hetawal, A.; Luo, S.; Janardhanan, P.; Kannan, V.; Rymarquis, L.A.; Nobuta, K.; German, R.; et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008, 26, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Rfam. Available online: http://www.sanger.ac.uk/resources/databases/rfam.html (accessed on 9 January 2013).
- Kozomara, A.; Griffiths-Jones, S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar] [CrossRef] [PubMed]
- MIREAP program. Available online: https://sourceforge.net/projects/mireap/ (accessed on 19 December 2012).
- Yin, Z.; Li, C.; Han, X.; Shen, F. Identification of conserved microRNAs and their target genes in tomato (Lycopersicon esculentum). Gene 2008, 414, 60–66. [Google Scholar] [CrossRef] [PubMed]
- MFOLD software. Available online: http://mfold.rna.albany.edu/ (accessed on 15 June 2013).
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef] [PubMed]
- Varkonyi-Gasic, E.; Wu, R.; Wood, M.; Walton, E.F.; Hellens, R.P. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 2007, 3, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macovei, A.; Tuteja, N. MicroRNAs targeting DEAD-box helicases are involved in salinity stress response in rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.H.; Pan, X.P.; Cox, S.B.; Cobb, G.P.; Anderson, T.A. Evidence that miRNAs are different from other RNAs. Cell. Mol. Life Sci. 2006, 63, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Unver, T.; Namuth-Covert, D.M.; Budak, H. Review of current methodological approaches for characterizing microRNAs in plants. Int. J. Plant Genom. 2009, 2009, 262463. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, J.S.; Kim, H.; Nason, J.D.; Wing, R.A.; Wendel, J.F. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 2006, 16, 1252–1261. [Google Scholar] [CrossRef] [PubMed]
- Kavi Kishor, P.B.; Sreenivasulu, N. Is proline accumulation per se correlated with stress tolerance or is proline homeostasis a more critical issue? Plant Cell Environ. 2014, 37, 300–311. [Google Scholar] [CrossRef] [PubMed]
- Parida, A.K.; Das, A.B. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; James, R.A.; Lauchli, A. Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 2006, 57, 1025–1043. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Stewart, C.N.; Taki, F.A.; He, Q.; Liu, H.; Zhang, B. High-throughput deep sequencing shows that microRNAs play important roles in switchgrass responses to drought and salinity stress. Plant Biotechnol. J. 2014, 12, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Barrera-Figueroa, B.E.; Gao, L.; Wu, Z.; Zhou, X.; Zhu, J.; Jin, H.; Liu, R.; Zhu, J.K. High throughput sequencing reveals novel and abiotic stress-regulated microRNAs in the inflorescences of rice. BMC Plant Biol. 2012, 12, 132. [Google Scholar] [CrossRef] [PubMed]
- Frazier, T.P.; Sun, G.; Burklew, C.E.; Zhang, B. Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Mol. Biotechnol. 2011, 49, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Junghans, U.; Polle, A.; Duchting, P.; Weiler, E.; Kuhlman, B.; Gruber, F.; Teichmann, T. Adaptation to high salinity in poplar involves changes in xylem anatomy and auxin physiology. Plant Cell Environ. 2006, 29, 1519–1531. [Google Scholar] [CrossRef] [PubMed]
- Mockaitis, K.; Estelle, M. Auxin receptors and plant development: A new signaling paradigm. Annu. Rev. Cell Dev. Biol. 2008, 24, 55–80. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.C.; Bartel, D.P.; Bartel, B. MicroRNA-directed regulation of ArabidopsisAUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 2005, 17, 1360–1375. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.F.; Tian, Q.; Reed, J.W. ArabidopsismicroRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 2006, 133, 4211–4218. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.H.; Han, S.J.; Yoon, E.K.; Lee, W.S. Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res. 2006, 34, 1892–1899. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 1999, 96, 5844–5849. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 2003, 15, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Fahlgren, N.; Montgomery, T.A.; Howell, M.D.; Allen, E.; Dvorak, S.K.; Alexander, A.L.; Carrington, J.C. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 2006, 16, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.K.; Yang, J.H.; Lim, J.; Kim, S.H.; Kim, S.K.; Lee, W.S. Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res. 2010, 38, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Hunter, C.; Willmann, M.R.; Wu, G.; Yoshikawa, M.; de la Luz Gutierrez-Nava, M.; Poethig, S.R. Trans-acting siRNA-mediated repression of RTTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 2006, 133, 2973–2981. [Google Scholar] [CrossRef] [PubMed]
- Nemhauser, J.L.; Feldman, L.J.; Zambryski, P.C. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 2000, 127, 3877–3888. [Google Scholar] [PubMed]
- Pekker, I.; Alvarez, J.P.; Eshed, Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 2005, 17, 2899–2910. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 2005, 17, 444–463. [Google Scholar] [CrossRef] [PubMed]
- Schruff, M.C.; Spielman, M.; Tiwari, S.; Adams, S.; Fenby, N.; Scott, R.J. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 2006, 133, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E.; et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef] [PubMed]
- Harper, R.M.; Stowe-Evans, E.L.; Luesse, D.R.; Muto, H.; Tatematsu, K.; Watahiki, M.K.; Yamamoto, K.; Liscum, E. The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 2000, 12, 757–770. [Google Scholar] [CrossRef] [PubMed]
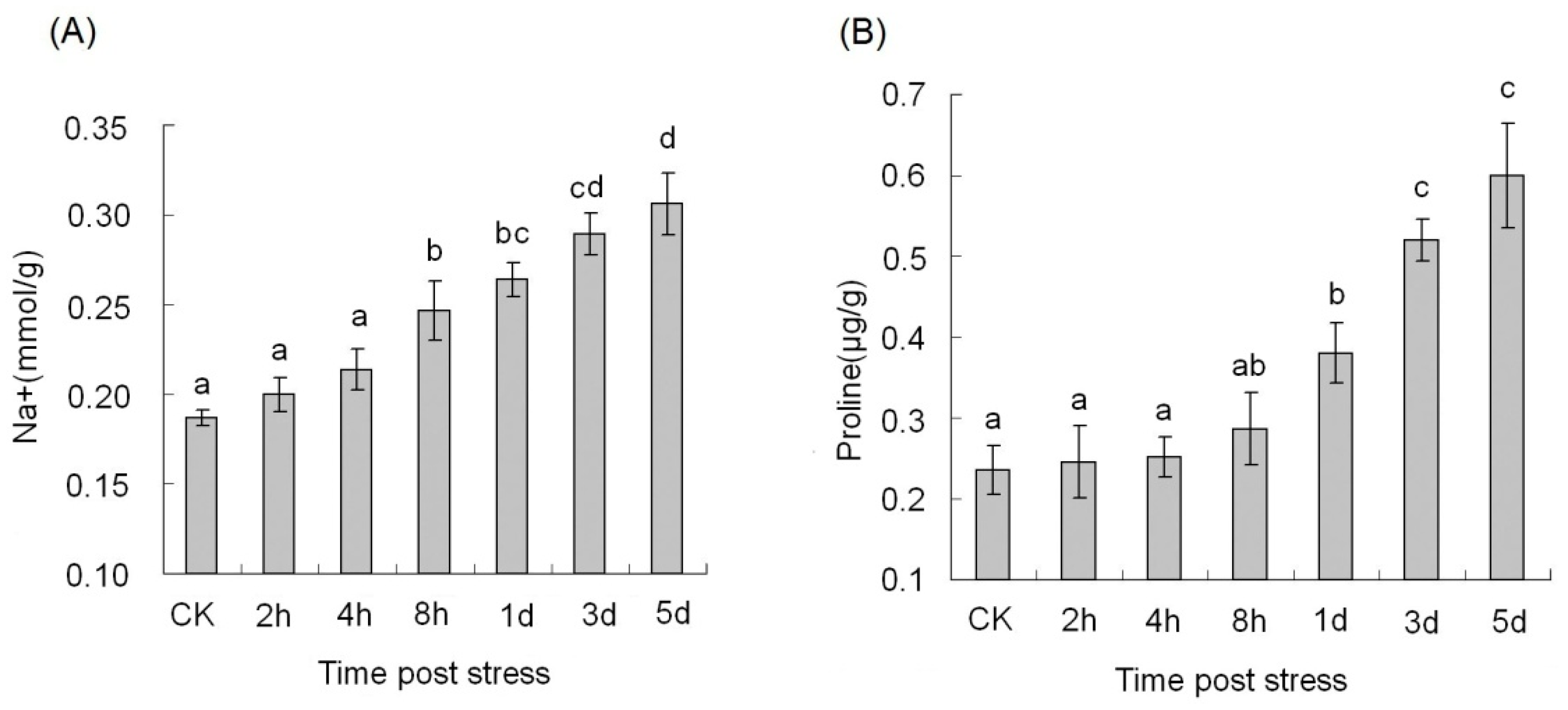
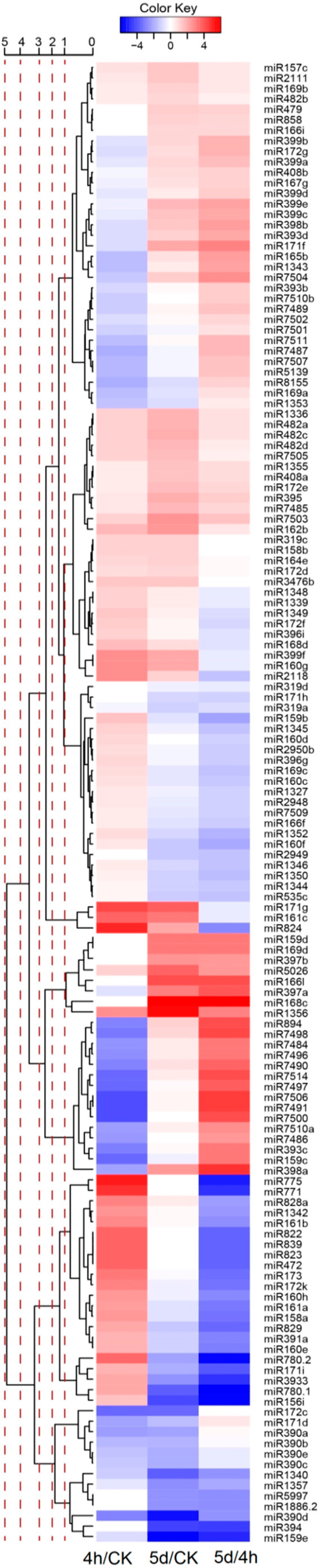
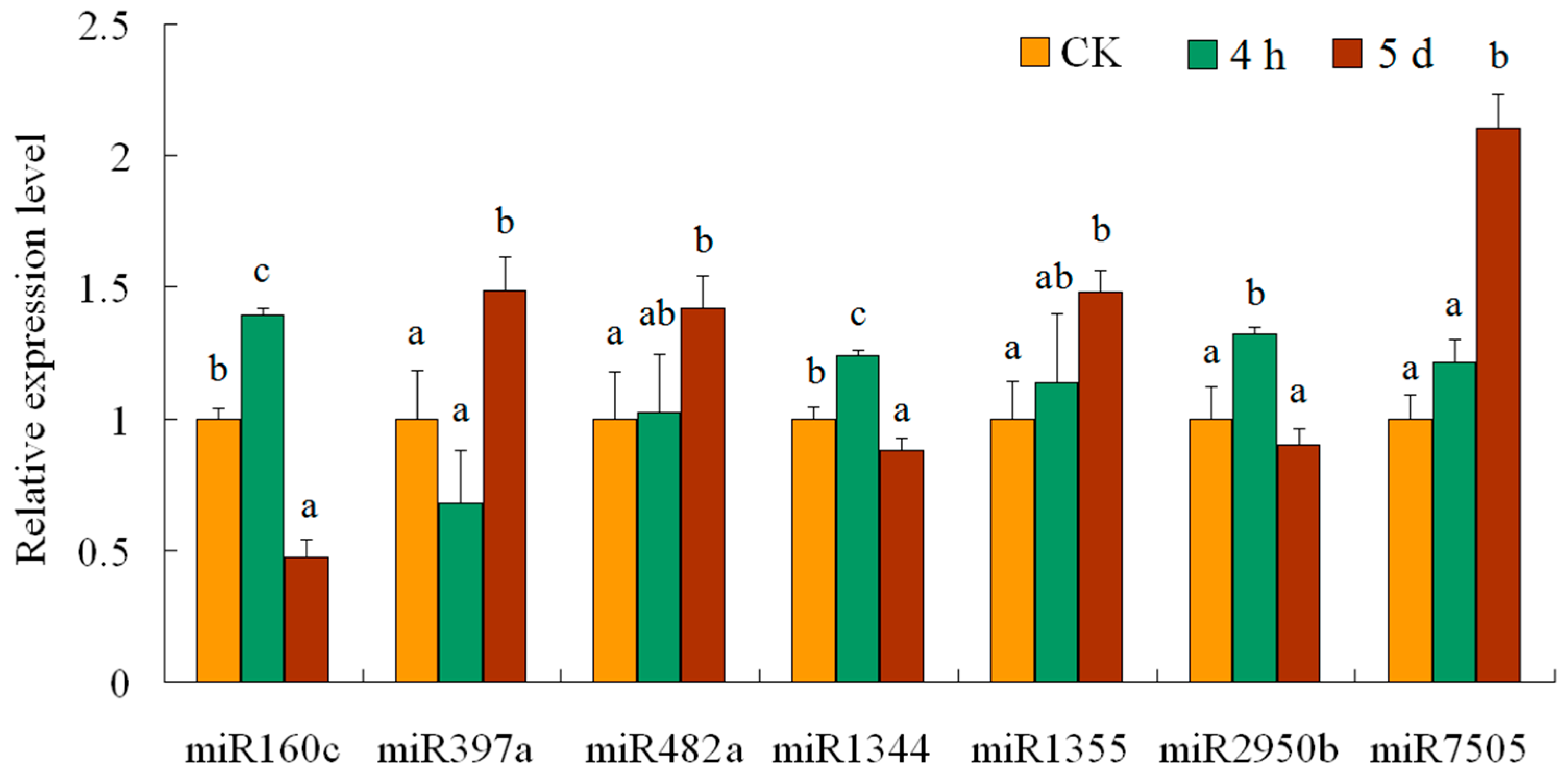
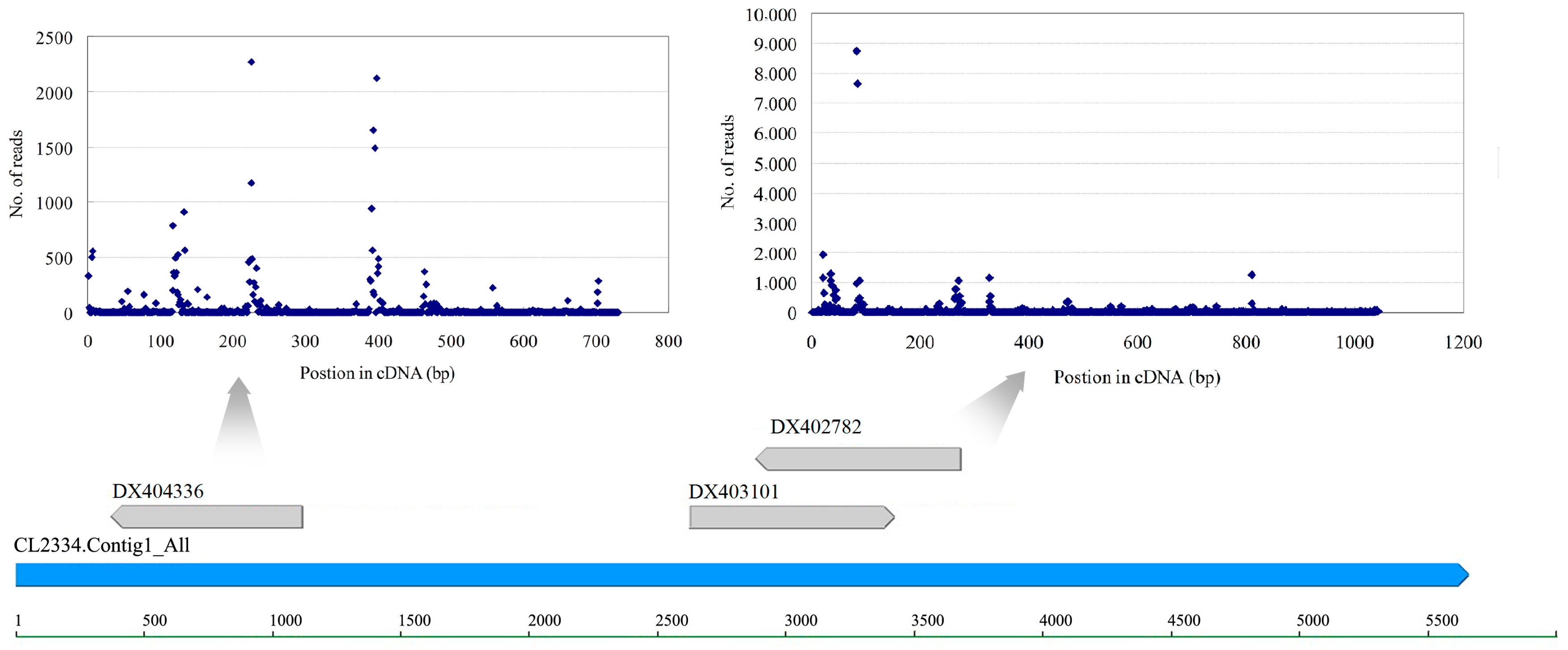



| Category | Small RNA | Degradome | Transcriptome | ||||
|---|---|---|---|---|---|---|---|
| CK | 4 h | 5 d | CK | 4 h | 5 d | ||
| Total reads | 17,533,903 | 16,853,769 | 22,982,882 | 22,766,839 | 30,703,662 | 20,880,679 | 57,194,024 |
| High quality | 17,389,916 | 16,704,868 | 22,900,931 | 22,744,209 | 30,684,331 | 20,837,655 | 54,261,421 |
| Clean reads | 17,026,070 | 15,731,477 | 22,590,391 | 22,653,372 | 30,649,472 | 20,685,800 | 52,170,788 |
| Unigene sequences | 4,331,028 | 3,703,594 | 4,802,868 | 864,720 | 1,217,757 | 1,171,577 | 81,114 |
| Name | Mature miRNA Sequence | Gene ID | LM (nt) | Arm | LP (nt) | G+C(%) | miRNA* Sequence | ΔG kcal/mol | MFEIs |
|---|---|---|---|---|---|---|---|---|---|
| miR1327 | TGAATATTGTTAAAGTAGAAA | Unigene15286 | 21 | 3′ | 122 | 27.87 | TCTACTTTAACAATATTCATA | −43.30 | 1.27 |
| miR1335 | TGAAGCTGCCACATGATCTAG | Unigene16386 | 21 | 5′ | 103 | 46.60 | AGATCATGCTGGCAGCTTCAAC | −50.20 | 1.05 |
| miR1336 | TCTTTCCTACTCCTCCCATTCC | CL1064.Contig1 | 22 | 3′ | 105 | 39.05 | AAGTGGGATGGGTGGAAAGATT | −44.50 | 1.09 |
| miR1337 | AATGGAGGAGTTGGAAAGATT | Unigene26231 | 21 | 5′ | 95 | 28.42 | TTTCCAATTCCTCCCATTCCAC | −36.70 | 1.36 |
| miR1338 | AATGAGTTAGGCGAGAGGTTC | CL5909.Contig2 | 21 | 3′ | 169 | 31.36 | CTCTGGTTTAACTCATTTGTA | −51.10 | 0.96 |
| miR1339 | AAAAGCAATGAAGAACGACCT | CL5831.Contig1 | 21 | 3′ | 102 | 38.24 | GCTGCGTTCTTCATTGCTTAACA | −41.30 | 1.06 |
| miR1340 | ACCGGCCGGGGGACGGACTGGG | CL6985.Contig2 | 22 | 3′ | 119 | 65.55 | CAGTCCCGAACCCGTCAGCTGC | −62.60 | 0.80 |
| miR1341 | CTTTTTATAGGATAGGGACTG | Unigene30026 | 21 | 3′ | 103 | 29.13 | GTTCCTATCTTATTAAAAAGAA | −37.10 | 1.24 |
| miR1342 | TGAAGCTGCAAGATGATCTGA | CL8098.Contig4 | 21 | 5′ | 262 | 34.35 | AGATCATGCTGGCAGCTTCAAC | −57.70 | 0.64 |
| miR1343 | CAGAGATCGTCAAAGCATATC | CL1118.Contig5 | 21 | 5′ | 94 | 37.23 | TATGCTTTGACGATCTCTGAT | −60.90 | 1.74 |
| miR1344 | TTTGCATGAACTAGGAGACGT | Unigene10644 | 21 | 3′ | 105 | 54.29 | CGGCTGCTAGTTCATGGATGCC | −45.60 | 0.80 |
| miR1345 | AAGCGGAAGCTGAATTAGTTG | Unigene14674 | 21 | 3′ | 76 | 43.42 | GAAACTAATCGAGCTCCGTTTGA | −26.00 | 0.79 |
| miR1346 | GCTGCCATCTCATGCATTCGG | Unigene20900 | 21 | 5′ | 165 | 44.85 | CGATGCATGGCATGGGAGCACCA | −59.10 | 0.80 |
| miR1347 | TACAGCTTTAGAAATCATCCCT | Unigene13628 | 22 | 5′ | 109 | 29.36 | GGATGATTTCTAAAGCTCTAGA | −48.40 | 1.51 |
| miR1348 | AGTGTCTGGGTGGTGTAGTTGGT | Unigene4349 | 23 | 5′ | 83 | 50.60 | CCATTCGAACACGGGCTCAGACATTT | −23.90 | 0.65 |
| miR1349 | TAACTTGTCTTCGCCCTTCTC | Unigene8023 | 21 | 3′ | 201 | 34.83 | GAAGTGCATGGCAAGTTAGA | −48.70 | 0.70 |
| miR1350 | TGGCACGGCTCAATCAAATTA | Unigene7762 | 21 | 5′ | 89 | 38.20 | ATTTGATTGAGCCATTCCAAC | −34.60 | 1.02 |
| miR1351 | ACTCATAATTTAGCAAAGTCG | Unigene8111 | 21 | 3′ | 129 | 28.68 | TTGCTGGATTATGAGTCTAAT | −61.40 | 1.66 |
| miR1352 | AATGCTTGAGGTGATAGGTTCA | Unigene14827 | 22 | 5′ | 137 | 50.37 | AACCATTGCCTCAAGCACTTG | −58.80 | 0.85 |
| miR1353 | AAGGCAAAGGAAGAAAAGAGTGA | CL9674.Contig1 | 23 | 5′ | 105 | 38.10 | ACTCTTTTTTTCCTGCCTTGC | −45.90 | 1.15 |
| miR1354 | AAAGTGGATGAAATTTTTAGC | Unigene26528 | 21 | 3′ | 159 | 36.48 | TAAAAATTTCATTCATTTCTA | −66.10 | 1.14 |
| miR1355 | ATGCACTGCCTCTTCCCTGGC | Unigene10226 | 21 | 3′ | 102 | 53.92 | AACAGGCTGAGCATGGATGGA | −43.40 | 0.79 |
| miR1356 | AACTGTGAAGCTATAAGGTAT | Unigene19434 | 21 | 5′ | 110 | 37.27 | ACTTTTGTTTCACATTAC | −23.90 | 0.65 |
| miR1357 | AAGCTGTTGATGGCCGGCATGA | CL9715.Contig1 | 22 | 5′ | 91 | 46.15 | ATGCCTATCATCAGGAGACTCT | −30.30 | 0.72 |
| miRNA Family | Target Gene | Alignment Score | Alignment Range | Cleavage Site | Category | Annotation | Abundance | ||
|---|---|---|---|---|---|---|---|---|---|
| CK | 4 h | 5 d | |||||||
| miR160 | Unigene5547 (CotAD_09605) | 4 | 378–398 | 389 | 2 | ARF 10 | 361 | 254 | 207 |
| CL11606.Contig2 (CotAD_08070) | 4 | 198–218 | 208 | 2 | ARF16 | 361 | 54 | 207 | |
| CL11606.Contig1 (CotAD_76501) | 2.5 | 345–366 | 357 | 2 | ARF16 | 2794 | 233 | 2249 | |
| CL1202.Contig4 (CotAD_55881) | 1 | 497–518 | 509 | 2 | ARF17 | 420 | 83 | 394 | |
| CL1202.Contig2 (CotAD_36520) | 2 | 2–22 | 12 | 3 | ARF17 | 476 | 83 | 307 | |
| miR164 | CL1985.Contig3 (CotAD_58864) | 4.5 | 19–39 | 30 | 2 | NAC | 0 | 61 | 0 |
| CL1985.Contig5 (CotAD_43532) | 4.5 | 19–39 | 30 | 2 | NAC | 0 | 61 | 0 | |
| miR165 | CL795.Contig6 (CotAD_15207) | 4 | 93–113 | 103 | 3 | Class III HD-Zip protein 4 | 111 | 0 | 63 |
| miR167 | Unigene18486 (CotAD_40344) | 4.5 | 3–23 | 15 | 2 | ARF6 | 45 | 0 | 125 |
| miR169 | CL599.Contig2 (CotAD_06772) | 2 | 1095–1115 | 1105 | 3 | Nuclear transcription factor Y subunit A-3 | 151 | 6 | 88 |
| miR172 | CL799.Contig8 (CotAD_29570) | 0 | 761–782 | 773 | 2 | AP2-EREBP | 189 | 104 | 277 |
| miR319 | CL11426.Contig1 (CotAD_73356) | 2 | 361–381 | 373 | 2 | MYB | 84 | 295 | 398 |
| miR396 | Unigene30805 (CotAD_25614) | 2 | 2–22 | 12 | 2 | GRF | 0 | 5 | 45 |
| miR828 | Unigene27368 (CotAD_00308) | 1 | 1–22 | 13 | 0 | MYB | 510 | 131 | 1060 |
| miR894 | CL861.Contig1 (CotAD_07539) | 4 | 2507–2528 | 2521 | 4 | Leucine Rich Repeat Kinase (LRRK) | 0 | 3 | 35 |
| miR1327 | CL4730.Contig1 | 3.5 | 782–803 | 795 | 3 | Simple Sequence Repeat Marker, mRNA sequence (SSR) | 0 | 5 | 72 |
| miR1344 | CL10222.Contig1 | 4 | 429–450 | 438 | 4 | Uncharacterized protein | 47 | 2 | 46 |
| miR1355 | CL9524.Contig2 (CotAD_06422) | 3.5 | 55–76 | 62 | 3 | Copper binding protein 6 (BCP6) | 58 | 0 | 14 |
| miR8155 | CL11661.Contig1 (CotAD_51244) | 4 | 735–753 | 741 | 4 | AUX/IAA | 105 | 2 | 61 |
| miR5083 | CL11334.Contig1 (CotAD_47266) | 3.5 | 334–356 | 341 | 2 | Unknown protein | 51 | 2 | 43 |
| CL5215.Contig1 (CotAD_47976) | 3.5 | 1524–1546 | 1536 | 3 | Nucleosome assembly protein | 73 | 1 | 13 | |
| miR7484 | CL11292.Contig4 (CotAD_04736) | 4 | 2–24 | 17 | 2 | Predicted protein | 0 | 2 | 16 |
| miR7502 | Unigene26285 (CotAD_22251) | 3.5 | 11–32 | 23 | 4 | Trihelix | 0 | 1 | 0 |
| CL503.Contig1 (CotAD_21495) | 4.5 | 18–39 | 30 | 4 | Allene Oxide Cyclase (AOC) | 0 | 12 | 0 | |
| miR7505 | Unigene19150 (CotAD_63955) | 4 | 533–554 | 545 | 3 | NADH-ubiquinone oxidoreductase subunit 9 (NAD9) | 732 | 2 | 951 |
| miR7508 | Unigene9225 (CotAD_26989) | 4.5 | 1–21 | 12 | 2 | Pectin Methylesterase (PME2) | 313 | 8 | 650 |
| CL11591.Contig1 (CotAD_08283) | 4.5 | 1034–1054 | 1045 | 4 | Translocon-associated protein subunit alpha | 72 | 4 | 46 | |
| CL1882.Contig3 (CotAD_18999) | 4 | 16–36 | 28 | 4 | Zinc finger family protein, | 0 | 6 | 3 | |
| miR7512 | CL6.Contig3 (CotAD_04930) | 4.5 | 13–35 | 26 | 4 | Uncharacterized protein | 0 | 2 | 0 |
| CL6.Contig4 (CotAD_04930) | 4.5 | 13–35 | 26 | 4 | Uncharacterized protein | 0 | 2 | 0 | |
| Unigene19759 | 4 | 12–31 | 23 | 4 | Alpha/beta-Hydrolases superfamily protein isoform | 0 | 1 | 0 | |
| Name | Sequences | Abundance | Genomic Position | ||||
|---|---|---|---|---|---|---|---|
| CK | 4 h | 5 d | G. raimondii | G. arboreum | G. hirsutum | ||
| TAS3a D8(+) | TTCTTGACCTTGTAAGGCCTT | 2 | 2 | 1 | Chr9: 41,462,977–41,462,936 | scaffold13100:41–82 | At_chr3: 23,630,524–23,630,565 |
| TAS3a D7(+) | TTCTTGACCTTGTAAGACCCC | 228 | 122 | 91 | Dt_chr8: 37,518,786–37,518,827 | ||
| TAS3b D8(+) | TTCTTGACCTTGTAAGACCTT | 816 | 897 | 541 | scaffold493: 67,555–67,595 | Ca8:60,478,108–60,478,068 | Dt_chr11: 54,832,692–54,832,651 |
| TAS3b D7(+) | TTCTTGACCTTGTAAGACCCA | 11,718 | 11,060 | 7524 | At_chr11: 5,189,802–5,189,761 | ||
| Name | G. raimondii | G. arboreum | G. hirsutum | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene identifier | Target Sites | Gene Identifier | Target Sites | Gene Identifier | Target Sites | ||||
| ARF2 | Cotton_D_10022860
Cotton_D_10033064 | 1328–1351
1331–1352 | Cotton_A_03644
Cotton_A_01955 | 1325–1346
1331–1352 | CotAD_43230
CotAD_76410 CotAD_58764 | 1307–1329
1331–1353 1331–1353 | |||
| ARF3 | Cotton_D_10031398
Cotton_D_10033714 | 1254–1275
1269–1290 | 1464–1485
1479–1500 | Cotton_A_11311
Cotton_A_03933 | 1272–1293
1320–1341 | 1482–1503
1530–1551 | CotAD_00650
CotAD_64060 CotAD_72124 | 1254–1275
1281–1303 1104–1125 | 1464–1485
1491–1512 1314–1335 |
| ARF4 | Cotton_D_10015852
Cotton_D_10019010 | 1368–1389
1350–1371 | 1575–1596
1554–1575 | Cotton_A_01738
Cotton_A_11048 | 1197–1218
1359–1380 | 1404–1425
1563–1584 | CotAD_13694
CotAD_54309 CotAD_12422 | 1368–1389
1350–1371 1359–1380 | 1575–1596
1554–1575 1563–1584 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, Z.; Han, X.; Li, Y.; Wang, J.; Wang, D.; Wang, S.; Fu, X.; Ye, W. Comparative Analysis of Cotton Small RNAs and Their Target Genes in Response to Salt Stress. Genes 2017, 8, 369. https://doi.org/10.3390/genes8120369
Yin Z, Han X, Li Y, Wang J, Wang D, Wang S, Fu X, Ye W. Comparative Analysis of Cotton Small RNAs and Their Target Genes in Response to Salt Stress. Genes. 2017; 8(12):369. https://doi.org/10.3390/genes8120369
Chicago/Turabian StyleYin, Zujun, Xiulan Han, Yan Li, Junjuan Wang, Delong Wang, Shuai Wang, Xiaoqiong Fu, and Wuwei Ye. 2017. "Comparative Analysis of Cotton Small RNAs and Their Target Genes in Response to Salt Stress" Genes 8, no. 12: 369. https://doi.org/10.3390/genes8120369




