Current Research on Non-Coding Ribonucleic Acid (RNA)
Abstract
:1. Introduction
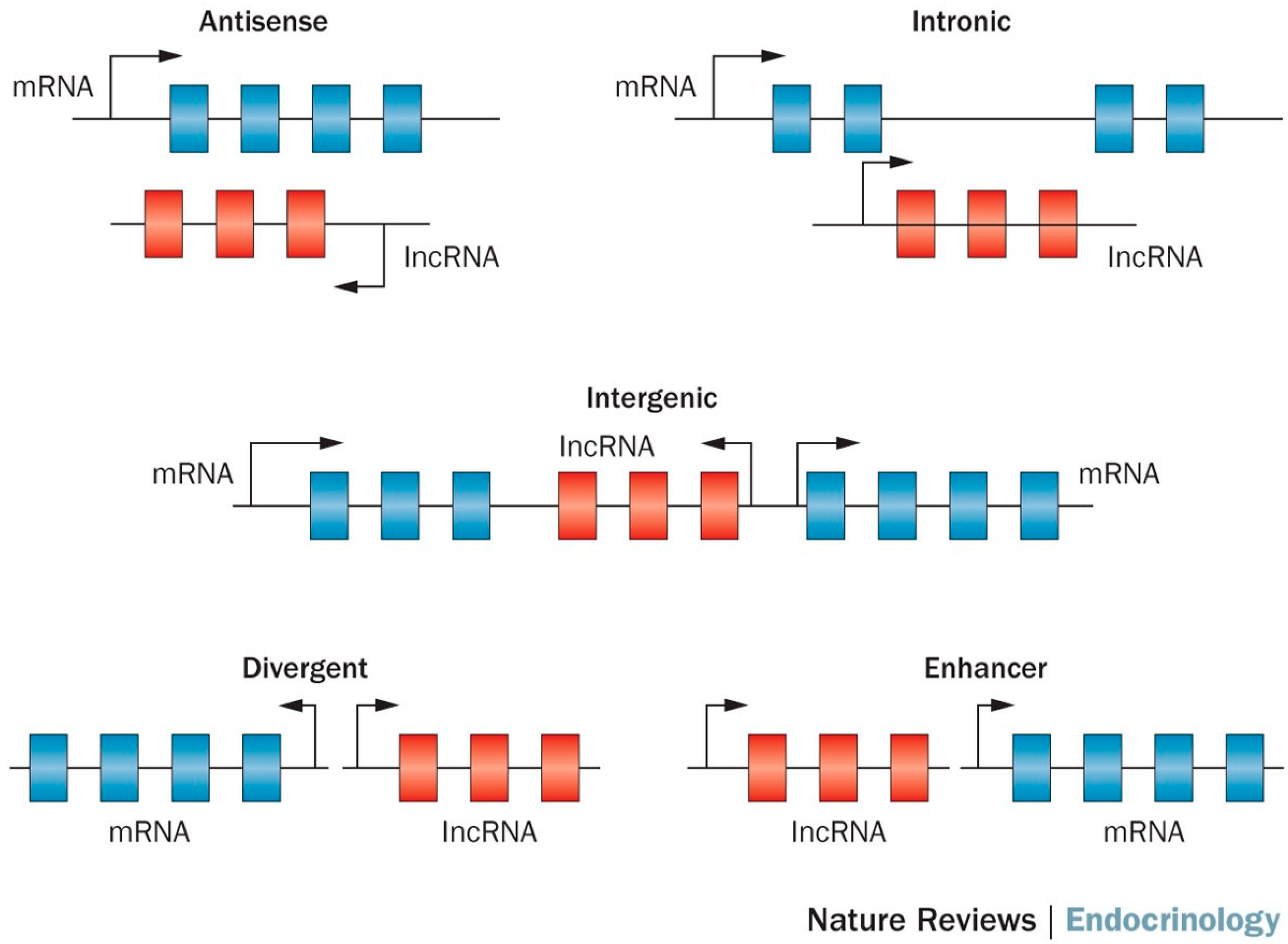
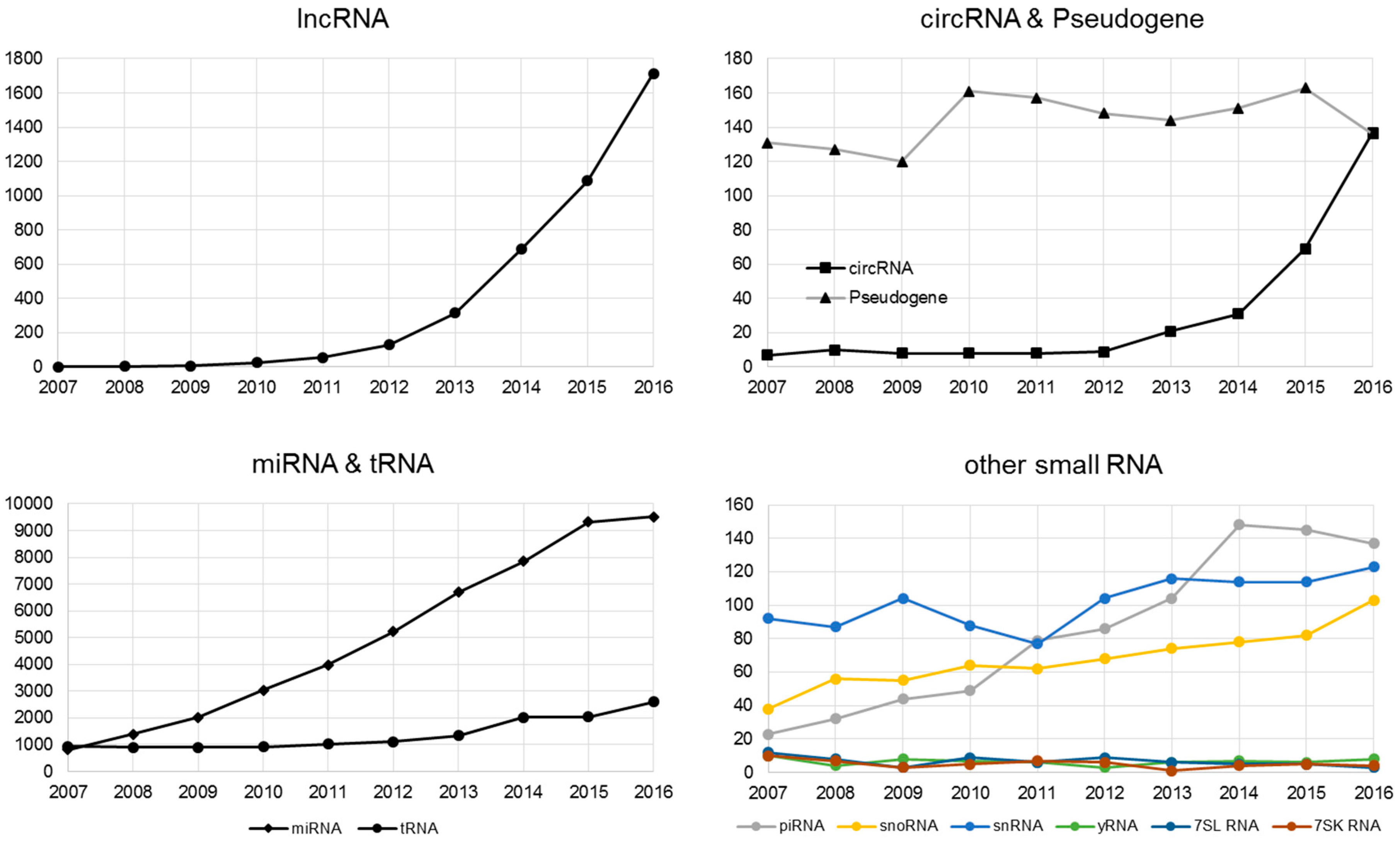
2. lncRNA
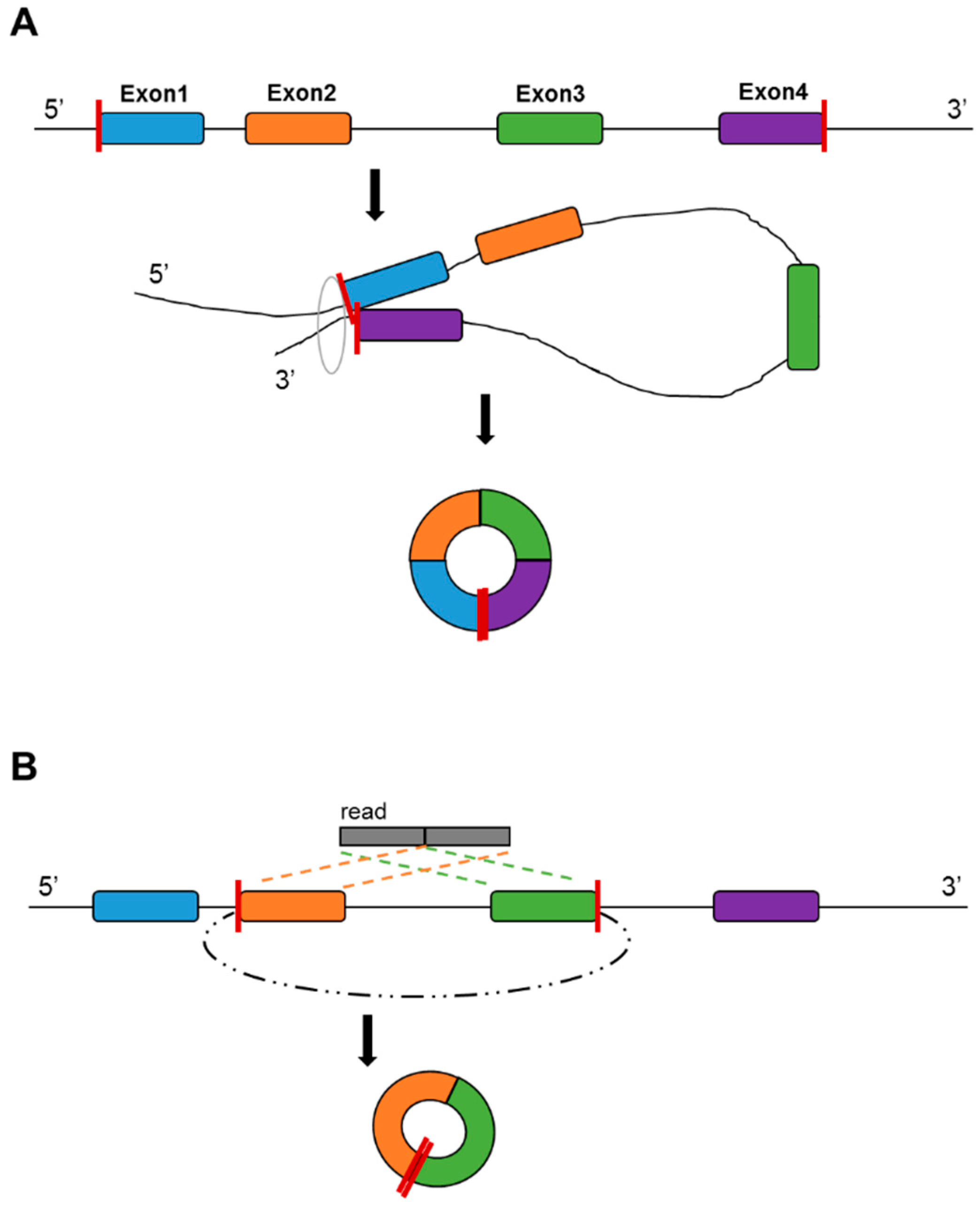
3. Circular RNA (circRNA)
4. Pseudogenes
5. Small RNA
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Freedman, M.L.; Monteiro, A.N.; Gayther, S.A.; Coetzee, G.A.; Risch, A.; Plass, C.; Casey, G.; De Biasi, M.; Carlson, C.; Duggan, D.; et al. Principles for the post-GWAS functional characterization of cancer risk loci. Nat. Genet. 2011, 43, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Blattler, A.; Yao, L.; Witt, H.; Guo, Y.; Nicolet, C.M.; Berman, B.P.; Farnham, P.J. Global loss of DNA methylation uncovers intronic enhancers in genes showing expression changes. Genome Biol. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P.; Bernstein, B.E.; Birney, E.; Dunham, I.; Green, E.D.; Gunter, C.; Snyder, M. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar]
- Eddy, S.R. The C-value paradox, junk DNA and encode. Curr. Biol. 2012, 22. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.K.; Jiang, L. Can ENCODE tell us how much junk DNA we carry in our genome? Biochem. Biophys. Res. Commun. 2013, 430, 1340–1343. [Google Scholar] [CrossRef] [PubMed]
- Graur, D.; Zheng, Y.; Price, N.; Azevedo, R.B.; Zufall, R.A.; Elhaik, E. On the immortality of television sets: “Function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol. Evol. 2013, 5, 578–590. [Google Scholar] [CrossRef] [PubMed]
- Rands, C.M.; Meader, S.; Ponting, C.P.; Lunter, G. 8.2% of the human genome is constrained: variation in rates of turnover across functional element classes in the human lineage. PLoS Genet. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Perkel, J.M. Visiting “noncodarnia”. Biotechniques 2013, 54, 301, 303–304. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Knoll, M.; Lodish, H.F.; Sun, L. Long non-coding RNAs as regulators of the endocrine system. Nat. Rev. Endocrinol. 2015, 11, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar] [PubMed]
- Dinger, M.E.; Amaral, P.P.; Mercer, T.R.; Mattick, J.S. Pervasive transcription of the eukaryotic genome: Functional indices and conceptual implications. Brief. Funct. Genom. Proteom. 2009, 8, 407–423. [Google Scholar] [CrossRef] [PubMed]
- Amaral, P.P.; Mattick, J.S. Noncoding RNA in development. Mamm. Genome 2008, 19, 454–492. [Google Scholar] [CrossRef] [PubMed]
- Dinger, M.E.; Amaral, P.P.; Mercer, T.R.; Pang, K.C.; Bruce, S.J.; Gardiner, B.B.; Askarian-Amiri, M.E.; Ru, K.; Solda, G.; Simons, C.; et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008, 18, 1433–1445. [Google Scholar] [CrossRef] [PubMed]
- Schoeftner, S.; Blasco, M.A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008, 10, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Sunkin, S.M.; Mehler, M.F.; Mattick, J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 2008, 105, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, L.H.; Spieker, T.; Koschmieder, S.; Schaffers, S.; Humberg, J.; Jungen, D.; Bulk, E.; Hascher, A.; Wittmer, D.; Marra, A.; et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J. Thorac. Oncol. 2011, 6, 1984–1992. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Hammerle, M.; Eissmann, M.; Hsu, J.; Kim, Y.; Hung, G.; Revenko, A.; Arun, G.; Stentrup, M.; Gross, M.; et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013, 73, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liu, S.; Cai, G.; Kong, L.; Zhang, T.; Ren, Y.; Wu, Y.; Mei, M.; Zhang, L.; Wang, X. Long non coding RNA MALAT1 promotes tumor growth and metastasis by inducing epithelial-mesenchymal transition in oral squamous cell carcinoma. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Bhan, A.; Hussain, I.; Ansari, K.I.; Bobzean, S.A.; Perrotti, L.I.; Mandal, S.S. Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long noncoding RNA HOTAIR in vitro and in vivo. J. Steroid Biochem. Mol. Biol. 2014, 141, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Bhan, A.; Hussain, I.; Ansari, K.I.; Kasiri, S.; Bashyal, A.; Mandal, S.S. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J. Mol. Biol. 2013, 425, 3707–3722. [Google Scholar] [CrossRef] [PubMed]
- Hajjari, M.; Salavaty, A. Hotair: An oncogenic long non-coding RNA in different cancers. Cancer Biol. Med. 2015, 12, 1–9. [Google Scholar] [PubMed]
- Zhong, D.N.; Luo, Y.H.; Mo, W.J.; Zhang, X.; Tan, Z.; Zhao, N.; Pang, S.M.; Chen, G.; Rong, M.H.; Tang, W. High expression of long noncoding HOTAIR correlated with hepatocarcinogenesis and metastasis. Mol. Med. Rep. 2017. [Google Scholar] [CrossRef]
- Wang, J.; Cao, L.; Wu, J.; Wang, Q. Long non-coding RNA SNHG1 regulates NOB1 expression by sponging miR-326 and promotes tumorigenesis in osteosarcoma. Int. J. Oncol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Gioia, R.; Drouin, S.; Ouimet, M.; Caron, M.; St-Onge, P.; Richer, C.; Sinnett, D. LncRNAs downregulated in childhood acute lymphoblastic leukemia modulate apoptosis, cell migration, and DNA damage response. Oncotarget 2017, 8, 80645–80650. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Gao, G.; Cao, Y. Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis. Markers 2016. [Google Scholar] [CrossRef] [PubMed]
- Yarmishyn, A.A.; Kurochkin, I.V. Long noncoding RNAs: A potential novel class of cancer biomarkers. Front. Genet. 2015, 6, 145. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Rulli, S.; Lader, E. Serum lncRNA detection as potential biomarker of lung cancer. FASEB J. 2015, 29. [Google Scholar]
- Li, Y.; Wen, X.; Wang, L.; Sun, X.; Ma, H.; Fu, Z.; Li, L. LncRNA ZEB1-AS1 predicts unfavorable prognosis in gastric cancer. Surg. Oncol. 2017, 26, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.Y.; Kim, G.; Park, S.Y.; Lee, Y.R.; Kwon, S.H.; Kim, H.S.; Yoon, J.S.; Lee, J.S.; Kweon, Y.-O.; Ha, H.T.; et al. Clinical significance of lncRNA-ATB expression in human hepatocellular carcinoma. Oncotarget 2017, 8, 78588–78597. [Google Scholar] [CrossRef] [PubMed]
- Quek, X.C.; Thomson, D.W.; Maag, J.L.; Bartonicek, N.; Signal, B.; Clark, M.B.; Gloss, B.S.; Dinger, M.E. LncRNAdb v2.0: Expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015, 43. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, Z.; Wang, D.; Qiu, C.; Liu, M.; Chen, X.; Zhang, Q.; Yan, G.; Cui, Q. LncRNADisease: A database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013, 41. [Google Scholar] [CrossRef] [PubMed]
- Volders, P.J.; Verheggen, K.; Menschaert, G.; Vandepoele, K.; Martens, L.; Vandesompele, J.; Mestdagh, P. An update on LNCipedia: A database for annotated human lncRNA sequences. Nucleic Acids Res. 2015, 43, 4363–4364. [Google Scholar] [CrossRef] [PubMed]
- Bhartiya, D.; Pal, K.; Ghosh, S.; Kapoor, S.; Jalali, S.; Panwar, B.; Jain, S.; Sati, S.; Sengupta, S.; Sachidanandan, C.; et al. lncRNome: a comprehensive knowledgebase of human long noncoding RNAs. Database (Oxford) 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Josset, L.; Tchitchek, N.; Gralinski, L.E.; Ferris, M.T.; Eisfeld, A.J.; Green, R.R.; Thomas, M.J.; Tisoncik-Go, J.; Schroth, G.P.; Kawaoka, Y.; et al. Annotation of long non-coding RNAs expressed in collaborative cross founder mice in response to respiratory virus infection reveals a new class of interferon-stimulated transcripts. RNA Biol. 2014, 11, 875–890. [Google Scholar] [CrossRef] [PubMed]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Crowe, M.L.; Grimmond, S.M.; Mattick, J.S. NRED: A database of long noncoding RNA expression. Nucleic Acids Res. 2009, 37. [Google Scholar] [CrossRef] [PubMed]
- Velculescu, V.E.; Zhang, L.; Vogelstein, B.; Kinzler, K.W. Serial analysis of gene expression. Science 1995, 270, 484–487. [Google Scholar] [CrossRef] [PubMed]
- Shiraki, T.; Kondo, S.; Katayama, S.; Waki, K.; Kasukawa, T.; Kawaji, H.; Kodzius, R.; Watahiki, A.; Nakamura, M.; Arakawa, T.; et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. USA 2003, 100, 15776–15781. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, Z.; Bailey, T.L.; Perkins, A.C.; Tallack, M.R.; Xu, Z.; Liu, H. Prediction of novel long non-coding RNAs based on RNA-Seq data of mouse Klf1 knockout study. BMC Bioinform. 2012, 13, 331. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Shishkin, A.A.; Zhu, X.P.; Kadri, S.; Maza, I.; Guttman, M.; Hanna, J.H.; Regev, A.; Garber, M. Evolutionary analysis across mammals reveals distinct classes of long non-coding RNAs. Genome Biol. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Cottrell, K.A.; Djuranovic, S. Urb-RIP - an adaptable and efficient approach for immunoprecipitation of RNAs and associated RNAs/proteins. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhao, S.; Sheng, Q.; Guo, M.; Lehmann, B.; Pietenpol, J.; Samuels, D.C.; Shyr, Y. RNAseq by total RNA library identifies additional RNAs compared to poly(a) RNA library. BioMed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Kapranov, P.; Drenkow, J.; Dike, S.; Brubaker, S.; Patel, S.; Long, J.; Stern, D.; Tammana, H.; Helt, G.; et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 2005, 308, 1149–1154. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Chen, H.; Wang, T.; Zhang, W.; Ru, G.; Lang, J. Expression profile analysis of microRNAs in prostate cancer by next-generation sequencing. Prostate 2015, 75, 500–516. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wu, J.; Zhao, S.; Ye, F.; Su, Y.; Clark, T.; Sheng, Q.; Lehmann, B.; Shu, X.O.; Cai, Q. RNA sequencing of formalin-fixed, paraffin-embedded specimens for gene expression quantification and data mining. Int. J. Genom. 2016, 2016, 9837310. [Google Scholar] [CrossRef] [PubMed]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermuller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef] [PubMed]
- Hon, C.C.; Ramilowski, J.A.; Harshbarger, J.; Bertin, N.; Rackham, O.J.; Gough, J.; Denisenko, E.; Schmeier, S.; Poulsen, T.M.; Severin, J.; et al. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature 2017, 543, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Hinrichs, A.S.; Karolchik, D.; Baertsch, R.; Barber, G.P.; Bejerano, G.; Clawson, H.; Diekhans, M.; Furey, T.S.; Harte, R.A.; Hsu, F. The UCSC genome browser database: update 2006. Nucleic Acids Res. 2006, 34, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015, 10, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.; Chen, C.Y.; Chuang, T.J. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip. Rev. RNA 2015, 6, 563–579. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.Z.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats RNA 2013, 19, 426. RNA 2013, 19, 426. [Google Scholar]
- Bahn, J.H.; Zhang, Q.; Li, F.; Chan, T.M.; Lin, X.; Kim, Y.; Wong, D.T.W.; Xiao, X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 2015, 61, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Koh, W.; Pan, W.; Gawad, C.; Fan, H.C.; Kerchner, G.A.; Wyss-Coray, T.; Blumenfeld, Y.J.; El-Sayed, Y.Y.; Quake, S.R. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc. Natl. Acad. Sci. USA 2014, 111. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.W.; Li, H.M.; Qing, X.R.; Huang, D.H.; Li, H.G. Identification and characterization of human testis derived circular RNAs and their existence in seminal plasma. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Nigro, J.M.; Cho, K.R.; Fearon, E.R.; Kern, S.E.; Ruppert, J.M.; Oliner, J.D.; Kinzler, K.W.; Vogelstein, B. Scrambled exons. Cell 1991, 64, 607–613. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [PubMed]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of circRNAs. Mol. Cell 2017, 66. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 2017, 66, 22–37. [Google Scholar] [CrossRef] [PubMed]
- Qin, M.L.; Liu, G.; Huo, X.S.; Tao, X.M.; Sun, X.M.; Ge, Z.H.; Yang, J.; Fan, J.; Liu, L.; Qin, W.X. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016, 16, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liang, W.; Zhang, P.; Chen, J.; Qian, H.; Zhang, X.; Xu, W. Circular RNAs: Emerging cancer biomarkers and targets. J. Exp. Clin. Cancer Res. 2017, 36, 152. [Google Scholar] [CrossRef] [PubMed]
- You, X.T.; Vlatkovic, I.; Babic, A.; Will, T.; Epstein, I.; Tushev, G.; Akbalik, G.; Wang, M.T.; Glock, C.; Quedenau, C.; et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015, 18, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Kohlmaier, A.; Teupser, D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell. Mol. Life Sci. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Singh, D.; Zeng, Z.; Coleman, S.J.; Huang, Y.; Savich, G.L.; He, X.; Mieczkowski, P.; Grimm, S.A.; Perou, C.M.; et al. MapSplice: Accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010, 38, 178. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, S.; Otto, C.; Doose, G.; Tanzer, A.; Langenberger, D.; Christ, S.; Kunz, M.; Holdt, L.M.; Teupser, D.; Hackermuller, J.; et al. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Westholm, J.O.; Miura, P.; Olson, S.; Shenker, S.; Joseph, B.; Sanfilippo, P.; Celniker, S.E.; Graveley, B.R.; Lai, E.C. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014, 9, 1966–1980. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, J.; Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. [Google Scholar] [CrossRef] [PubMed]
- Szabo, L.; Morey, R.; Palpant, N.J.; Wang, P.L.; Afari, N.; Jiang, C.; Parast, M.M.; Murry, C.E.; Laurent, L.C.; Salzman, J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.J.; Wu, C.S.; Chen, C.Y.; Hung, L.Y.; Chiang, T.W.; Yang, M.Y. NCLscan: Accurate identification of non-co-linear transcripts (fusion, trans-splicing and circular RNA) with a good balance between sensitivity and precision. Nucleic Acids Res. 2016, 44. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Metge, F.; Dieterich, C. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics 2016, 32, 1094–1096. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Zhang, N.; Han, P.; Moon, B.S.; Lai, R.K.; Wang, K.; Lu, W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016, 44. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Xiang, Y.; Xia, S.; Liu, H.; Wang, J.; Ozguc, F.M.; Lei, L.; Kong, R.; Diao, L.; He, C.; et al. CircView: A visualization and exploration tool for circular RNAs. Brief. Bioinform. 2017. [Google Scholar] [CrossRef] [PubMed]
- Glazar, P.; Papavasileiou, P.; Rajewsky, N. circBase: A database for circular RNAs. RNA 2014, 20, 1666–1670. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Han, P.; Zhou, T.; Guo, X.; Song, X.; Li, Y. CircRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci. Rep. 2016, 6, 34985. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, S.; Das, S.; Sen, R.; Basak, P.; Chakrabarti, J. Circ2Traits: A comprehensive database for circular RNA potentially associated with disease and traits. Front. Genet. 2013, 4, 283. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Li, J.R.; Sun, C.H.; Andrews, E.; Chao, R.F.; Lin, F.M.; Weng, S.L.; Hsu, S.D.; Huang, C.C.; Cheng, C.; et al. CircNet: A database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016, 44. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42. [Google Scholar] [CrossRef] [PubMed]
- Chu, Q.; Zhang, X.; Zhu, X.; Liu, C.; Mao, L.; Ye, C.; Zhu, Q.H.; Fan, L. PlantcircBase: A database for plant circular RNAs. Mol. Plant 2017, 10, 1126–1128. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Feng, J.; Lei, L.; Hu, J.; Xia, L.; Wang, J.; Xiang, Y.; Liu, L.; Zhong, S.; Han, L.; et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief. Bioinform. 2016. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.A.; Gu, Z.P.; Clark, R.A.; Reinert, K.; Samonte, R.V.; Schwartz, S.; Adams, M.D.; Myers, E.W.; Li, P.W.; Eichler, E.E. Recent segmental duplications in the human genome. Science 2002, 297, 1003–1007. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.A.; Eichler, E.E. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat. Rev. Genet. 2006, 7, 552–564. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.M.; Gerstein, M. Studying genomes through the aeons: Protein families, pseudogenes and proteome evolution. J. Mol. Biol. 2002, 318, 1155–1174. [Google Scholar] [CrossRef]
- Zhang, Z.D.; Frankish, A.; Hunt, T.; Harrow, J.; Gerstein, M. Identification and analysis of unitary pseudogenes: Historic and contemporary gene losses in humans and other primates. Genome Biol. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Gerstein, M.B. A computational approach for identifying pseudogenes in the ENCODE regions. Genome Biol. 2006, 7 Suppl 1. [Google Scholar] [CrossRef] [PubMed]
- Valdes, C.; Capobianco, E. Methods to detect transcribed pseudogenes: RNA-Seq discovery allows learning through features. Methods Mol. Biol. 2014, 1167, 157–183. [Google Scholar] [PubMed]
- Khurana, E.; Lam, H.Y.K.; Cheng, C.; Carriero, N.; Cayting, P.; Gerstein, M.B. Segmental duplications in the human genome reveal details of pseudogene formation. Nucleic Acids Res. 2010, 38, 6997–7007. [Google Scholar] [CrossRef] [PubMed]
- Dahia, P.L.M.; FitzGerald, M.G.; Zhang, X.; Marsh, D.J.; Zheng, Z.M.; Pietsch, T.; von Deimling, A.; Haluska, F.G.; Haber, D.A.; Eng, C. A highly conserved processed PTEN pseudogene is located on chromosome band 9p21. Oncogene 1998, 16, 2403–2406. [Google Scholar] [CrossRef] [PubMed]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar] [CrossRef] [PubMed]
- Karreth, F.A.; Reschke, M.; Ruocco, A.; Ng, C.; Chapuy, B.; Leopold, V.; Sjoberg, M.; Keane, T.M.; Verma, A.; Ala, U.; et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell 2015, 161, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Karro, J.E.; Yan, Y.; Zheng, D.; Zhang, Z.; Carriero, N.; Cayting, P.; Harrrison, P.; Gerstein, M. Pseudogene.org: A comprehensive database and comparison platform for pseudogene annotation. Nucleic Acids Res. 2007, 35. [Google Scholar] [CrossRef] [PubMed]
- Lam, H.Y.K.; Khurana, E.; Fang, G.; Cayting, P.; Carriero, N.; Cheung, K.-H.; Gerstein, M.B. Pseudofam: The pseudogene families database. Nucleic Acids Res. 2009, 37. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Morin, R.D.; O’Connor, M.D.; Griffith, M.; Kuchenbauer, F.; Delaney, A.; Prabhu, A.L.; Zhao, Y.J.; McDonald, H.; Zeng, T.; Hirst, M.; et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2009, 19, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. mirBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Berezikov, E.; Robine, N.; Samsonova, A.; Westholm, J.O.; Naqvi, A.; Hung, J.H.; Okamura, K.; Dai, Q.; Bortolamiol-Becet, D.; Martin, R.; et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011, 21, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, R.; Vaucheret, H.; Trejo, J.; Bartel, D.P. A diverse and evolutionarily fluid set of micrornas in Arabidopsis thaliana. Genes Dev. 2006, 20, 3407–3425. [Google Scholar] [CrossRef] [PubMed]
- Westholm, J.O.; Ladewig, E.; Okamura, K.; Robine, N.; Lai, E.C. Common and distinct patterns of terminal modifications to mirtrons and canonical microRNAs. RNA 2012, 18, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Larter, C.Z.; Yeh, M.M. Animal models of NASH: Getting both pathology and metabolic context right. J. Gastroenterol. Hepatol. 2008, 23. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- van Rooij, E.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Hill, J.; Olson, E.N. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 2007, 316, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Calado, D.P.; Galler, G.; Thai, T.H.; Patterson, H.C.; Wang, J.; Rajewsky, N.; Bender, T.P.; Rajewsky, K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 2007, 131, 146–159. [Google Scholar] [CrossRef] [PubMed]
- Guarnieri, D.J.; DiLeone, R.J. MicroRNAs: A new class of gene regulators. Ann. Med. 2008, 40, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Zhang, J.; Tang, P.; Tu, N.; Wang, K.; Wu, G. Overexpression of mir185 inhibits autophagy and apoptosis of dopaminergic neurons by regulating the ampk/mtor signaling pathway in parkinson’s disease. Mol. Med. Rep. 2017. [Google Scholar] [CrossRef] [PubMed]
- Casanova-Salas, I.; Rubio-Briones, J.; Calatrava, A.; Mancarella, C.; Masia, E.; Casanova, J.; Fernandez-Serra, A.; Rubio, L.; Ramirez-Backhaus, M.; Arminan, A.; et al. Identification of miR-187 and miR-182 as biomarkers of early diagnosis and prognosis in patients with prostate cancer treated with radical prostatectomy. J. Urol. 2014, 192, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Lupold, S.E. MicroRNA expression and function in prostate cancer: A review of current knowledge and opportunities for discovery. Asian J. Androl. 2016, 18, 559–567. [Google Scholar] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene 2006, 25, 6202–6210. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Silva, M.R.; Cabrera-Cabrera, F.; Guida, M.C.; Cayota, A. Hints of tRNA-derived small RNAs role in RNA silencing mechanisms. Genes 2012, 3, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Feng, J.; Liu, Q.; Sun, F.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009, 583, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Harada, F.; Dahlberg, J.E. Specific cleavage of tRNA by nuclease S1. Nucleic Acids Res. 1975, 2, 865–871. [Google Scholar] [CrossRef] [PubMed]
- Nekrasov, M.P.; Ivshina, M.P.; Chernov, K.G.; Kovrigina, E.A.; Evdokimova, V.M.; Thomas, A.A.M.; Hershey, J.W.B.; Ovchinnikov, L.P. The mRNA-binding protein YB-1 (p50) prevents association of the eukaryotic initiation factor eIF4G with mRNA and inhibits protein synthesis at the initiation stage. J. Biol. Chem. 2003, 278, 13936–13943. [Google Scholar] [CrossRef] [PubMed]
- Bakowska-Zywicka, K.; Kasprzyk, M.; Twardowski, T. tRNA-derived short RNAs bind to Saccharomyces cerevisiae ribosomes in a stress-dependent manner and inhibit protein synthesis in vitro. FEMS Yeast Res. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, S.; Ivanov, P.; Hu, G.F.; Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009, 185, 35–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Bosompem, A.; Mohan, S.; Erdogan, B.; Ye, F.; Vickers, K.C.; Sheng, Q.; Zhao, S.; Li, C.I.; Su, P.F.; et al. Transfer RNA detection by small RNA deep sequencing and disease association with myelodysplastic syndromes. BMC Genom. 2015, 16, 727. [Google Scholar] [CrossRef] [PubMed]
- Abbott, J.A.; Francklyn, C.S.; Robey-Bond, S.M. Transfer RNA and human disease. Front. Genet. 2014, 5, 158. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.; Ivanov, P. tRNA fragments in human health and disease. FEBS Lett. 2014, 588, 4297–4304. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Lee, I.; Lee, Y.S.; Bao, X. Small non-coding transfer RNA-derived RNA fragments (tRFs): Their biogenesis, function and implication in human diseases. Genom.Inform. 2015, 13, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Mishima, E.; Inoue, C.; Saigusa, D.; Inoue, R.; Ito, K.; Suzuki, Y.; Jinno, D.; Tsukui, Y.; Akamatsu, Y.; Araki, M.; et al. Conformational change in transfer RNA is an early indicator of acute cellular damage. J. Am. Soc. Nephrol. 2014, 25, 2316–2326. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Qin, Y.; Clark, W.C.; Dai, Q.; Yi, C.; He, C.; Lambowitz, A.M.; Pan, T. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 2015, 12, 835–837. [Google Scholar] [CrossRef] [PubMed]
- Seto, A.G.; Kingston, R.E.; Lau, N.C. The coming of age for Piwi proteins. Mol. Cell 2007, 26, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Ross, R.J.; Weiner, M.M.; Lin, H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature 2014, 505, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.W.; Anderson, C.; Marshall, E.A.; Minatel, B.C.; Enfield, K.S.; Saprunoff, H.L.; Lam, W.L.; Martinez, V.D. Piwi-interacting RNAs in cancer: emerging functions and clinical utility. Mol. Cancer 2016, 15, 5. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhang, Z.; Gerstein, M.B.; Zheng, D. Small RNAs originated from pseudogenes: cis- or trans-acting? PLoS Comput. Biol. 2009, 5. [Google Scholar] [CrossRef] [PubMed]
- Parrish, N.F.; Fujino, K.; Shiromoto, Y.; Iwasaki, Y.W.; Ha, H.; Xing, J.; Makino, A.; Kuramochi-Miyagawa, S.; Nakano, T.; Siomi, H.; et al. piRNAs derived from ancient viral processed pseudogenes as transgenerational sequence-specific immune memory in mammals. RNA 2015, 21, 1691–1703. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.T.; Langley, A.R.; Christov, C.P.; Kheir, E.; Shafee, T.; Gardiner, T.J.; Krude, T. Dynamic interaction of Y RNAs with chromatin and initiation proteins during human DNA replication. J. Cell Sci. 2011, 124, 2058–2069. [Google Scholar] [CrossRef] [PubMed]
- Hizir, Z.; Bottini, S.; Grandjean, V.; Trabucchi, M.; Repetto, E. RNY (YRNA)-derived small RNAs regulate cell death and inflammation in monocytes/macrophages. Cell Death Dis. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Valadkhan, S.; Gunawardane, L.S. Role of small nuclear RNAs in eukaryotic gene expression. Essays Biochem. 2013, 54, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Henry, R.W.; Mittal, V.; Ma, B.; Kobayashi, R.; Hernandez, N. SNAP19 mediates the assembly of a functional core promoter complex (SNAPc) shared by RNA polymerases II and III. Genes Dev. 1998, 12, 2664–2672. [Google Scholar] [CrossRef] [PubMed]
- Falaleeva, M.; Pages, A.; Matuszek, Z.; Hidmi, S.; Agranat-Tamir, L.; Korotkov, K.; Nevo, Y.; Eyras, E.; Sperling, R.; Stamm, S. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 2016, 113. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.M.; Levinson, W.E.; Sullivan, D.; Fanshier, L.; Quintrell, N.; Jackson, J. The low molecular weight RNAs of Rous sarcoma virus. II. The 7 S RNA. Virology 1970, 42, 927–937. [Google Scholar] [CrossRef]
- Shan, S.O.; Walter, P. Co-translational protein targeting by the signal recognition particle. FEBS Lett. 2005, 579, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Abell, B.M.; Pool, M.R.; Schlenker, O.; Sinning, I.; High, S. Signal recognition particle mediates post-translational targeting in eukaryotes. EMBO J. 2004, 23, 2755–2764. [Google Scholar] [CrossRef] [PubMed]
- Peterlin, B.M.; Brogie, J.E.; Price, D.H. 7SK snRNA: A noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip. Rev. RNA 2012, 3, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Capece, V.; Garcia Vizcaino, J.C.; Vidal, R.; Rahman, R.U.; Pena Centeno, T.; Shomroni, O.; Suberviola, I.; Fischer, A.; Bonn, S. Oasis: Online analysis of small RNA deep sequencing data. Bioinformatics 2015, 31, 2205–2207. [Google Scholar] [CrossRef] [PubMed]
- Vitsios, D.M.; Enright, A.J. Chimira: Analysis of small RNA sequencing data and microRNA modifications. Bioinformatics 2015, 31, 3365–3367. [Google Scholar] [CrossRef] [PubMed]
- Baras, A.S.; Mitchell, C.J.; Myers, J.R.; Gupta, S.; Weng, L.C.; Ashton, J.M.; Cornish, T.C.; Pandey, A.; Halushka, M.K. miRge - a multiplexed method of processing small RNA-seq data to determine microRNA entropy. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Strickland, S.A.; Mohan, S.; Li, S.; Bosompem, A.; Vickers, K.C.; Zhao, S.; Sheng, Q.; Kim, A.S. MicroRNAs and tRNA-derived fragments predict the transformation of myelodysplastic syndromes to acute myeloid leukemia. Leuk. Lymphoma 2017, 58, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Pantano, L.; Estivill, X.; Marti, E. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucleic Acids Res. 2010, 38. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, L.F.; Christoff, A.P.; Margis, R. isomiRID: A framework to identify microRNA isoforms. Bioinformatics 2013, 29, 2521–2523. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zang, Q.; Zhang, H.; Ban, R.; Yang, Y.; Iqbal, F.; Li, A.; Shi, Q. DeAnniso: A tool for online detection and annotation of isomirs from small RNA sequencing data. Nucleic Acids Res. 2016, 44. [Google Scholar] [CrossRef] [PubMed]
- Selitsky, S.R.; Sethupathy, P. tDRmapper: Challenges and solutions to mapping, naming, and quantifying tRNA-derived RNAs from human small RNA-sequencing data. BMC Bioinform. 2015, 16. [Google Scholar] [CrossRef] [PubMed]


| Name | Link | Dataset | Organism | PMID |
|---|---|---|---|---|
| ChIPBase v2.0 | http://rna.sysu.edu.cn/chipbase/ | transcriptional regulatory networks of ncRNAs and protein-coding genes | 10 species | 27924033 |
| DIANA-LncBase v2 | http://www.microrna.gr/LncBase/ | miRNA-lncRNA interactions | human and mouse | 26612864 |
| LNCipedia 4.1 | http://www.lncipedia.org/ | annotated lncRNA transcripts | human | 25378313 |
| lncRNAdb v2.0 | http://www.lncrnadb.org/ | functional lncRNAs | 68 species | 25332394 |
| LncRNADisease | http://www.cuilab.cn/lncrnadisease/ | lncRNA-disease associations | human | 23175614 |
| LncRNAWiki | http://lncrna.big.ac.cn/ | lncRNA knowledgebase | human | 25399417 |
| lncRNome | http://genome.igib.res.in/lncRNome/ | lncRNA knowledgebase | human | 23846593 |
| MONOCLdb | http://www.monocldb.org/ | lncRNAs expressed in collaborative cross founder mice in response to respiratory virus infection | mouse | 24922324 |
| NONCODE (v5.0) | http://www.noncode.org/ | lncRNA knowledgebase | 17 species | 26586799 |
| NRED | http://nred.matticklab.com/ | lncRNA expression | human and mouse | 18829717 |
| Tools | Language | Prerequisites | Input | PMID |
|---|---|---|---|---|
| find_circ | Python | Python (2.7) (numpy, pysam) Bowtie2 SAMtools | reads in fastq | 23446348 |
| MapSplice2 | Python | g++ (≥4.3.3) Python (≥2.4.3) SAMtools Bowtie | reads in fasta/fastq | 20802226 |
| Segemehl | C | NA | reads in fasta | 1975021224512684 |
| circExplorer2 | Python | Python (≥2.7) (pysam ≥ 0.8.4, pybedtools, pandas, docopt, and scipy) TopHat (≥2.0.9) Cufflinks (≥2.1.1) BEDTools genePredToGtf gtfToGenePred bedGraphToBigWig (optional) bedToBigBed (optional) STAR (≥2.4.0j) MapSplice (≥2.1.9) BWA (≥0.6.2-r126) segemehl (≥0.2.0) | reads in fastq | 27365365 |
| circRNA_Finder | Perl | Perl awk STAR (versions 2.4.1c) samtools | reads in fasta | 25544350 |
| CIRI | Perl | Perl | alignment in SAM | 2558336528334140 |
| ACFS | Perl | BWA-0.7.3a Perl BLAT (optional) | reads in fasta | 27929140 |
| KNIFE | Perl, Python & R | Bowtie2 (≥2.2.1) Bowtie (≥0.12.7) Perl Python 2.7.5 (numpy, scipy) R 3.0.2 (data.table) SAMtools | reads in fastq | 26076956 |
| NCLscan | Python & C++ | Python 2.7 BEDTools SAMtools BLAT BWA | reads in fasta | 26442529 |
| DCC | Python | Python (pysam, pandas, numpy, and HTSeq) | alignment in BAM | 26556385 |
| UROBORUS | Perl | TopHat SAMtools Bowtie1/Bowtie2 | alignment in SAM | 26873924 |
| Name | Link | Dataset | Organism | PMID |
|---|---|---|---|---|
| circBase | http://www.circbase.org/ | merged and unified data sets of circRNAs | Human, Mouse, Worm, fly, and coelacanth | 25234927 |
| circRNADb | http://202.195.183.4:8000/circrnadb/circRNADb.php | exonic circRNAs | Human | 27725737 |
| Circ2Traits | http://gyanxet-beta.com/circdb/ | disease-circRNA association | Human | 24339831 |
| CircNet | http://circnet.mbc.nctu.edu.tw/ | tissue-specific circRNA expression profiles and circRNA–miRNA-gene regulatory networks | Human | 26450965 |
| circRNABase (in starBase v2.0) | http://202.116.90.187/mirCircRNA.php | miRNA-circRNA interactions | Human, Mouse, and worm | 24297251 |
| PlantcircBase | http://ibi.zju.edu.cn/plantcircbase/ | plant circular RNA Database | Rice, Mouse-ear cress, corn, Tomato, and Barley | 28315753 |
| Name | Link | Organism | PMID |
|---|---|---|---|
| ComiR | http://www.benoslab.pitt.edu/comir/ | human, mouse, worm and fly | 23703208 |
| MBSTAR | http://www.isical.ac.in/~bioinfo_miu/MBStar30.htm | human | 25614300 |
| miRDB | http://www.mirdb.org/ | human, mouse, rat, dog and chicken | 25378301 |
| miRmap | http://mirmap.ezlab.org/ | human, mouse, cow, opossum, chicken, chimpanzee, zebrafish and other | 23716633 |
| mirPath v.3 | http://www.microrna.gr/miRPathv3 | human, mouse, rat, worm, fly, chicken and zebrafish | 25977294 |
| miRNA_Targets | http://mamsap.it.deakin.edu.au/mirna_targets/ | human, mouse, chicken, cow, worm, fly and zebrafish | 22940442 |
| miRSystem | http://mirsystem.cgm.ntu.edu.tw/ | human and mouse | 22870325 |
| miRTar2GO | http://www.mirtar2go.org/ | human (cell type specific) | 27903911 |
| miRwalk2.0 | http://zmf.umm.uni-heidelberg.de/mirwalk2 | 15 species | 26226356 |
| MR-microT | http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=mrmicrot/index | human, mouse and fly | 22285563 |
| PITA | https://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html | human, mouse, fly and worm | 17893677 |
| psRNATarget | http://plantgrn.noble.org/psRNATarget/ | plant | 21622958 |
| STarMir | http://sfold.wadsworth.org/starmir.html | mammalian | 24803672 |
| TAPIR | http://bioinformatics.psb.ugent.be/webtools/tapir | 10 plant species | 20430753 |
| TargetScan v7.1 | http://www.targetscan.org/vert_71/ | human, mouse, worm, fly and fish | 26267216 |
| Tools4miRs | https://tools4mirs.org/ | any | 27153626 |
| Name | Link | Description | Organism | PMID |
|---|---|---|---|---|
| ChIPBase v2.0 | http://rna.sysu.edu.cn/chipbase/ | regulatory networks of ncRNAs and protein-coding genes | human | 27924033 |
| deepBase v2.0 | http://biocenter.sysu.edu.cn/deepBase/ | small RNAs, LncRNAs and circular RNAs | 19 species | 26590255 |
| DIANA-TarBase v7.0 | http://www.microrna.gr/tarbase | experimentally validated miRNA:gene interactions | 24 species | 25416803 |
| doRiNA v2.0 | http://dorina.mdc-berlin.de/ | RNA interactions in post-transcriptional regulation | human, mouse, fly and worm | 25416797 |
| dPORE | http://cbrc.kaust.edu.sa/dpore | SNPs on the regulation of miRNAs and predicted TFBSs | human | 21326606 |
| Firefly Discovery Engine | https://www.fireflybio.com/portal/search | published miRNA papers | 6 species | NA |
| HOCTAR2 | http://hoctar.tigem.it/ | miRNA target and expression | human | 21435384 |
| mESAdb | http://konulab.fen.bilkent.edu.tr/mirna/ | sequences and expression of miRNA | human, mouse and zebrafish | 21177657 |
| microRNA.org | http://www.microrna.org | targets and expression | human, mouse, rat, fly and worm | 18158296 |
| miRBase | http://www.mirbase.org/ | miRNA sequences and annotation | 206 species | 24275495 |
| miRCancer | http://mircancer.ecu.edu/ | miRNA Cancer Association | human | 23325619 |
| miRDB | http://www.mirdb.org/ | targets and functional annotations | human, mouse, rat, dog and chicken | 25378301 |
| miRecords | http://miRecords.umn.edu/miRecords | animal miRNA-target interactions | 9 species | 18996891 |
| miRGate | http://mirgate.bioinfo.cnio.es | miRNA-mRNA targets | human, mouse and rat | 25858286 |
| miRGator v3.0 | http://mirgator.kobic.re.kr/ | expression profiles and target relationships | human | 23193297 |
| miRGen v2.0 | http://www.microrna.gr/mirgen | cell-line-specific miRNA TSSs and TF binding sites | human and mouse | 26586797 |
| miRmine | http://guanlab.ccmb.med.umich.edu/mirmine | miRNA expression profiles | human | 28108447 |
| miRNAMap v2.0 | http://miRNAMap.mbc.nctu.edu.tw/ | experimentally verified microRNAs and miRNA target genes | human, mouse, rat and other metazoan genomes | 18029362 |
| miRNEST v2.0 | http://mirnest.amu.edu.pl | collection of animal, plant and virus microRNA data | 544 species | 24243848 |
| miROrtho | http://cegg.unige.ch/mirortho | miRNA gene candidates | 45 species | 18927110 |
| miRSel | http://services.bio.ifi.lmu.de/mirsel | literature derived miRNA-gene association | human, mouse and rat | 20233441 |
| miRTarBase v7.0 | http://miRTarBase.mbc.nctu.edu.tw/ | experimentally validated miRNA-target interactions | 23 species | 29126174 |
| miRWalk v2.0 | http://zmf.umm.uni-heidelberg.de/mirwalk2 | predicted and experimentally verified miRNA-target interactions | 15 species | 26226356 |
| PMRD | http://bioinformatics.cau.edu.cn/PMRD | plant microRNA | 21 plant species | 19808935 |
| PolymiRTS Database | http://compbio.uthsc.edu/miRSNP/ | DNA variations in miRNA seed regions and miRNA target sites | human and mouse | 24163105 |
| Psmir | http://www.bio-bigdata.com/Psmir/ | potential associations between small molecules and miRNAs | human | 26759061 |
| RepTar | http://reptar.ekmd.huji.ac.il | predicted miRNA target | human and mouse | 21149264 |
| SonamiR DB v2.0 | http://compbio.uthsc.edu/SomamiR/ | cancer somatic mutations in miRNAs and their target sites | human | 26578591 |
| StarBase v2.0 | http://starbase.sysu.edu.cn/ | RNA-RNA and protein-RNA interaction networks from CLIP-Seq data | human, mouse and worm | 24297251 |
| STarMirDB | http://sfold.wadsworth.org/starmirDB.php | miRNA binding sites | human, mouse and worm | 27144897 |
| TargetScan v7.1 | http://www.targetscan.org/vert_71/ | miRNA targets | human, mouse, worm, fly and fish | 26267216 |
| TMREC | http://bioinfo.hrbmu.edu.cn/TMREC/ | transcription factor and miRNA regulatory cascades in human diseases | human | 25932650 |
| TransmiR | http://cmbi.bjmu.edu.cn/transmir | transcription factor–microRNA regulation | 14 species | 19786497 |
| VIRmiRNA | http://crdd.osdd.net/servers/virmirna | experimental viral miRNA and their targets | 68 viruses | 25380780 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Samuels, D.C.; Zhao, S.; Xiang, Y.; Zhao, Y.-Y.; Guo, Y. Current Research on Non-Coding Ribonucleic Acid (RNA). Genes 2017, 8, 366. https://doi.org/10.3390/genes8120366
Wang J, Samuels DC, Zhao S, Xiang Y, Zhao Y-Y, Guo Y. Current Research on Non-Coding Ribonucleic Acid (RNA). Genes. 2017; 8(12):366. https://doi.org/10.3390/genes8120366
Chicago/Turabian StyleWang, Jing, David C. Samuels, Shilin Zhao, Yu Xiang, Ying-Yong Zhao, and Yan Guo. 2017. "Current Research on Non-Coding Ribonucleic Acid (RNA)" Genes 8, no. 12: 366. https://doi.org/10.3390/genes8120366






