Humanized-Single Domain Antibodies (VH/VHH) that Bound Specifically to Naja kaouthia Phospholipase A2 and Neutralized the Enzymatic Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. N. kaouthia Venom and Horse Immune Serum against N. kaouthia Venom
2.2. Humanized-Camel VH/VHH Phage Display Library
2.3. Phage Bio-Panning for Selecting Phage Clones that Displayed P3- and P5-Bound VH/VHH from the Phage Library
2.4. Screening of the Transformed E. coli Clones that Could Express VH/VHH
2.5. Determination of Specific Binding of the VH/VHH to the P3 and P5 PLA2
2.6. Determination of the Restriction Fragment Length Polymorphism (RFLP) of the vh/vhh Sequences
2.7. Amino Acid Sequences, Immunoglobulin Frameworks (FRs) and Complementarity Determining Regions (CDRs) of the VH/VHH
2.8. Large Scale Production and Purification of the VH/VHH
2.9. PLA2 Enzymatic Assay and Neutralization of the Enzymatic Activity by Humanized-VH/VHH and Horse Anti-N. kaouthia Venom
2.10. Homology Modeling and Molecular Docking for Determination of the Interface Binding between the VH/VHH and the P3 and P5 PLA2
3. Results
3.1. PLA2 Fractions from N. kaouthia Venom
3.2. Enzymatic Activity of the P3 and P5 from N. kaouthia Venom
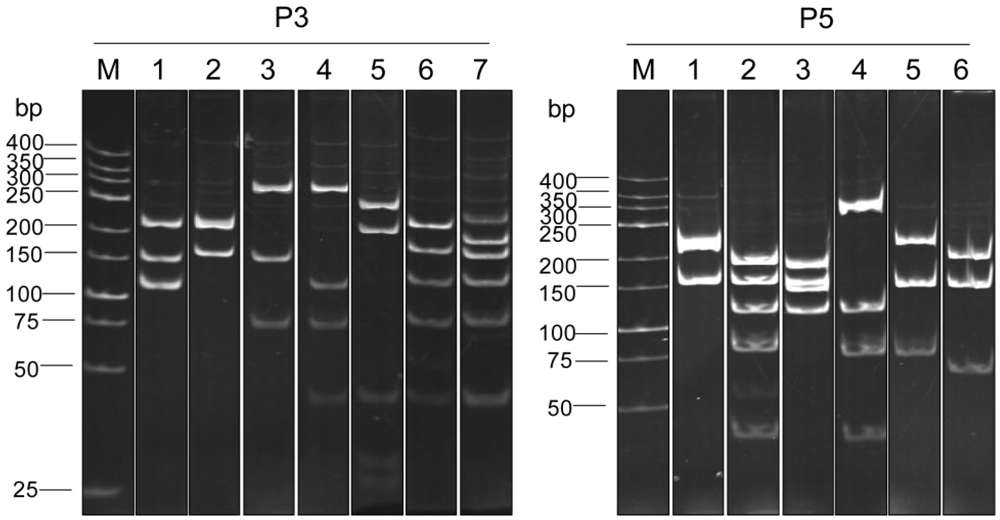
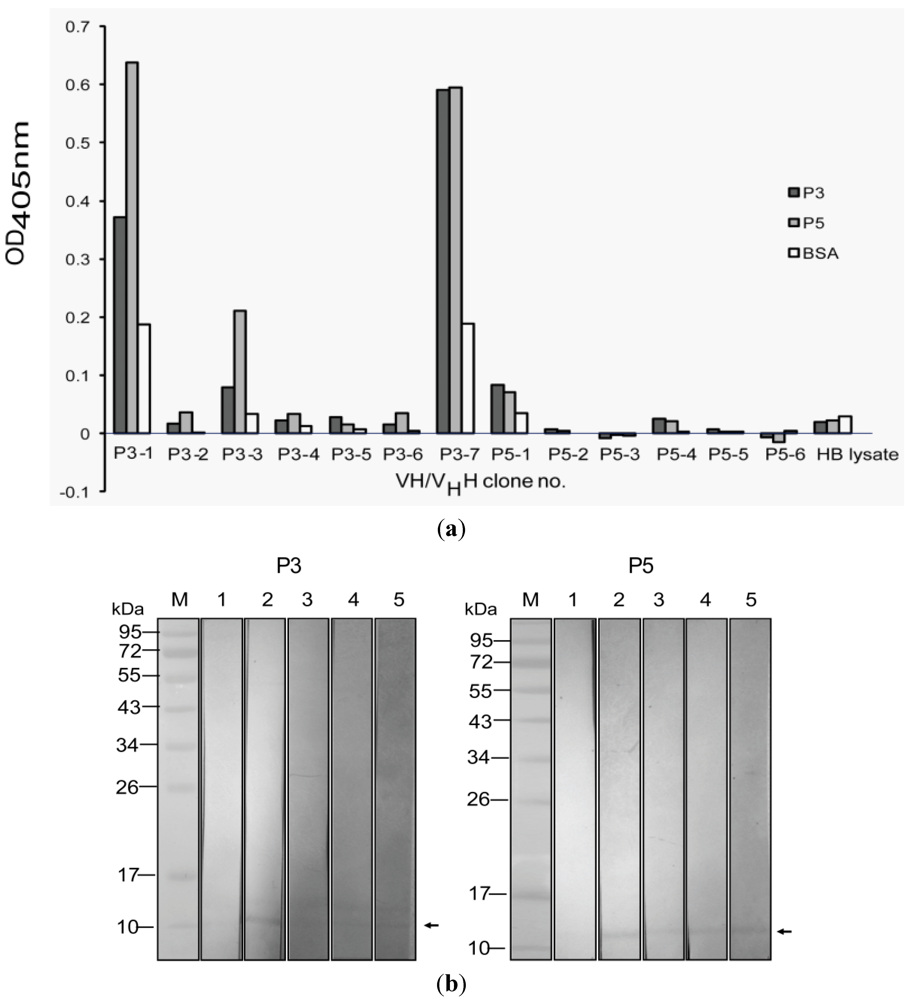
3.3. Selection of Phage Clones that Displayed P3/P5-bound VH/VHH and Characterization of the VH/VHH

| VH/VHH clone number | Closest human V region | Percent amino acid identity with human FRs | ||
|---|---|---|---|---|
| FR1 | FR2 | FR3 | ||
| VHH-P3-1 | Z27054 IGHV3-66*02 | 92.0 | 70.6 | 78.9 |
| VHH-P3-3 | Z27054 IGHV3-66*02 | 84.0 | 58.8 | 84.2 |
| VH-P3-7 | HM855939 | 92.0 | 88.2 | 84.2 |
3.4. VH/VHH-Mediated Inhibition of PLA2 Enzymatic Activity

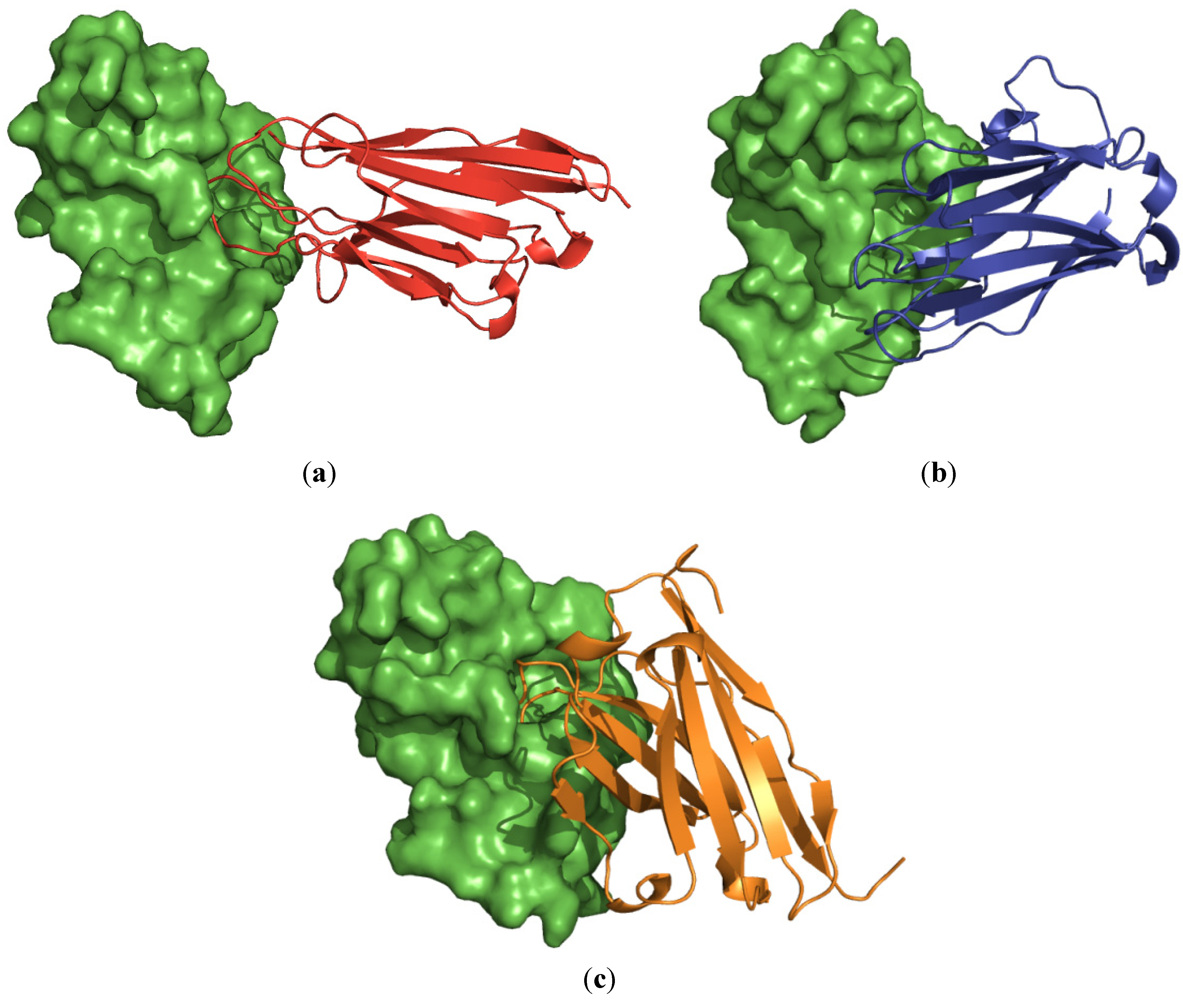
3.5. Interface Binding of the VH/VHH and the PLA2
4. Discussions
5. Conclusions
Acknowledgments
Conflict of Interest
References
- Kulkeaw, K.; Chaicumpa, W.; Sakolvaree, Y.; Tongtawe, P.; Tapchaisri, P. Proteome and immunome of the venom of the Thai cobra, Naja kaouthia. Toxicon 2007, 49, 1026–1041. [Google Scholar] [CrossRef]
- Mebs, D.; Claus, I. Amino acid sequences and toxicities of snake venom components. In Snake Toxins; Harvey, A.L., Ed.; Pegamon Press: New York, NY, USA, 1991; pp. 425–447. [Google Scholar]
- Armugam, A.; Earnest, L.; Chung, M.C.; Copalakrishnakone, P.; Tan, C.H.; Tan, N.H.; Jeyaseelan, K. Cloning and characterization of cDNAs encoding three isoforms of phospholipase A2 in Malayan spitting cobra (Naja naja sputatrix) venom. Toxicon 1997, 35, 27–37. [Google Scholar] [CrossRef]
- Warrell, D.A. Snake bite. Lancet 2010, 375, 77–88. [Google Scholar] [CrossRef]
- Doley, R.; Mukherjee, A.K. Purification and characterization of an anticoagulant phospholipase A2 from Indian monocled cobra (Naja kaouthia) venom. Toxicon 2003, 41, 81–91. [Google Scholar] [CrossRef]
- Dennis, E.A. Diversity of group types, regulation, and function of phospholipase A2. J. Biol. Chem. 1994, 269, 13057–13060. [Google Scholar]
- Bingham, C.O., III; Austen, K.F. Phospholipase A2 enzymes in eicosanoid generation. Proc. Assoc. Am. Phys. 1999, 111, 516–524. [Google Scholar] [CrossRef]
- Doley, R.; King, G.F.; Mukherjee, A.K. Differential hydrolysis of erythrocyte and mitochondrial membrane phospholipids by two phospholipase A2 isoenzymes (NK-PLA2-I and NK-PLA2-II) from venom of the Indian monocle cobra, Naja kaouthia. Arch. Biochem. Biophys. 2004, 425, 1–13. [Google Scholar] [CrossRef]
- Evans, J.; Ownby, C.L. Neutralization of edema, hemorrhage and myonecrosis induced by North American crotalid venoms in simulated first-aid treatments. Toxicon 1999, 37, 633–650. [Google Scholar] [CrossRef]
- Samy, R.P.; Gopalakrishnakone, P.; Chow, V.T.K. Therapeutic application of natural inhibitors against snake venom phospholipase A2. Bioinformation 2012, 8, 48–57. [Google Scholar]
- Kini, R.M. Anticoagulant proteins from snake venoms: Structure, function and mechanism. Biochem. J. 2006, 397, 377–387. [Google Scholar] [CrossRef]
- Khandelwal, G.; Katx, K.D.; Brooks, D.E.; Gonzales, S.M.; Ulishney, C.D. Naja kaouthia: Two cases of Asiatic cobra envenomations. J. Emerg. Med. 2007, 2, 171–174. [Google Scholar]
- Sekhar, C.C.; Chakrabarty, D. Fibrinolytic toxin from Indian monocle cobra (Naja kaouthia) venom. J. Biosci. 2011, 36, 355–361. [Google Scholar] [CrossRef]
- Zhang, H.L.; Xu, S.J.; Wang, Q.Y.; Song, S.Y.; Shu, Y.Y.; Lin, Z.J. Structure of a cardiotoxic phospholipase A(2) from Ophiophagus hannah with the “pancreatic loop”. J. Struct. Biol. 2002, 138, 207–215. [Google Scholar] [CrossRef]
- Lizano, S.; Angulo, Y.; Lomonte, B.; Fox, J.W.; Lambeau, G.; Lazdunski, M.; Guyierrez, M.G. Two phospholipase A2 inhibitors from the plasma of Cerrophidion (Bothrops) godmani which selectively inhibit two different group-II phospholipase A2 myotoxins from its own venom: Isolation, molecular cloning and biological properties. Biochem. J. 2000, 346, 631–639. [Google Scholar] [CrossRef]
- Alzogaray, V.; Danquah, W.; Aguirre, A.; Urrutia, M.; Berguer, P.; García Véscovi, E.; Haag, F.; Koch-Nolte, F.; Goldbaum, F.A. Single-domain llama antibodies as specific intracellular inhibitors of SpvB, the actin ADP-ribosylating toxin of Salmonella typhimurium. FASEB J. 2011, 25, 526–534. [Google Scholar]
- Harmsen, M.M.; de Haard, H.J. Properties, production and applications of camelid single-domain antibody fragments. Appl. Microbiol. Biotechnol. 2007, 7, 13–22. [Google Scholar]
- Lauwereys, M.; Arbabi Ghahroudi, M.; Desmyter, A.; Kinne, J.; Hölzer, W.; de Genst, E.; Wyns, L.; Muyldermans, S. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J. 1998, 17, 3512–3520. [Google Scholar]
- Thanongsaksrikul, J.; Srimanote, P.; Maneewatch, S.; Choowongkomon, K.; Tapchaisri, P.; Makino, S.; Kurazono, H.; Chaicumpa, W. A VHH that neutralizes the zinc metalloproteinase activity of botulinum neurotoxin type A. J. Biol. Chem. 2010, 285, 9657–9666. [Google Scholar]
- Thanongsaksrikul, J.; Chaicumpa, W. Botulinum neurotoxin and botulinum: A novel therapeutic approach. Toxins 2011, 3, 469–488. [Google Scholar] [CrossRef]
- Karlsson, E.; Amberg, H.; Waker, D. Isolation of the principal neurotoxins of two Naja naja subspecies. Eur. J. Biochem. 1971, 21, 1–16. [Google Scholar] [CrossRef]
- Kulkeaw, K.; Sakolvaree, Y.; Srimanote, P.; Tongtawe, P.; Maneewatch, S.; Sookrung, N.; Tungtrongchitr, A.; Tapchaisri, P.; Kurazono, H.; Chaicumpa, W. Human monoclonal ScFv neutralize lethal Thai cobra, Naja kaouthia, venom. J. Proteomics 2009, 72, 270–282. [Google Scholar] [CrossRef]
- IMGT Home page. IMGT Tools: IMGT/V-QUEST. Available online: http://www.imgt.org (accessed on 19 May 2012).
- Poungpair, O.; Pootong, A.; Maneewatch, S.; Srimanote, P.; Tongtawe, P.; Songserm, T.; Tapchaisri, P.; Chaicumpa, W. A human single chain transbody specific to matrix protein (M1) interferes with the replication of influenza A virus. Bioconjug. Chem. 2010, 21, 1134–1141. [Google Scholar] [CrossRef]
- Kini, R.M.; Evans, H.J. A model to explain the pharmacological effects of snake venom phospholipases A2. Toxicon 1989, 27, 613–635. [Google Scholar] [CrossRef]
- Kulkeaw, K. Production of Human Anti-Naja kaouthia Venom by Antibody Phage Display Library. Thammasat University: Pathumthani, Thailand, 2008. [Google Scholar]
- Gu, L.; Wang, Z.; Song, S.; Shu, Y.; Lin, Z. Crystal structures of an acidic phospholipase A2 from the venom of Naja kaouthia. Toxicon 2002, 40, 917–922. [Google Scholar] [CrossRef]
- Kang, T.S.; Georgieva, D.; Genov, N.; Murakami, M.T.; Sinha, M.; Kumar, R.P.; Kaur, P.; Kumar, S.; Dey, S.; Sharma, S.; et al. Enzymatic toxins from snake venom: Structural characterization and mechanism of catalysis. FEBS J. 2011, 278, 4544–4576. [Google Scholar]
Supplementary Files
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Chavanayarn, C.; Thanongsaksrikul, J.; Thueng-in, K.; Bangphoomi, K.; Sookrung, N.; Chaicumpa, W. Humanized-Single Domain Antibodies (VH/VHH) that Bound Specifically to Naja kaouthia Phospholipase A2 and Neutralized the Enzymatic Activity. Toxins 2012, 4, 554-567. https://doi.org/10.3390/toxins4070554
Chavanayarn C, Thanongsaksrikul J, Thueng-in K, Bangphoomi K, Sookrung N, Chaicumpa W. Humanized-Single Domain Antibodies (VH/VHH) that Bound Specifically to Naja kaouthia Phospholipase A2 and Neutralized the Enzymatic Activity. Toxins. 2012; 4(7):554-567. https://doi.org/10.3390/toxins4070554
Chicago/Turabian StyleChavanayarn, Charnwit, Jeeraphong Thanongsaksrikul, Kanyarat Thueng-in, Kunan Bangphoomi, Nitat Sookrung, and Wanpen Chaicumpa. 2012. "Humanized-Single Domain Antibodies (VH/VHH) that Bound Specifically to Naja kaouthia Phospholipase A2 and Neutralized the Enzymatic Activity" Toxins 4, no. 7: 554-567. https://doi.org/10.3390/toxins4070554





