Immunogenicity Characterization of Two Ancient Wheat α-Gliadin Peptides Related to Coeliac Disease
Abstract
:1. Introduction
2. Results and Discussion
ELISA
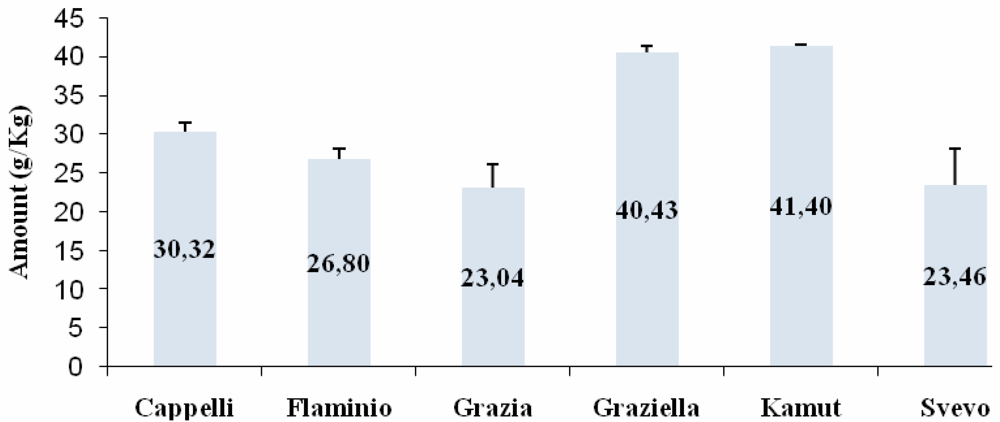
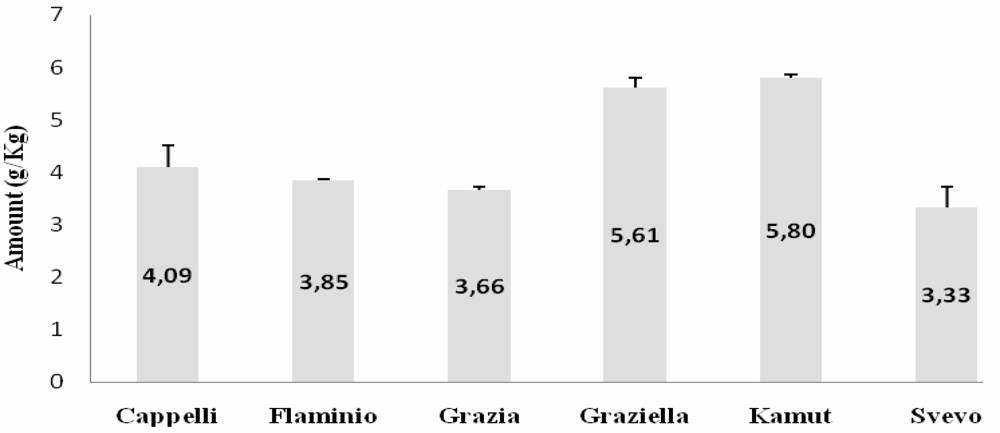
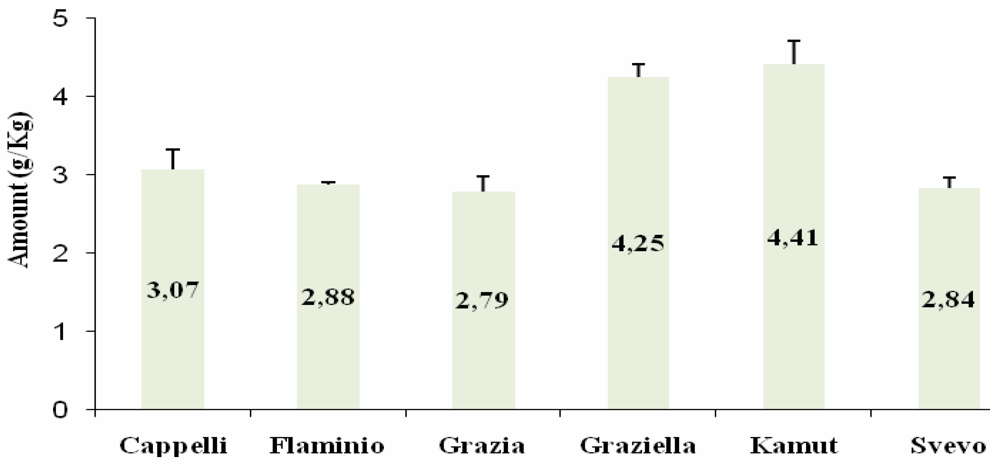
Western Blot

Α-Gliadin Gene Sequences Description and Variability
| Wheat accessions | Full-ORF | Pseudogenes | Total |
|---|---|---|---|
| Senatore Cappelli | 2 (GQ999806-GQ999807) | 3 (GQ999818-GQ999820) | 5 |
| Flaminio | 2 (GQ999808-GQ999809) | 1 (GQ999821) | 4 |
| Grazia | 2 (GQ999810-GQ999811) | 2 (GQ999822-GQ999823) | 4 |
| Graziella | 2 (GQ999812-GQ999813) | 2 (GQ999824-GQ999825) | 4 |
| Kamut | 2 (GQ999814-GQ999815) | 3 (GQ999829-GQ999831) | 6 |
| Svevo | 2 (GQ999816-GQ999817) | 3 (GQ999826-GQ999828) | 5 |
| Total | 12 | 14 | 26 |

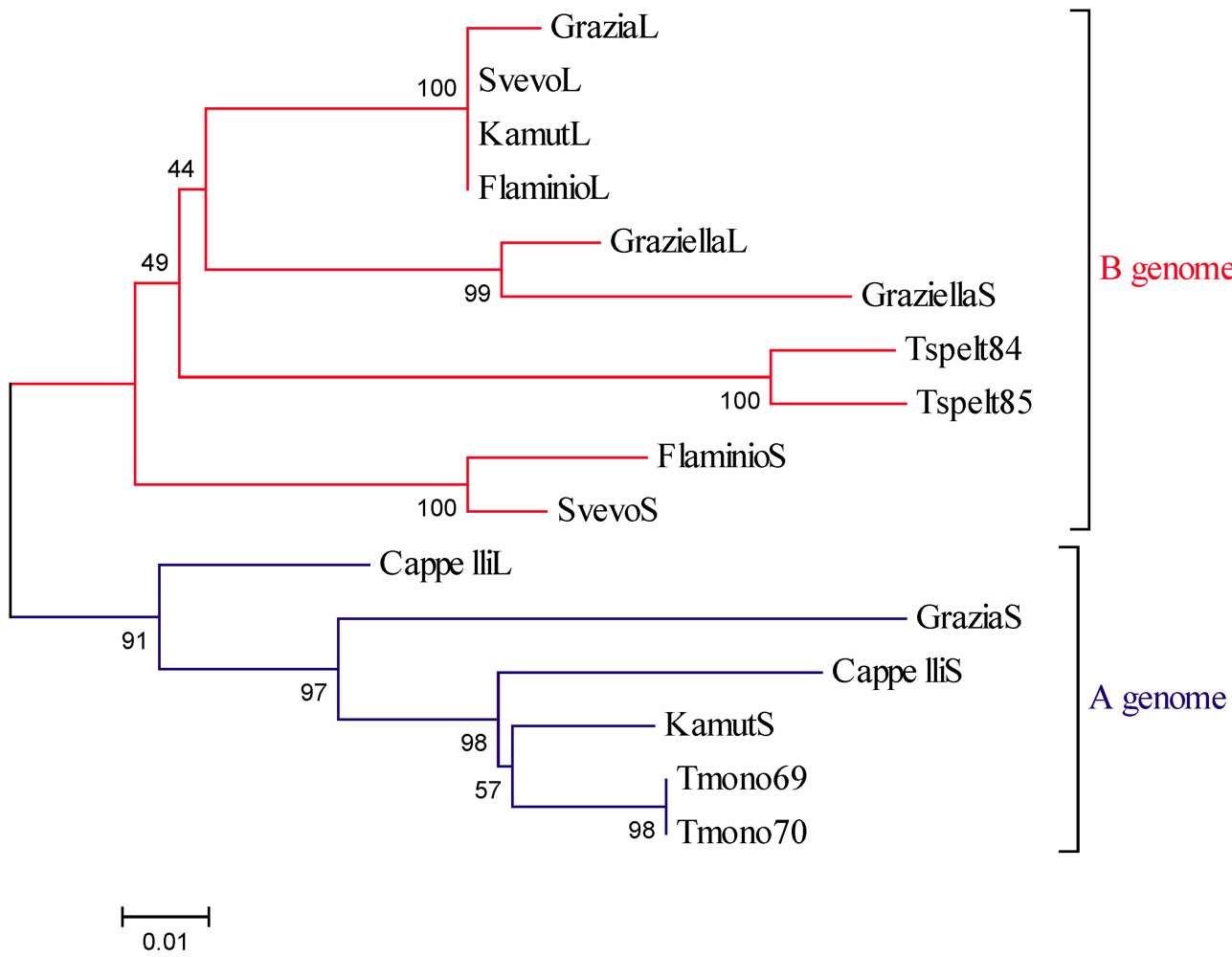
3. Experimental Section
Wheat Accessions
Gliadin Extraction
Monoclonal Antibodies
Enzyme-Linked ImmunoSorbent Assay (ELISA)
SDS-PAGE and Western Blot
DNA Extraction, Amplification, Cloning and Sequencing
- forward primer: 5’- ATG AAG ACC TTT CTC ATC C – 3’
- reverse primer: 5’- YYA GTT RGT ACC GAA GAT GCC – 3’
4. Conclusions
Acknowledgements
References
- Van Heel, D.A.; West, J. Recent advances in celiac disease. Gut 2006, 55, 1037–1046. [Google Scholar]
- Maki, M.; Collin, P. Coeliac disease. Lancet 1997, 349, 1755–1759. [Google Scholar]
- Martucci, S.; Biagi, F.; Di Sabatino, A.; Corazza, G.R. Coeliac disease. Digest. Liver Dis. 2002, 34, S150–153. [Google Scholar]
- Stepniak, D.; Koning, F. Celiac disease - Sandwiched between innate and adaptive immunity. Hum. Immunol. 2006, 67, 460–468. [Google Scholar]
- Sollid, L.M.; Lie, B.A. Celiac disease genetics: current concepts and practical applications. Clin. Gastroenterol. Hepatol. 2005, 3, 843–851. [Google Scholar]
- Kagnoff, M.F. Celiac disease: pathogenesis of a model immunogenetic disease. J. Clin. Invest. 2007, 117, 41–49. [Google Scholar]
- Stepniak, D.; Vader, L.W.; Kooy, Y.; van Veelen, P.A.; Moustakas, A.; Papandreou, N.A.; Eliopoulos, E.; Drijfhout, J.W.; Papadopoulos, G.K.; Koning, F. T-cell recognition of HLADQ2-bound gluten peptides can be influenced by an N-terminal proline at p-1. Immunogenetics 2005, 57, 8–15. [Google Scholar]
- Dewar, D.H.; Amato, M.; Ellis, H.J.; Pollock, E.L.; Gonzales-Cinca, N.; Engel, W.; Wieser, H.; Ciclitira, P.J. The toxicity of high molecular weight glutenin subunits of wheat to patients with coeliac disease. Eur. J. Gastroenterol. Hepatol. 2006, 18, 483–491. [Google Scholar]
- Shan, L.; Molberg, O.; Parrot, I.; Hausch, F.; Filiz, F.; Gray, G.M.; Sollid, L.M.; Khosla, C. Structural basis for gluten intolerance in celiac sprue. Science 2002, 297, 2275–2279. [Google Scholar]
- Sturgess, R.; Day, P.; Ellis, H.J.; Lundin, K.E.; Gjertsen, H.A.; Kontakou, M.; Ciclitira, P.J. Wheat peptide challenge in coeliac disease. Lancet 1994, 343, 758–761. [Google Scholar]
- Kim, C.Y.; Quarsten, H.; Bergseng, E.; Khosla, C.; Sollid, L.M. Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc. Natl. Acad. Sci. USA 2004, 101, 4175–4179. [Google Scholar]
- van De Wal, Y.; Kooy, Y.M.C.; van Veelen, P.; Vader, W.; August, S.A.; Drijfhout, J.W.; Peña, S.A.; Koning, F. Glutenin is involved in the gluten-driven mucosal T cell response. Eur. J. Immunol. 1999, 29, 3133–3139. [Google Scholar]
- Vader, W.; Kooy, Y.; van Veelen, P.; De Ru, A.; Harris, D.; Benckhuijsen, W.; Peña, S.; Mearin, L.; Drijfhout, J.W.; Koning, F. The gluten response in children with celiac disease is directed toward multiple gliadin and glutenin peptides. Gastroenterology 2002, 122, 1729–1737. [Google Scholar]
- Maiuri, L.; Ciacci, C.; Auricchio, S.; Brown, W.; Quaratino, S.; Londei, M. Interleukin 15 mediates epithelial changes in celiac disease. Gastroenterology 2000, 119, 996–1006. [Google Scholar]
- Arentz-Hansen, H.; Korner, R.; Molberg, O.; Quarsten, H.; Vader, W.; Kooy, Y.M.C.; Lundin, K.E.A.; Koning, F.; Roepstorff, P.; Sollid, L.M.; Mc-Adam, S. The intestinal T cell response to α-gliadin in adult celiac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J. Exp. Med. 2000, 191, 603–612. [Google Scholar]
- Shidrawi, R.G.; Day, P.; Przemioslo, R.; Ellis, H.J.; Nelufer, J.M.; Ciclitira, P.J. In vitro toxicity of gluten peptides in coeliac disease assessed by organ culture. Scand. J. Gastroenterol. 1995, 30, 758–763. [Google Scholar] [CrossRef] [PubMed]
- Maiuri, L.; Ciacci, C.; Ricciardelli, I.; Vacca, L.; Raia, V.; Auricchio, S.; Picard, J.; Osman, M.; Quaratino, S.; Londei, M. Association between innate response to gliadin and activation of pathogenic T cells in coeliac disease. Lancet 2003, 362, 30–37. [Google Scholar]
- Molberg, O.; Uhlen, A.K.; Jensen, T.; Flaete, N.S.; Fleckenstein, B.; Arentz-Hansen, H.; Raki, M.; Lundin, K.E.; Sollid, L.M. Mapping of gluten T-cell epitopes in the bread wheat ancestors: implications for celiac disease. Gastroenterology 2005, 128, 393–401. [Google Scholar]
- Spaenij-Dekking, L.; Kooy-Winkelaar, Y.; van Veelen, P.; Drijfhout, J.W.; Jonker, H.; van Soest, L.; Smulders, M.J.; Bosch, D.; Gilissen, L.J.; Koning, F. Natural variation in toxicity of wheat: potential for selection of nontoxic varieties for celiac disease patients. Gastroenterology 2005, 129, 797–806. [Google Scholar]
- Gu, Y.Q.; Crossman, C.; Kong, X.; Luo, M.; You, F.M.; Coleman-Derr, D.; Dubcovsky, J.; Anderson, O.D. Genomic organization of the complex α-gliadin gene loci in wheat. Theor. Appl. Genet. 2004, 109, 648–657. [Google Scholar]
- van Herpen, T.W.J.M.; Goryunova, S.V.; van der Schoot, J.; Mitreva, M.; Salentijn, E.; Vorst, O.; Schenk, M.F.; van Veelen, P.A.; Koning, F.; van Soest, L.J.M.; Vosman, B.; Bosch, D.; Hamer, R.J.; Gilissen, L.J.W.J.; Smulders, M.J.M. Alpha-gliadin genes from the A, B, and D genomes of wheat contain different sets of celiac disease epitopes. BMC Genomics 2006, 7, 1–13. [Google Scholar] [PubMed]
- Anderson, O.D.; Greene, F.C. The α-gliadin gene family. II DNA and protein sequence variation, subfamily structure and origins of pseudogenes. Theor. Appl. Genet. 1997, 95, 59–65. [Google Scholar] [CrossRef]
- King, J.L.; Jukes, T.H. Non-Darwinian evolution. Science 1969, 164, 788–798. [Google Scholar]
- Nei, M. Selectionism and neutralism in molecular evolution. Mol. Biol. Evol. 2005, 22, 2318–2342. [Google Scholar]
- Valdes, I.; Garcia, E.; Llorente, M.; Mendez, E. Innovative approach to low-level gluten determination in foods using a novel sandwich enzyme-linked immunosorbent assay protocol. Eur. J. Gastroenterol. Hepatol. 2003, 15, 465–474. [Google Scholar]
- Ellis, H.J.; Rosen-Bronson, S.; O’Reilly, N.; Ciclitira, P.J. Measurement of gluten using a monoclonal antibody to a celiac toxic peptide of A-gliadin. Gut 1998, 43, 190–195. [Google Scholar]
- Ciclitira, P.J.; Dewar, D.H.; Suligoj, T.; O’Sullivan, C.K.; Ellis, H.J. Monoclonal antibodies to the immunodominant gliadin peptide. In Proceedings of the 12th International Celiac Symposium, New York, NY, USA, November 2006. (2009, in press).
- Delves, P.J. Antibody Applications; Wiley & Sons Ltd: Chichester, UK, 1995; pp. 29–35. [Google Scholar]
- Delves, P.J. Antibody Applications; Wiley & Sons Ltd: Chichester, UK, 1995; pp. 99–114. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Hall, T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar]
- Librado, P.; Rozas, J. DnaSp v.5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Zuckerkandl, E.; Pauling, L. Evolutionary divergence and convergence in protein. In Evolving Genes and Proteins; Bryson, V., Vogel, H.J., Eds.; Academic Press: New York, NY, USA, 1965; pp. 97–166. [Google Scholar]
- Saitou, N.; Nei, M. The neighbour-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar]
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Gregorini, A.; Colomba, M.; Ellis, H.J.; Ciclitira, P.J. Immunogenicity Characterization of Two Ancient Wheat α-Gliadin Peptides Related to Coeliac Disease. Nutrients 2009, 1, 276-290. https://doi.org/10.3390/nu1020276
Gregorini A, Colomba M, Ellis HJ, Ciclitira PJ. Immunogenicity Characterization of Two Ancient Wheat α-Gliadin Peptides Related to Coeliac Disease. Nutrients. 2009; 1(2):276-290. https://doi.org/10.3390/nu1020276
Chicago/Turabian StyleGregorini, Armando, Mariastella Colomba, H. Julia Ellis, and Paul J. Ciclitira. 2009. "Immunogenicity Characterization of Two Ancient Wheat α-Gliadin Peptides Related to Coeliac Disease" Nutrients 1, no. 2: 276-290. https://doi.org/10.3390/nu1020276




