Implications of the Nucleocapsid and the Microenvironment in Retroviral Reverse Transcription
Abstract
:1. Retroviruses as mobile elements in nature
2. Reverse transcription of the genomic RNA
Forewords
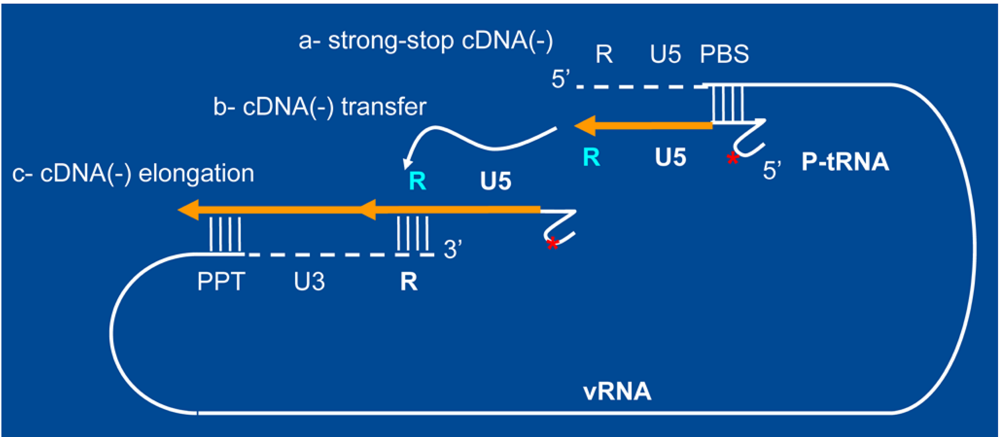
b- Primer tRNA annealing
d- Transfer of sscDNA
e-f- Minus strand cDNA synthesis
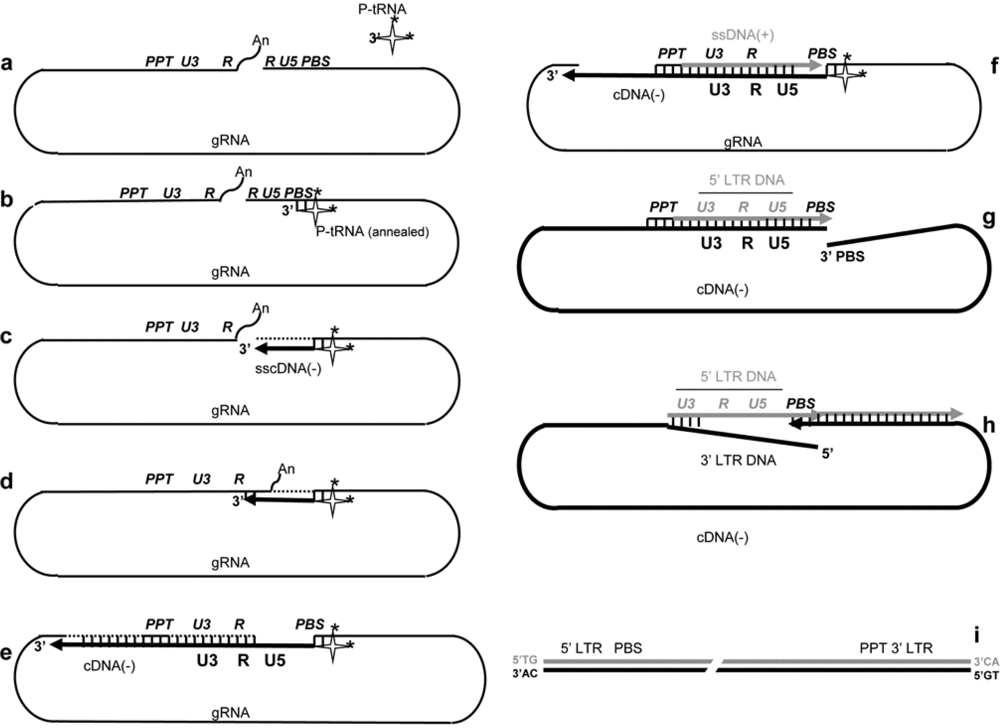
g- Initiation of plus-strand DNA synthesis
h- Plus-strand DNA transfer
3. Reverse transcription of subgenomic and cellular RNAs
b. Incorporation of cellular RNAs into virions
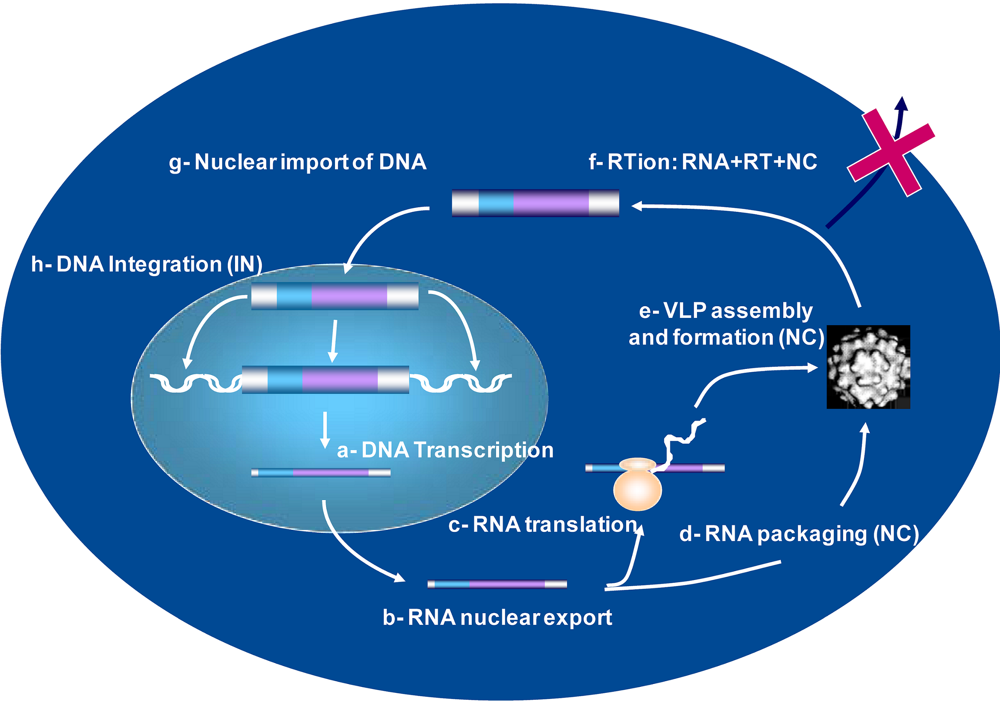
4. Viral DNA Synthesis during retrovirus assembly
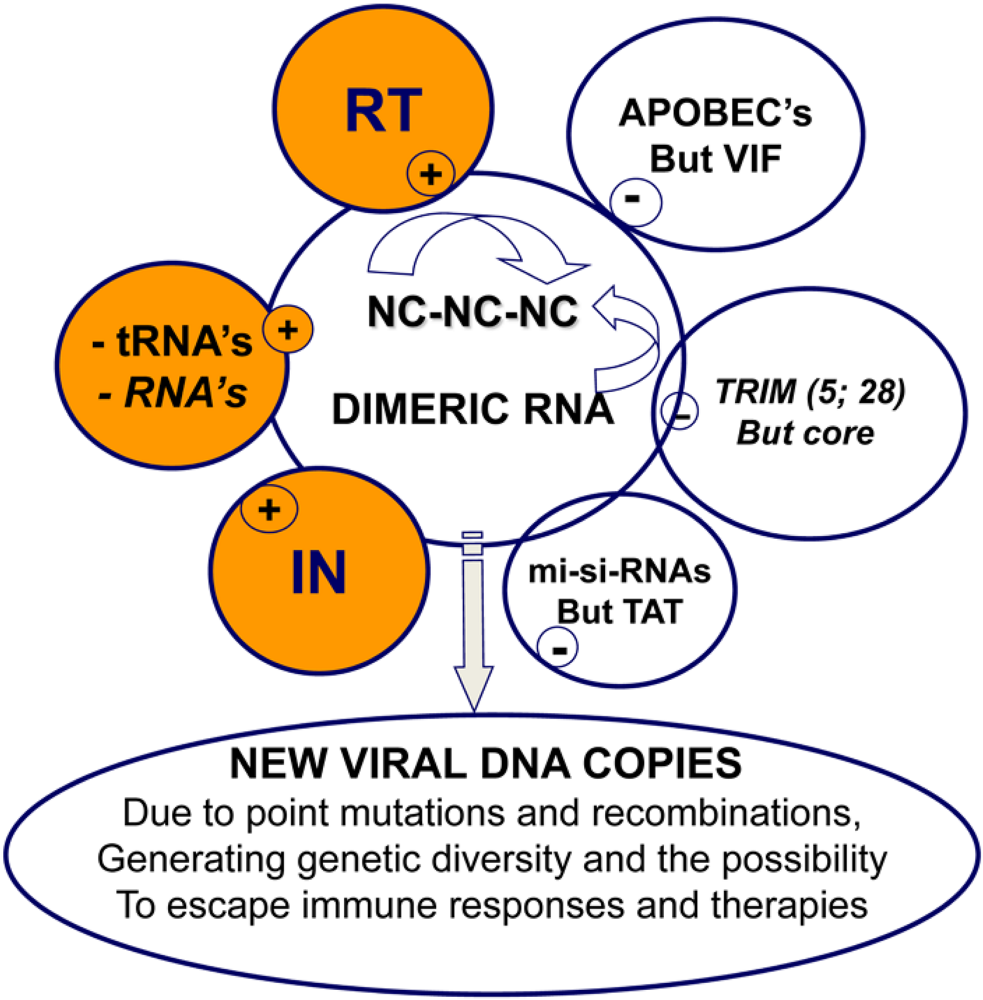
5. Retrovirus assembly and the control of reverse transcription
6. Reverse transcription in primary cells
7. Conclusion
Acknowledgments
References
- Baltimore, D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature 1970, 226, 1209–1211. [Google Scholar] [CrossRef] [PubMed]
- Gilboa, E.; Mitra, S.W.; Goff, S.; Baltimore, D. A detailed model of reverse transcription and tests of crucial aspects. Cell 1979, 18, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Coffin, J.M. Retroviridae and their replication. In Virology, 2nd; Fields B.N.;, Knipe, D.M.; Chanock R.M.;, Hirsch, M.S.; Melnick J.L.;, Monath, T.P.; Roizman, B., Eds.; Raven Press Ltd: New York, NY, USA, 1990. [Google Scholar]
- Coffin, J.M. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J. Gen. Virol. 1979, 42, 1–26. [Google Scholar] [CrossRef]
- Mougel, M.; Houzet, L.; Darlix, J.L. When is it time for reverse transcription to start and go? Retrovirology 2009, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Temin, H.M.; Mizutani, S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 1970, 226, 1211–1213. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, S.; Boettiger, D.; Temin, H.M. A DNA-dependent DNA polymerase and a DNA endonuclease in virions of Rous sarcoma virus. Nature 1970, 228, 424–427. [Google Scholar] [CrossRef] [PubMed]
- Delelis, O.; Carayon, K.; Saib, A.; Deprez, E.; Mouscadet, J.F. Integrase and integration: biochemical activities of HIV-1 integrase. Retrovirology 2008, 5, 114. [Google Scholar] [CrossRef] [PubMed]
- Lewinski, M.K.; Bushman, F.D. Retroviral DNA integration--mechanism and consequences. Adv. Genet. 2005, 55, 147–181. [Google Scholar] [PubMed]
- Goff, S.P. Host factors exploited by retroviruses. Nat. Rev. Microbiol. 2007, 5, 253–263. [Google Scholar] [CrossRef]
- Jern, P.; Coffin, J.M. Effects of retroviruses on host genome function. Annu. Rev. Genet. 2008, 42, 709–732. [Google Scholar] [CrossRef] [PubMed]
- Wolf, D.; Goff, S.P. Host restriction factors blocking retroviral replication. Annu. Rev. Genet. 2008, 42, 143–163. [Google Scholar] [CrossRef] [PubMed]
- Negre, D.; Duisit, G.; Mangeot, P.E.; Moullier, P.; Darlix, J.L.; Cosset, F.L. Lentiviral vectors derived from simian immunodeficiency virus. Curr. Top. Microbiol. Immunol. 2002, 261, 53–74. [Google Scholar] [PubMed]
- Beauregard, A.; Curcio, M.J.; Belfort, M. The take and give between retrotransposable elements and their hosts. Annu. Rev. Genet. 2008, 42, 587–617. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, F.X.; Wilhelm, M.; Gabriel, A. Reverse transcriptase and integrase of the Saccharomyces cerevisiae Ty1 element. Cytogenet. Genome Res. 2005, 110, 269–287. [Google Scholar] [CrossRef]
- Wilhelm, M.; Wilhelm, F.X. Reverse transcription of retroviruses and LTR retrotransposons. Cell. Mol. Life Sci. 2001, 58, 1246–1262. [Google Scholar] [CrossRef] [PubMed]
- Han, J.S.; Boeke, J.D. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? Bioessays 2005, 27, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Nikolaitchik, O.; Singh, J.; Wright, A.; Bencsics, C.E.; Coffin, J.M.; Ni, N.; Lockett, S.; Pathak, V.K.; Hu, W.S. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. U S A 2009, 106, 13535–13540. [Google Scholar] [CrossRef] [PubMed]
- Chertova, E.; Chertov, O.; Coren, L.V .; Roser, J.D.; Trubey, C.M.; Bess Jr., J.W.; Sowder 2nd, R. C.; Barsov, E.; Hood, B.L.; Fisher, R.J.; Nagashima, K.; Conrads, T.P.; Veenstra, T.D.; Lifson, J.D.; Ott, D.E. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 2006, 80, 9039–9052. [Google Scholar] [CrossRef] [PubMed]
- Darlix, J.L.; Garrido, J.L.; Morellet, N.; Mely, Y.; de Rocquigny, H. Properties, functions, and drug targeting of the multifunctional nucleocapsid protein of the human immunodeficiency virus. Adv. Pharmacol. 2007, 55, 299–346. [Google Scholar] [PubMed]
- Darlix, J.L.; Lapadat-Tapolsky, M.; de Rocquigny, H.; Roques, B.P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 1995, 254, 523–537. [Google Scholar] [CrossRef] [PubMed]
- Maeda, N.; Fan, H.; Yoshikai, Y. Oncogenesis by retroviruses: old and new paradigms. Rev. Med. Virol. 2008, 18, 387–405. [Google Scholar] [CrossRef] [PubMed]
- Iordanskiy, S.N.; Bukrinsky, M.I. Analysis of viral and cellular proteins in HIV-1 reverse transcription complexes by co-immunoprecipitation. Methods Mol. Biol. 2009, 485, 121–134. [Google Scholar] [PubMed]
- Levin, J.G.; Guo, J.; Rouzina, I.; Musier-Forsyth, K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005, 80, 217–286. [Google Scholar] [PubMed]
- Rein, A.; Henderson, L.E.; Levin, J.G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998, 23, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, B. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 1996, 54, 1–34. [Google Scholar] [PubMed]
- Kleiman, L.; Halwani, R.; Javanbakht, H. The selective packaging and annealing of primer tRNALys3 in HIV-1. Curr. HIV Res. 2004, 2, 163–175. [Google Scholar] [CrossRef]
- Molling, K.; Bolognesi, D.P.; Bauer, H.; Busen, W.; Plassmann, H.W.; Hausen, P. Association of viral reverse transcriptase with an enzyme degrading the RNA moiety of RNA-DNA hybrids. Nat. New Biol. 1971, 234, 240–243. [Google Scholar] [CrossRef] [PubMed]
- Klarmann, G.J.; Hawkins, M.E.; Le Grice, S.F. Uncovering the complexities of retroviral ribonuclease H reveals its potential as a therapeutic target. AIDS Rev. 2002, 4, 183–194. [Google Scholar] [PubMed]
- Allain, B.; Lapadat, T.M.; Berlioz, C.; Darlix, J.L. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. Embo J. 1994, 13, 973–981. [Google Scholar] [PubMed]
- Munch, J.; Rucker, E.; Standker, L.; Adermann, K.; Goffinet, C.; Schindler, M.; Wildum, S.; Chinnadurai, R.; Rajan, D.; Specht, A.; Gimenez-Gallego, G.; Sanchez, P.C.; Fowler, D.M.; Koulov, A.; Kelly, J.W.; Mothes, W.; Grivel, J.C.; Margolis, L.; Keppler, O.T.; Forssmann, W.G.; Kirchhoff, F. Semen-derived amyloid fibrils drastically enhance HIV infection. Cell 2007, 131, 1059–1071. [Google Scholar] [CrossRef] [PubMed]
- Tsuchihashi, Z.; Brown, P.O. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1994, 68, 5863–5870. [Google Scholar] [PubMed]
- Godet, J.; de Rocquigny, H.; Raja, C.; Glasser, N.; Ficheux, D.; Darlix, J.L.; Mely, Y. During the early phase of HIV-1 DNA synthesis, nucleocapsid protein directs hybridization of the TAR complementary sequences via the ends of their double-stranded stem. J. Mol. Biol. 2006, 356, 1180–1192. [Google Scholar] [CrossRef] [PubMed]
- Lener, D.; Tanchou, V.; Roques, B.P.; Le Grice, S.F.; Darlix, J.L. Involvement of HIV-I nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J Biol Chem 1998, 273, 33781–33786. [Google Scholar] [CrossRef] [PubMed]
- Jacob, D.T.; DeStefano, J.J. A new role for HIV nucleocapsid protein in modulating the specificity of plus strand priming. Virology 2008, 378, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Post, K.; Kankia, B.; Gopalakrishnan, S.; Yang, V.; Cramer, E.; Saladores, P.; Gorelick, R.J.; Guo, J.; Musier-Forsyth, K.; Levin, J.G. Fidelity of plus-strand priming requires the nucleic acid chaperone activity of HIV-1 nucleocapsid protein. Nucleic Acids Res. 2009, 37, 1755–1766. [Google Scholar] [CrossRef] [PubMed]
- Abbondanzieri, E.A.; Bokinsky, G.; Rausch, J.W.; Zhang, J.X.; Le Grice, S.F.; Zhuang, X. Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature 2008, 453, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Auxilien, S.; Keith, G.; Le Grice, S.F.; Darlix, J.L. Role of post-transcriptional modifications of primer tRNALys,3 in the fidelity and efficacy of plus strand DNA transfer during HIV-1 reverse transcription. J. Biol. Chem. 1999, 274, 4412–4420. [Google Scholar] [CrossRef] [PubMed]
- Ramalanjaona, N.; de Rocquigny, H.; Millet, A.; Ficheux, D.; Darlix, J.L.; Mely, Y. Investigating the mechanism of the nucleocapsid protein chaperoning of the second strand transfer during HIV-1 DNA synthesis. J. Mol. Biol. 2007, 374, 1041–1053. [Google Scholar] [CrossRef] [PubMed]
- Buckman, J.S.; Bosche, W.J.; Gorelick, R.J. Human immunodeficiency virus type 1 nucleocapsid zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J. Virol. 2003, 77, 1469–1480. [Google Scholar] [CrossRef] [PubMed]
- Carteau, S.; Gorelick, R.J.; Bushman, F.D. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 1999, 73, 6670–6679. [Google Scholar] [PubMed]
- Ciuffi, A.; Bushman, F.D. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 2006, 22, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Ciuffi, A.; Llano, M.; Poeschla, E.; Hoffmann, C.; Leipzig, J.; Shinn, P.; Ecker, J.R.; Bushman, F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005, 11, 1287–1289. [Google Scholar] [CrossRef] [PubMed]
- Di Santo, R.; Costi, R.; Roux, A.; Miele, G.; Crucitti, G.C.; Iacovo, A.; Rosi, F.; Lavecchia, A.; Marinelli, L.; Di Giovanni, C.; Novellino, E.; Palmisano, L.; Andreotti, M.; Amici, R.; Galluzzo, C.M.; Nencioni, L.; Palamara, A.T.; Pommier, Y.; Marchand, C. Novel quinolinonyl diketo acid derivatives as HIV-1 integrase inhibitors: design, synthesis, and biological activities. J. Med. Chem. 2008, 51, 4744–4750. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.; Vogt, V.M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 1995, 69, 6487–6497. [Google Scholar] [PubMed]
- Muriaux, D.; Mirro, J.; Harvin, D.; Rein, A. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. U S A 2001, 98, 5246–5251. [Google Scholar] [CrossRef] [PubMed]
- Zuber, G.; McDermott, J.; Karanjia, S.; Zhao, W.; Schmid, M.F.; Barklis, E. Assembly of retrovirus capsid-nucleocapsid proteins in the presence of membranes or RNA. J. Virol. 2000, 74, 7431–7441. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Hu, J.; Russell, R.S.; Kameoka, M.; Wainberg, M.A. Spliced human immunodeficiency virus type 1 RNA is reverse transcribed into cDNA within infected cells. AIDS Res. Hum. Retroviruses 2004, 20, 203–211. [Google Scholar] [PubMed]
- Houzet, L.; Paillart, J.C.; Smagulova, F.; Maurel, S.; Morichaud, Z.; Marquet, R.; Mougel, M. HIV controls the selective packaging of genomic, spliced viral and cellular RNAs into virions through different mechanisms. Nucleic Acids Res. 2007, 35, 2695–2704. [Google Scholar] [CrossRef] [PubMed]
- Houzet, L.; Morichaud, Z.; Mougel, M. Fully-spliced HIV-1 RNAs are reverse transcribed with similar efficiencies as the genomic RNA in virions and cells, but more efficiently in AZT-treated cells. Retrovirology 2007, 4, 30. [Google Scholar] [CrossRef]
- Maurel, S.; Houzet, L.; Garcia, E.L.; Telesnitsky, A.; Mougel, M. Characterization of a natural heterodimer between MLV genomic RNA and the SD' retroelement generated by alternative splicing. RNA 2007, 13, 2266–2276. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, R.; Fisher, J.; Goff, S.P. RNA packaging. Curr. Top. Microbiol. Immunol. 1996, 214, 177–218. [Google Scholar] [PubMed]
- Katz, R.A.; Terry, R.W.; Skalka, A.M. A conserved cis-acting sequence in the 5' leader of avian sarcoma virus RNA is required for packaging. J. Virol. 1986, 59, 163–167. [Google Scholar] [PubMed]
- Russell, R.S.; Roldan, A.; Detorio, M.; Hu, J.; Wainberg, M.A.; Liang, C. Effects of a single amino acid substitution within the p2 region of HIV-1 on packaging of spliced viral RNA. J. Virol. 2003, 77, 12986–12995. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Barklis, E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J. Virol. 1995, 69, 5716–5722. [Google Scholar] [PubMed]
- Housset, V.; Darlix, J.L. Mutations at the capsid-nucleocapsid cleavage site of gag polyprotein of Moloney murine leukemia virus abolish virus infectivity. C. R. Acad. Sci. III 1996, 319, 81–89. [Google Scholar] [PubMed]
- Sinck, L.; Richer, D.; Howard, J.; Alexander, M.; Purcell, D.F.; Marquet, R.; Paillart, J.C. In vitro dimerization of human immunodeficiency virus type 1 (HIV-1) spliced RNAs. RNA 2007, 13, 2141–2150. [Google Scholar] [CrossRef] [PubMed]
- Houzet, L.; Battini, J.; Bernard, E.; Thibert, V.; Mougel, M. A new retroelement constituted by a natural alternatively spliced RNA of murine replication-competent retroviruses. EMBO J. 2003, 22, 4866–4875. [Google Scholar] [CrossRef] [PubMed]
- Adkins, B.; Hunter, T. Identification of a packaged cellular mRNA in virions of rous sarcoma virus. J. Virol. 1981, 39, 471–480. [Google Scholar] [PubMed]
- Aronoff, R.; Linial, M. Specificity of retroviral RNA packaging. J Virol 1991, 65, 71–80. [Google Scholar] [PubMed]
- Ikawa, Y.; Ross, J.; Leder, P. An association between globin messenger RNA and 60S RNA derived from Friend leukemia virus. Proc. Natl. Acad. Sci. U S A 1974, 71, 1154–1158. [Google Scholar] [CrossRef] [PubMed]
- Gallis, B.; Linial, M.; Eisenman, R. An avian oncovirus mutant deficient in genomic RNA: characterization of the packaged RNA as cellular messenger RNA. Virology 1979, 94, 146–161. [Google Scholar] [CrossRef] [PubMed]
- Linial, M.; Medeiros, E.; Hayward, W.S. An avian oncovirus mutant (SE 21Q1b) deficient in genomic RNA: biological and biochemical characterization. Cell 1978, 15, 1371–1381. [Google Scholar] [CrossRef] [PubMed]
- Rulli Jr., S.J.; Hibbert, C.S.; Mirro, J.; Pederson, T.; Biswal, S.; Rein, A. Selective and nonselective packaging of cellular RNAs in retrovirus particles . J. Virol. 2007, 81, 6623–6631. [Google Scholar] [CrossRef] [PubMed]
- Onafuwa-Nuga, A.A.; King, S.R.; Telesnitsky, A. Nonrandom packaging of host RNAs in moloney murine leukemia virus. J. Virol. 2005, 79, 13528–13537. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Wang, T.; Zhang, W.; Yu, X.F. Virion packaging determinants and reverse transcription of SRP RNA in HIV-1 particles. Nucleic Acids Res. 2007, 35, 7288–7302. [Google Scholar] [CrossRef] [PubMed]
- Giles, K.E.; Caputi, M.; Beemon, K.L. Packaging and reverse transcription of snRNAs by retroviruses may generate pseudogenes. RNA 2004, 10, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Garcia, E.L.; Onafuwa-Nuga, A.; Sim, S.; King, S.R.; Wolin, S.L.; Telesnitsky, A. Packaging of host mY RNAs by murine leukemia virus may occur early in Y RNA biogenesis. J. Virol. 2009, 83, 12526–12534. [Google Scholar] [CrossRef] [PubMed]
- Onafuwa-Nuga, A.; Telesnitsky, A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination . Microbiol. Mol. Biol. Rev. 2009, 73, 451–480. [Google Scholar] [CrossRef] [PubMed]
- Kleiman, L. tRNA(Lys3): the primer tRNA for reverse transcription in HIV-1. IUBMB Life 2002, 53, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Levin, J.G.; Seidman, J.G. Selective packaging of host tRNA's by murine leukemia virus particles does not require genomic RNA. J. Virol. 1979, 29, 328–335. [Google Scholar] [PubMed]
- Darlix, J.L.; Cristofari, G.; Rau, M.; Pechoux, C.; Berthoux, L.; Roques, B. Nucleocapsid protein of human immunodeficiency virus as a model protein with chaperoning functions and as a target for antiviral drugs. Adv. Pharmacol. 2000, 48, 345–372. [Google Scholar] [PubMed]
- Strebel, K.; Khan, M.A. APOBEC3G encapsidation into HIV-1 virions: which RNA is it? Retrovirology 2008, 5, 55. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Tian, C.; Zhang, W.; Sarkis, P.T.; Yu, X.F. Interaction with 7SL RNA but not with HIV-1 genomic RNA or P bodies is required for APOBEC3F virion packaging. J. Mol. Biol. 2008, 375, 1098–1112. [Google Scholar] [CrossRef] [PubMed]
- D'Souza, V.; Summers, M.F. How retroviruses select their genomes. Nat. Rev. Microbiol. 2005, 3, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Cimarelli, A.; Darlix, J.L. Assembling the human immunodeficiency virus type 1. Cell. Mol. Life Sci. 2002, 59, 1166–1184. [Google Scholar] [CrossRef] [PubMed]
- Freed, E.O. Viral late domains. J. Virol. 2002, 76, 4679–4687. [Google Scholar] [CrossRef] [PubMed]
- Freed, E.O. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 1998, 251, 1–15. [Google Scholar] [CrossRef]
- Ganser-Pornillos, B.K.; Yeager, M.; Sundquist, W.I. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 2008, 18, 203–217. [Google Scholar] [CrossRef] [PubMed]
- Briggs, J.A.; Riches, J.D.; Glass, B.; Bartonova, V.; Zanetti, G.; Krausslich, H.G. Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. U S A 2009, 106, 11090–11095. [Google Scholar] [CrossRef] [PubMed]
- Briggs, J.A.; Simon, M.N.; Gross, I.; Krausslich, H.G.; Fuller, S.D.; Vogt, V.M.; Johnson, M.C. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 2004, 11, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Moore, M.; Innes, D.; Leijendekker, R.; Leigh-Brown, A.; Benaroch, P.; Geuze, H. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 2002, 3, 718–729. [Google Scholar] [CrossRef] [PubMed]
- Basyuk, E.; Galli, T.; Mougel, M.; Blanchard, J.M.; Sitbon, M.; Bertrand, E. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev. Cell 2003, 5, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Grigorov, B.; Arcanger, F.; Roingeard, P.; Darlix, J.L.; Muriaux, D. Assembly of infectious HIV-1 in human epithelial and T-lymphoblastic cell lines. J. Mol. Biol. 2006, 359, 848–862. [Google Scholar] [CrossRef] [PubMed]
- Houzet, L.; Gay, B.; Morichaud, Z.; Briant, L.; Mougel, M. Intracellular assembly and budding of the Murine Leukemia Virus in infected cells. Retrovirology 2006, 3, 12. [Google Scholar] [CrossRef] [PubMed]
- Darlix, J.L.; Bromley, P.A.; Spahr, P.F. New procedure for the direct analysis of in vitro reverse transcription of Rous sarcoma virus RNA. J. Virol. 1977, 22, 118–129. [Google Scholar] [PubMed]
- Lori, F.; di Marzo Veronese, F.; de Vico, A.L.; Lusso, P.; Reitz Jr., M.S.; Gallo, R C. Viral DNA carried by human immunodeficiency virus type 1 virions . J. Virol. 1992, 66, 5067–5074. [Google Scholar] [PubMed]
- Trono, D. Partial reverse transcripts in virions from human immunodeficiency and murine leukemia viruses . J. Virol. 1992, 66, 4893–4900. [Google Scholar] [PubMed]
- Zhang, Y.; Qian, H.; Love, Z.; Barklis, E. Analysis of the assembly function of the human immunodeficiency virus type 1 gag protein nucleocapsid domain. J. Virol. 1998, 72, 1782–1789. [Google Scholar] [PubMed]
- Zhang, H.; Dornadula, G.; Pomerantz, R.J. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenviroments: an important stage for viral infection of nondividing cells. J. Virol. 1996, 70, 2809–2824. [Google Scholar] [PubMed]
- Roan, N.R.; Greene, W.C. A seminal finding for understanding HIV transmission. Cell 2007, 131, 1044–1046. [Google Scholar] [CrossRef] [PubMed]
- Rothenberg, E.; Smotkin, D.; Baltimore, D.; Weinberg, R.A. In vitro synthesis of infectious DNA of murine leukaemia virus. Nature 1977, 269, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Borroto-Esoda, K.; Boone, L.R. Equine infectious anemia virus and human immunodeficiency virus DNA synthesis in vitro: characterization of the endogenous reverse transcriptase reaction. J. Virol. 1991, 65, 1952–1959. [Google Scholar] [PubMed]
- Roan, N.R.; Sowinski, S.; Munch, J.; Kirchhoff, F.; Greene, W.C. Aminoquinoline surfen inhibits the action of SEVI (semen-derived enhancer of viral infection) . J. Biol. Chem. 2010, 285, 1861–1869. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.A.; Gorelick, R.J. Nucleocapsid protein function in early infection processes . Virus Res. 2008. [Google Scholar]
- Houzet, L.; Morichaud, Z.; Didierlaurent, L.; Muriaux, D.; Darlix, J.L.; Mougel, M. Nucleocapsid mutations turn HIV-1 into a DNA-containing virus. Nucleic Acids Res. 2008, 36, 2311–2319. [Google Scholar] [CrossRef] [PubMed]
- Didierlaurent, L.; Houzet, L.; Morichaud, Z.; Darlix, J.L.; Mougel, M. The conserved N-terminal basic residues and zinc-finger motifs of HIV-1 nucleocapsid restrict the viral cDNA synthesis during virus formation and maturation. Nucleic Acids Res. 2008, 36, 4745–4753. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.A.; Bosche, W.J.; Shatzer, T.L.; Johnson, D.G.; Gorelick, R.J. Mutations in human immunodeficiency virus type 1 nucleocapsid protein zinc fingers cause premature reverse transcription. J. Virol. 2008, 82, 9318–9328. [Google Scholar] [CrossRef] [PubMed]
- Grigorov, B.; Decimo, D.; Smagulova, F.; Pechoux, C.; Mougel, M.; Muriaux, D.; Darlix, J.L. Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology 2007, 4, 54. [Google Scholar] [CrossRef] [PubMed]
- Brun, S.; Solignat, M.; Gay, B.; Bernard, E.; Chaloin, L.; Fenard, D.; Devaux, C.; Chazal, N.; Briant, L. VSV-G pseudotyping rescues HIV-1 CA mutations that impair core assembly or stability. Retrovirology 2008, 5, 57. [Google Scholar] [CrossRef] [PubMed]
- Dismuke, D.J.; Aiken, C. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J. Virol. 2006, 80, 3712–3720. [Google Scholar] [CrossRef] [PubMed]
- Forshey, B.M.; von Schwedler, U.; Sundquist, W.I.; Aiken, C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 2002, 76, 5667–5677. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Aiken, C. Nef enhances HIV-1 infectivity via association with the virus assembly complex. Virology 2008, 373, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Arhel, N.J.; Souquere-Besse, S.; Munier, S.; Souque, P.; Guadagnini, S.; Rutherford, S.; Prevost, M.C.; Allen, T.D.; Charneau, P. HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J. 2007, 26, 3025–3037. [Google Scholar] [CrossRef] [PubMed]
- Saphire, A.C.; Guan, T.; Schirmer, E.C.; Nemerow, G.R.; Gerace, L. Nuclear import of adenovirus DNA in vitro involves the nuclear protein import pathway and hsc70. J. Biol. Chem. 2000, 275, 4298–4304. [Google Scholar] [CrossRef] [PubMed]
- Ojala, P.M.; Sodeik, B.; Ebersold, M.W.; Kutay, U.; Helenius, A. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol. 2000, 20, 4922–4931. [Google Scholar] [CrossRef] [PubMed]
- Rabe, B.; Delaleau, M.; Bischof, A.; Foss, M.; Sominskaya, I.; Pumpens, P.; Cazenave, C.; Castroviejo, M.; Kann, M. Nuclear entry of hepatitis B virus capsids involves disintegration to protein dimers followed by nuclear reassociation to capsids . PLoS Pathog. 2009, 5, e1000563. [Google Scholar] [CrossRef] [PubMed]
- Stremlau, M.; Owens, C. M.; Perron, M.J.; Kiessling, M.; Autissier, P.; Sodroski, J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 2004, 427, 848–853. [Google Scholar] [CrossRef] [PubMed]
- Auewarakul, P.; Wacharapornin, P.; Srichatrapimuk, S.; Chutipongtanate, S.; Puthavathana, P. Uncoating of HIV-1 requires cellular activation. Virology 2005, 337, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.D.; Warmerdam, M.T.; Ferrell, S.S.; Benitez, R.; Greene, W.C. Intravirion generation of the C-terminal core domain of HIV-1 Nef by the HIV-1 protease is insufficient to enhance viral infectivity. Virology 1997, 234, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Riviere, L.; Darlix, J.L.; Cimarelli, A. Analysis of the viral elements required in the nuclear import of HIV-1 DNA . J. Virol. 84, 729–739. [CrossRef] [PubMed]
- Manganini, M.; Serafini, M.; Bambacioni, F.; Casati, C.; Erba, E.; Follenzi, A.; Naldini, L.; Bernasconi, S.; Gaipa, G.; Rambaldi, A.; Biondi, A.; Golay, J.; Introna, M. A human immunodeficiency virus type 1 pol gene-derived sequence (cPPT/CTS) increases the efficiency of transduction of human nondividing monocytes and T lymphocytes by lentiviral vectors. Hum. Gene Ther. 2002, 13, 1793–1807. [Google Scholar] [PubMed]
- Dvorin, J.D.; Bell, P.; Maul, G.G.; Yamashita, M.; Emerman, M.; Malim, M.H. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 2002, 76, 12087–12096. [Google Scholar] [CrossRef] [PubMed]
- Mbisa, J.L.; Delviks-Frankenberry, K.A.; Thomas, J.A.; Gorelick, R.J.; Pathak, V.K. Real-time PCR analysis of HIV-1 replication post-entry events. Methods Mol. Biol. 2009, 485, 55–72. [Google Scholar] [PubMed]
- Arfi, V.; Lienard, J.; Nguyen, X.N.; Berger, G.; Rigal, D.; Darlix, J.L.; Cimarelli, A. Characterization of the behavior of functional viral genomes during the early steps of human immunodeficiency virus type 1 infection. J. Virol. 2009, 83, 7524–7535. [Google Scholar] [CrossRef] [PubMed]
- Arfi, V.; Riviere, L.; Jarrosson-Wuilleme, L.; Goujon, C.; Rigal, D.; Darlix, J.L.; Cimarelli, A. Characterization of the early steps of infection of primary blood monocytes by human immunodeficiency virus type 1. J. Virol. 2008, 82, 6557–6565. [Google Scholar] [CrossRef] [PubMed]
- Skasko, M.; Weiss, K.K.; Reynolds, H.M.; Jamburuthugoda, V.; Lee, K.; Kim, B. Mechanistic differences in RNA-dependent DNA polymerization and fidelity between murine leukemia virus and HIV-1 reverse transcriptases. J. Biol. Chem. 2005, 280, 12190–12200. [Google Scholar] [CrossRef] [PubMed]
- Thomas, B.E.; Ramachandran, R.; Anitha, S.; Swaminathan, S. Feasibility of routine HIV testing among TB patients through a voluntary counselling and testing centre. Int. J. Tuberc. Lung Dis. 2007, 11, 1296–1301. [Google Scholar] [PubMed]
- HIV drug resistant mutations by drug class. Stanford HIV Drug resistance database . http://hivdb.stanford.edu (USA). [CrossRef]
- Bocharov, G.; Ford, N.J.; Edwards, J.; Breinig, T.; Wain-Hobson, S.; Meyerhans, A. A genetic-algorithm approach to simulating human immunodeficiency virus evolution reveals the strong impact of multiply infected cells and recombination. J. Gen. Virol. 2005, 86, 3109–3118. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.S.; and Temin, H.M. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci USA 1990, 87, 1556–1560. [Google Scholar] [CrossRef]
- Baird, H.A.; Gao, Y.; Galetto, R.; Lalonde, M.; Anthony, R.M.; Giacomoni, V.; Abreha, M.; Destefano, J.J.; Negroni, M.; Arts, E.J. Influence of sequence identity and unique breakpoints on the frequency of intersubtype HIV-1 recombination. Retrovirology 2006, 3, 91. [Google Scholar] [CrossRef] [PubMed]
- Goldschmidt, V.; Lisa, M.; Jenkins, L.; de Rocquigny, H.; Darlix, J.L; Mély, Y. The nucleocapsid protein of HIV-1 as a promising therapeutic target for antiviral drugs . HIV Therapy 2010. [Google Scholar]
- Bennasser, Y.; Yeung, M.L.; Benkirane, M.; Jeang, K.T. RNA interference and HIV-1: hits and misses. Curr. Opin. HIV AIDS 2006, 1, 208–11. [Google Scholar] [CrossRef]
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Share and Cite
Mougel, M.; Cimarelli, A.; Darlix, J.-L. Implications of the Nucleocapsid and the Microenvironment in Retroviral Reverse Transcription. Viruses 2010, 2, 939-960. https://doi.org/10.3390/v2040939
Mougel M, Cimarelli A, Darlix J-L. Implications of the Nucleocapsid and the Microenvironment in Retroviral Reverse Transcription. Viruses. 2010; 2(4):939-960. https://doi.org/10.3390/v2040939
Chicago/Turabian StyleMougel, Marylène, Andrea Cimarelli, and Jean-Luc Darlix. 2010. "Implications of the Nucleocapsid and the Microenvironment in Retroviral Reverse Transcription" Viruses 2, no. 4: 939-960. https://doi.org/10.3390/v2040939



