Complete Nucleotide Sequence and Molecular Characterization of Bacillus Phage TP21 and its Relatedness to Other Phages with the Same Name
Abstract
:1. Introduction
2. Results and Discussion
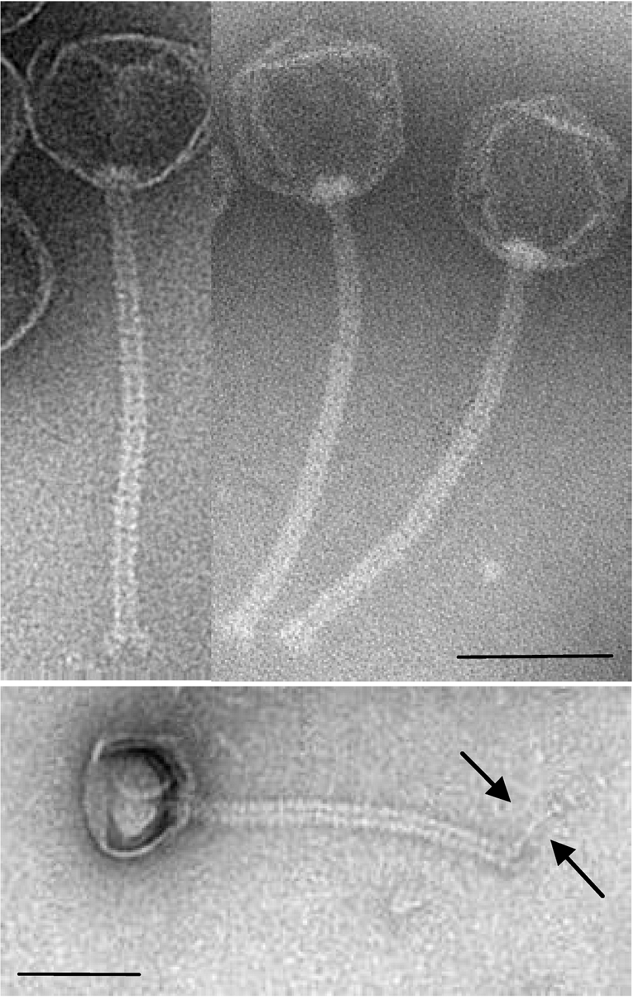
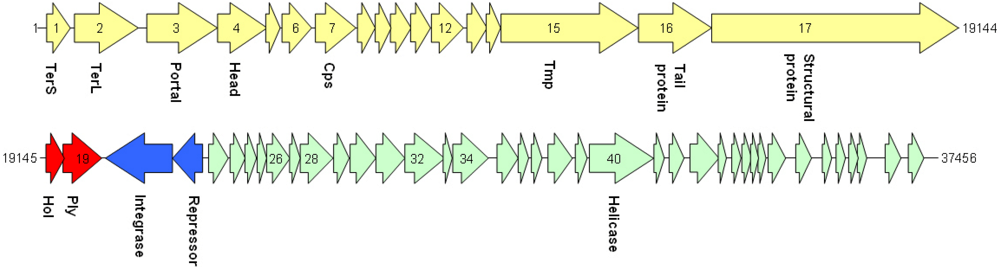
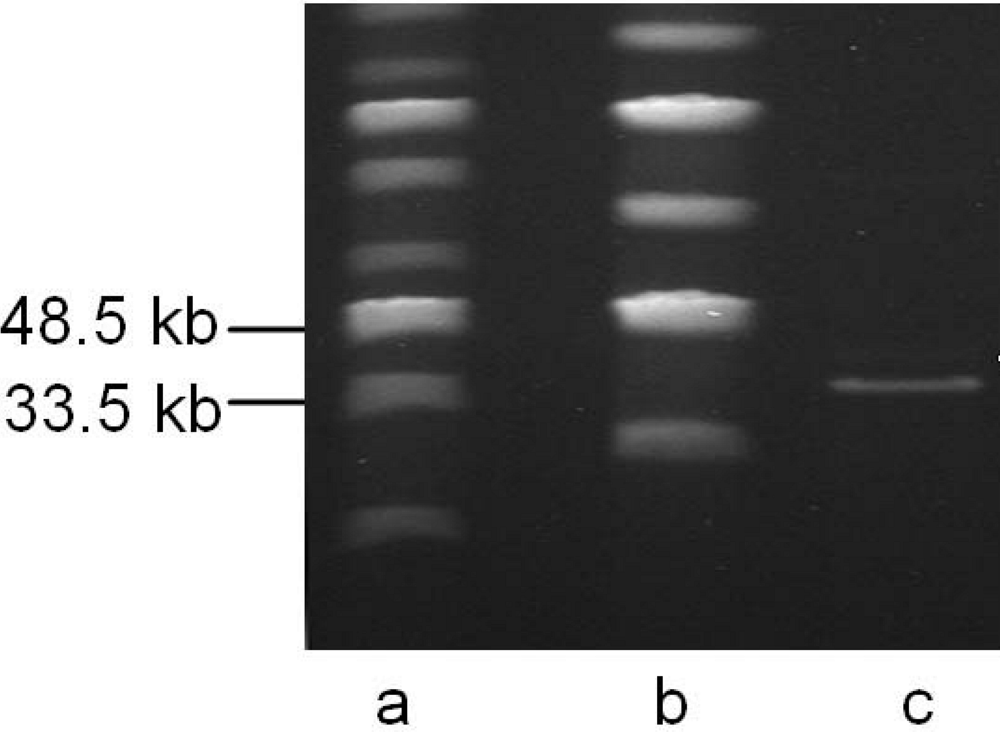
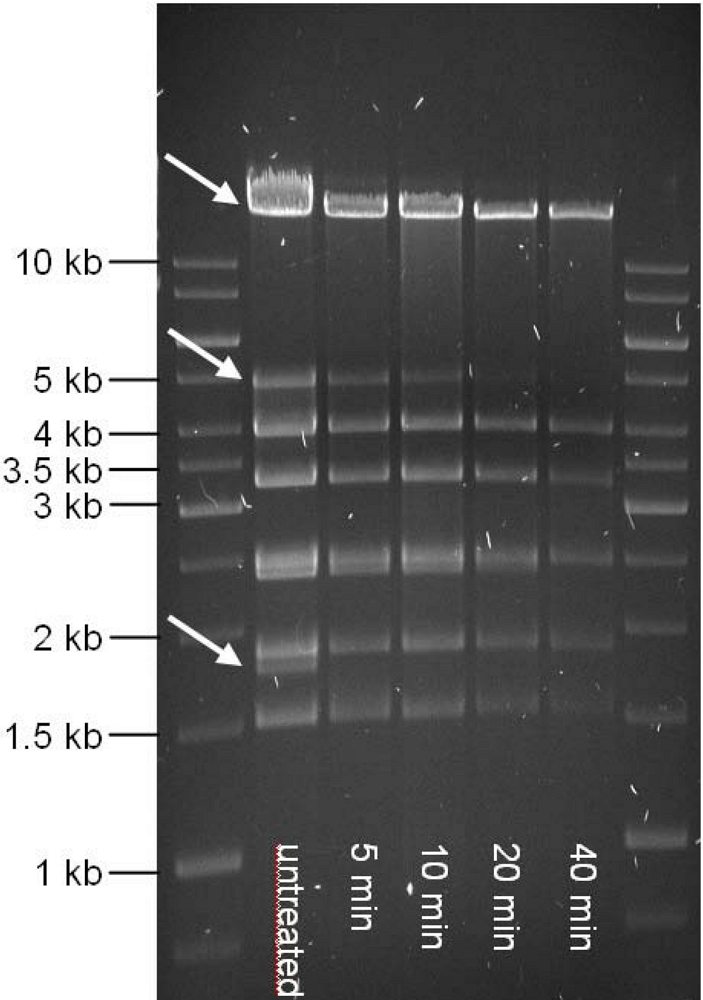
| cds | start | stop | MM (kDa) | pI | amino acid homologies (best hits) and deduced putative function |
|---|---|---|---|---|---|
| 1 | 10 | 489 | 17.8 | 6.6 | Phage terminase small subunit (various Streptococcus, Bacillus, Clostridium and Enterococcus prophages) |
| 2 | 808 | 2127 | 51.8 | 8.4 | Putative large terminase subunit (Staphylococcus phage CNPH82) |
| 3 | 2299 | 3771 | 56.8 | 5.0 | SPP1 family phage portal protein; ORF003 Staphylococcus phage 187 |
| 4 | 3775 | 4800 | 39.9 | 9.4 | Putative head protein (Clostridium phage phi CD119);SPP1 family phage head morphogenesis protein |
| 7 | 5798 | 6634 | 30.8 | 5.9 | Putative major head protein (various Clostridium prophages) |
| 9 | 7040 | 7372 | 12.4 | 9.0 | Putative phage head-tail adaptor (Bacillus prophages) |
| 10 | 7372 | 7788 | 15.4 | 10.7 | Phage protein, HK97 gp10 family (Bacilluscereus NVH0597-99 and others) |
| 15 | 9651 | 12494 | 99.9 | 5.3 | Phage tail tape measure protein, TP901 (Bacillus prophages) |
| 16 | 12506 | 14020 | 57.8 | 6.0 | Phage putative tail component (Bacilluscereus subsp. cytotoxis NVH 391-98); phage tail fiber protein (Bacillus phage Gamma) |
| 17 | 14024 | 19144 | 192.6 | 6.3 | Phage minor structural protein (Bacillus prophages) |
| 19 | 19500 | 20291 | 29.7 | 8.9 | Lysin (Bacillus phage IEBH); prophage LambdaBa01, N-acetylmuramoyl-L-alanine amidase |
| 20* | 20372 | 21754 | 53.7 | 8.9 | Recombinase (Bacillus and Paenibacillus prophages) |
| 21* | 21754 | 22386 | 23.9 | 8.9 | Transcriptional regulator (Bacillus thuringiensis serovar monterrey BGSC 4AJ1) |
| 22 | 22645 | 22893 | 9.8 | 9.4 | Cro-like protein, phage associated (Lactobacillus phage Sal2) |
| 32 | 26585 | 27400 | 32.5 | 8.9 | ORF016 (Staphylococcus phage 42E); primosome, DnaD subunit (Geobacillus sp.) |
| 34 | 27570 | 28313 | 29.2 | 9.2 | Rha family regulatory protein (Bacillusweihenstephanensis KBAB4); phage regulatory protein, Rha family (Paenibacillus sp. JDR-2) |
| 40 | 30404 | 31744 | 51.3 | 6.1 | Replicative DNA helicase (Bacillusthuringiensis serovar tochigiensis BGSC 4Y1) |
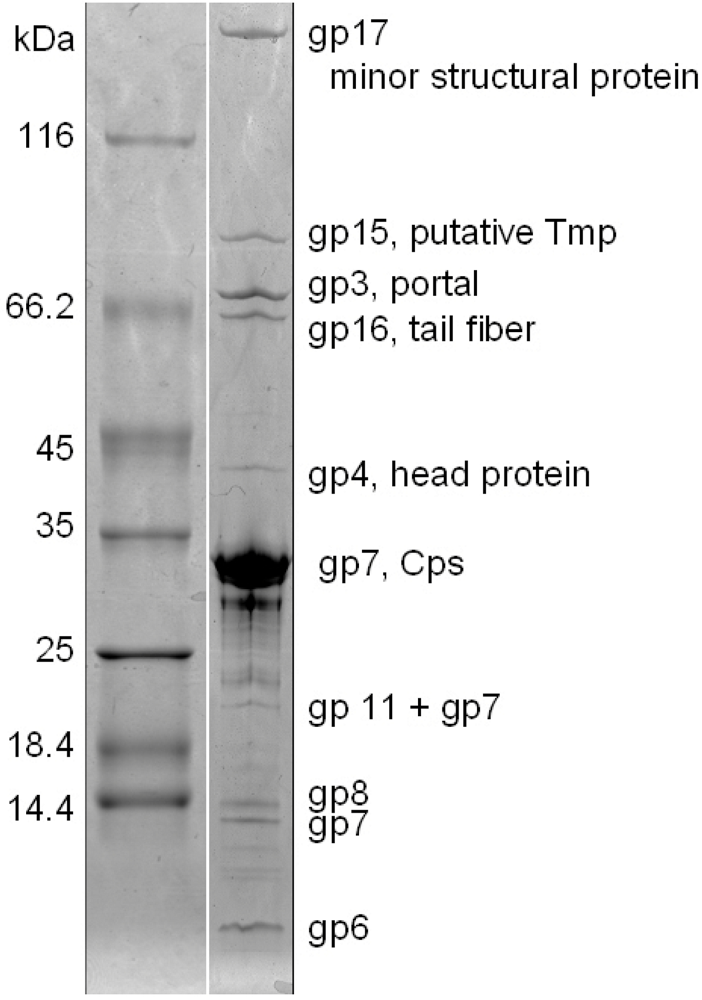
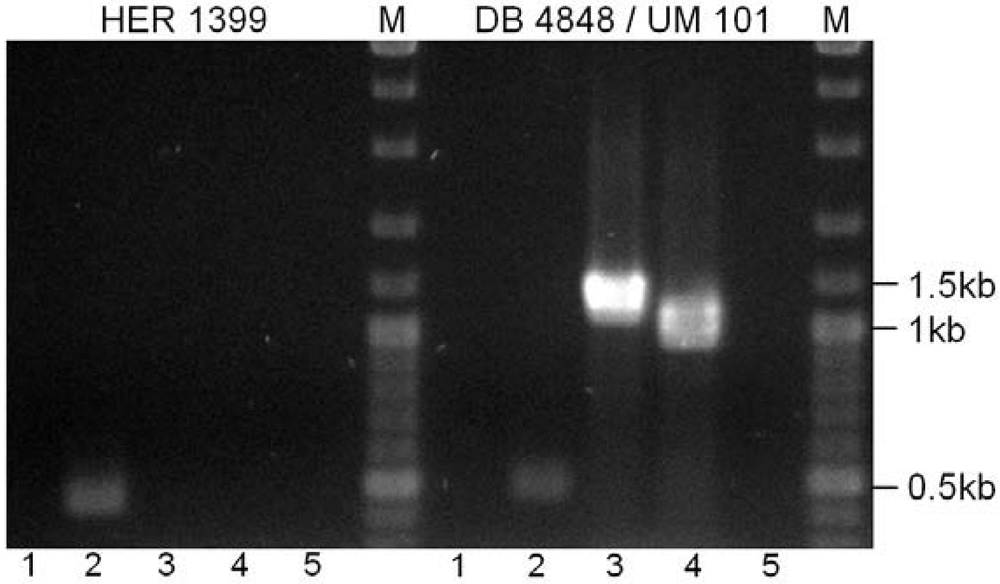
3. Experimental Section
3.1. Phage Propagation
3.2. Transmission Electron Microscopy
3.3. DNA Isolation and Sequencing
3.4. Peptide Mass Fingerprinting
3.5. Bal31 Nuclease Treatment, PCR and Electrophoresis
3.6. Bioinformatics
4. Conclusions
Acknowledgments
References
- Kolstø , A.B.; Tourasse, N.J.; Økstad , O.A. What sets Bacillus anthracis apart from other Bacillus species? Annu. Rev. Microbiol. 2009, 63, 451–476. [Google Scholar] [CrossRef] [PubMed]
- Stenfors Arnesen, L.P.; Fagerlund, A.; Granum, P.E. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 2008, 32, 579–606. [Google Scholar] [CrossRef] [PubMed]
- Schallmey, M.; Singh, A.; Ward, O.P. Developments in the use of Bacillus species for industrial production. Can. J. Microbiol. 2004, 50, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Rosas-García, N.M. Biopesticide production from Bacillus thuringiensis: an environmentally friendly alternative. Recent Pat. Biotechnol. 2009, 3, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.W.; Azizbekyan, R.R.; Bernier, R.L.; de Barjac, H.; Saindoux, S.; Valero, J.R.; Yu, M.X. Phage typing of Bacillus subtilis and B. thuringiensis. Res. Microbiol. 1995, 146, 643–657. [Google Scholar] [CrossRef] [PubMed]
- Fujinami, Y.; Hirai, Y.; Sakai, I.; Yoshino, M.; Yasuda, J. Sensitive detection of Bacillusanthracis using a binding protein originating from gamma-phage. Microbiol. Immunol. 2007, 51, 163–169. [Google Scholar] [PubMed]
- Matsuzaki, S.; Rashel, M.; Uchiyama, J.; Sakurai, S.; Ujihara, T.; Kuroda, M.; Ikeuchi, M.; Tani, T.; Fujieda, M.; Wakiguchi, H.; Imai, S. Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases. J. Infect. Chemother. 2005, 11, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Rosovitz, M.J.; Leppla, S.H. Virus deals anthrax a killer blow. Nature 2002, 418, 825–6. [Google Scholar] [CrossRef] [PubMed]
- He, N.-b.; Chen, J.-z.; Lin, C.-c. Six distinct types of bacteriophage attacking Bacillusthuringiensis. Acta Microbiologica Sinica 1978, 18, 220–224. [Google Scholar]
- Loessner, M.J.; Maier, S.K.; Daubek-Puza, H.; Wendlinger, G.; Scherer, S. Three Bacilluscereus bacteriophage endolysins are unrelated but reveal high homology to cell wall hydrolases from different bacilli. J. Bacteriol. 1997, 179, 2845–2851. [Google Scholar] [PubMed]
- Ruhfel, R.E.; Thorne, C.B. Physical and genetic characterisation of the Bacillus thuringiensis subsp. kurstaki HD-1 extrachromosomal temperate phage TP-21. In Abstracts of the Annual Meeting of the American Society for Microbiology; Washington, D.C., USA, 1998; Volume H-4, p. 145. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular CloningCold Spring Habour Laboratory Press: New York, NY, USA, 3rd ed; 2001. [Google Scholar]
- Klumpp, J.; Dorscht, J.; Lurz, R.; Bielmann, R.; Wieland, M.; Zimmer, M.; Calendar, R.; Loessner, M.J. Terminally Redundant, non-permuted Genome of Listeria Bacteriophage A511: a Model for the SPO1-like Myoviruses of Gram-Positive Bacteria. J. Bacteriol. 2008, 190, 5753–5765. [Google Scholar] [CrossRef] [PubMed]
- Steven, A.C.; Trus, B.L.; Maizel, J.V.; Unser, M.; Parry, D.A.; Wall, J.S.; Hainfeld, J.F.; Studier, F.W. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J. Mol. Biol. 1988, 200, 351–365. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.W. Tailed bacteriophages: the order caudovirales. Adv. Virus Res. 1998, 51, 135–201. [Google Scholar] [PubMed]
- Reuben, R.C.; Skalka, A. Identification of the site of interruption in relaxed circles producing during bacteriophage lambda DNA circle replication. J. Virol. 1977, 21, 673–682. [Google Scholar] [PubMed]
- Hoet, P.P.; Coene, M.M.; Cocito, C.G. Replication cycle of Bacillussubtilis hydroxymethyluracil-containing phages. Annu. Rev. Microbiol. 1992, 46, 95–116. [Google Scholar] [PubMed]
- Zimmer, M.; Scherer, S.; Loessner, M.J. Genomic analysis of Clostridiumperfringens bacteriophage phi3626, which integrates into guaA and possibly affects sporulation. J. Bacteriol. 2002, 184, 4359–4368. [Google Scholar] [CrossRef] [PubMed]
- Katsura, I. Determination of bacteriophage lambda tail length by a protein ruler. Nature 1987, 327, 73–75. [Google Scholar] [CrossRef] [PubMed]
- Loessner, M.J.; Inman, R.B.; Lauer, P.; Calendar, R. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeriamonocytogenes: implications for phage evolution. Mol. Microbiol. 2000, 35, 324–340. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, M.; Sattelberger, E.; Inman, R.B.; Calendar, R.; Loessner, M.J. Genome and proteome of Listeriamonocytogenes phage PSA: an unusual case for programmed + 1 translational frameshifting in structural protein synthesis. Mol. Microbiol. 2003, 50, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Eyer, L.; Pantucek, R.; Zdrahal, Z.; Konecna, H.; Kasparek, P.; Ruzickova, V.; Hernychova, L.; Preisler, J.; Doskar, J. Structural protein analysis of the polyvalent staphylococcal bacteriophage 812. Proteomics 2007, 7, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Low, L.Y.; Yang, C.; Perego, M.; Osterman, A.; Liddington, R.C. Structure and lytic activity of a Bacillusanthracis prophage endolysin. J. Biol. Chem. 2005, 280, 35433–35439. [Google Scholar] [CrossRef] [PubMed]
- Sonnhammer, E.L.; von Heijne, G.; Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. In Proceedings of the International Conference on Intelligent Systems for Molecular Biology; Montréal, Canada, 1998; Volume 6, pp. 175–182. [Google Scholar]
- Walter, T.M.; Aronson, A.I. Transduction of certain genes by an autonomously replicating Bacillusthuringiensis phage. Appl. Environ. Microbiol. 1991, 57, 1000–1005. [Google Scholar] [PubMed]
- Kropinski, A.M.; Prangishvili, D.; Lavigne, R. Position paper: The creation of a rational scheme for the nomenclature of viruses of Bacteria and Archaea. Environ. Microbiol. 2009, 11, 2775–2777. [Google Scholar] [CrossRef] [PubMed]
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Share and Cite
Klumpp, J.; Calendar, R.; Loessner, M.J. Complete Nucleotide Sequence and Molecular Characterization of Bacillus Phage TP21 and its Relatedness to Other Phages with the Same Name. Viruses 2010, 2, 961-971. https://doi.org/10.3390/v2040961
Klumpp J, Calendar R, Loessner MJ. Complete Nucleotide Sequence and Molecular Characterization of Bacillus Phage TP21 and its Relatedness to Other Phages with the Same Name. Viruses. 2010; 2(4):961-971. https://doi.org/10.3390/v2040961
Chicago/Turabian StyleKlumpp, Jochen, Richard Calendar, and Martin J. Loessner. 2010. "Complete Nucleotide Sequence and Molecular Characterization of Bacillus Phage TP21 and its Relatedness to Other Phages with the Same Name" Viruses 2, no. 4: 961-971. https://doi.org/10.3390/v2040961
APA StyleKlumpp, J., Calendar, R., & Loessner, M. J. (2010). Complete Nucleotide Sequence and Molecular Characterization of Bacillus Phage TP21 and its Relatedness to Other Phages with the Same Name. Viruses, 2(4), 961-971. https://doi.org/10.3390/v2040961




