Role of RNA Interference (RNAi) in the Moss Physcomitrella patens
Abstract
:1. Introduction
2. Physcomitrella patens Small RNAs
2.1. miRNAs (microRNAs)
2.2. ta-siRNAs (Trans-Acting Small Interfering RNAs)
2.3. ra-siRNAs (Repeat Associated Small Interfering RNAs)
2.4. Secondary siRNAs
3. Physcomitrella patens Homologues of RNAi Pathway Components
4. Physcomitrella patens and Epigenetic Modification
5. Physcomitrella patens and Autoregulation of miRNA Biogenesis
6. Physcomitrella patens and Artificial miRNAs
7. Conclusions and Future Prospects
References
- Vazquez, F. Arabidopsis endogenous small RNAs: Highways and byways. Trends Plant Sci 2006, 11, 460–468. [Google Scholar]
- Axtell, M.J.; Snyder, J.A.; Bartel, D.P. Common functions for diverse small RNAs of land plants. Plant Cell 2007, 19, 1750–1769. [Google Scholar]
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar]
- Lu, C.; Jeong, D.H.; Kulkarni, K.; Pillay, M.; Nobuta, K.; German, R.; Thatcher, S.R.; Maher, C.; Zhang, L.; Ware, D.; et al. Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs). Proc. Natl. Acad. Sci. USA 2008, 105, 4951–4956. [Google Scholar]
- Khraiwesh, B.; Arif, M.A.; Seumel, G.I.; Ossowski, S.; Weigel, D.; Reski, R.; Frank, W. Transcriptional control of gene expression by microRNAs. Cell 2010, 140, 111–122. [Google Scholar]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar]
- De Carvalho, F.; Gheysen, G.; Kushnir, S.; van Montagu, M.; Inze, D.; Castresana, C. Suppression of beta-1,3-glucanase transgene expression in homozygous plants. EMBO J 1992, 11, 2595–2602. [Google Scholar]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar]
- Napoli, C.; Lemieux, C.; Jorgensen, R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 1990, 2, 279–289. [Google Scholar]
- Romano, N.; Macino, G. Quelling: Transient inactivation of gene expression in neurospora crassa by transformation with homologous sequences. Mol. Microbiol 1992, 6, 3343–3353. [Google Scholar]
- Hamilton, A.J.; Baulcombe, D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 1999, 286, 950–952. [Google Scholar]
- Talmor-Neiman, M.; Stav, R.; Klipcan, L.; Buxdorf, K.; Baulcombe, D.C.; Arazi, T. Identification of trans-acting siRNAs in moss and an RNA-dependent RNA polymerase required for their biogenesis. Plant J 2006, 48, 511–521. [Google Scholar]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar]
- Peragine, A.; Yoshikawa, M.; Wu, G.; Albrecht, H.L.; Poethig, R.S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 2004, 18, 2368–2379. [Google Scholar]
- Rajagopalan, R.; Vaucheret, H.; Trejo, J.; Bartel, D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 2006, 20, 3407–3425. [Google Scholar]
- Vazquez, F.; Vaucheret, H.; Rajagopalan, R.; Lepers, C.; Gasciolli, V.; Mallory, A.C.; Hilbert, J.L.; Bartel, D.P.; Crete, P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 2004, 16, 69–79. [Google Scholar]
- Kenrick, P.; Crane, P. The origin and early evolution of plants on land. Nature 1997, 389, 33–39. [Google Scholar]
- Schaefer, D.G.; Zryd, J.P. Efficient gene targeting in the moss Physcomitrella patens. Plant J 1997, 11, 1195–1206. [Google Scholar]
- Rensing, S.A.; Lang, D.; Zimmer, A.D.; Terry, A.; Salamov, A.; Shapiro, H.; Nishiyama, T.; Perroud, P.F.; Lindquist, E.A.; Kamisugi, Y.; et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 2008, 319, 64–69. [Google Scholar]
- Fattash, I.; Voss, B.; Reski, R.; Hess, W.R.; Frank, W. Evidence for the rapid expansion of microRNA-mediated regulation in early land plant evolution. BMC Plant Biol 2007, 7, 13. [Google Scholar]
- Cho, S.H.; Addo-Quaye, C.; Coruh, C.; Arif, M.A.; Ma, Z.; Frank, W.; Axtell, M.J. Physcomitrella patens DCL3 is required for 22–24 nt siRNA accumulation, suppression of retrotransposon-derived transcripts, and normal development. PLoS Genet 2008, 4, e1000314. [Google Scholar]
- Axtell, M.J.; Bartel, D.P. Antiquity of microRNAs and their targets in land plants. Plant Cell 2005, 17, 1658–1673. [Google Scholar]
- Axtell, M.J.; Jan, C.; Rajagopalan, R.; Bartel, D.P. A two-hit trigger for siRNA biogenesis in plants. Cell 2006, 127, 565–577. [Google Scholar]
- Bezanilla, M.; Pan, A.; Quatrano, R.S. RNA interference in the moss Physcomitrella patens. Plant Physiol 2003, 133, 470–474. [Google Scholar]
- Bezanilla, M.; Perroud, P.F.; Pan, A.; Klueh, P.; Quatrano, R.S. An RNAi system in Physcomitrella patens with an internal marker for silencing allows for rapid identification of loss of function phenotypes. Plant Biol 2005, 7, 251–257. [Google Scholar]
- Khraiwesh, B.; Ossowski, S.; Weigel, D.; Reski, R.; Frank, W. Specific gene silencing by artificial microRNAs in Physcomitrella patens: An alternative to targeted gene knockouts. Plant Physiol 2008, 148, 684–693. [Google Scholar]
- Arazi, T.; Talmor-Neiman, M.; Stav, R.; Riese, M.; Huijser, P.; Baulcombe, D.C. Cloning and characterization of micro-RNAs from moss. Plant J 2005, 43, 837–848. [Google Scholar]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar]
- Park, W.; Li, J.; Song, R.; Messing, J.; Chen, X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol 2002, 12, 1484–1495. [Google Scholar]
- Yu, B.; Yang, Z.; Li, J.; Minakhina, S.; Yang, M.; Padgett, R.W.; Steward, R.; Chen, X. Methylation as a crucial step in plant microRNA biogenesis. Science 2005, 307, 932–935. [Google Scholar]
- Yang, Z.; Ebright, Y.W.; Yu, B.; Chen, X. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res 2006, 2, 667–675. [Google Scholar]
- Gan, J.; Tropea, J.E.; Austin, B.P.; Court, D.L.; Waugh, D.S.; Ji, X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell 2006, 124, 355–366. [Google Scholar]
- Hammond, S.M.; Bernstein, E.; Beach, D.; Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000, 404, 293–296. [Google Scholar]
- Noma, K.; Sugiyama, T.; Cam, H.; Verdel, A.; Zofall, M.; Jia, S.; Moazed, D.; Grewal, S.I. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet 2004, 36, 1174–1180. [Google Scholar]
- Hutvagner, G.; Simard, M.J. ARGONAUTE proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol 2008, 9, 22–32. [Google Scholar]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar]
- Morel, J.B.; Godon, C.; Mourrain, P.; Beclin, C.; Boutet, S.; Feuerbach, F.; Proux, F.; Vaucheret, H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 2002, 14, 629–639. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar]
- Baumberger, N.; Baulcombe, D.C. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 2005, 102, 11928–11933. [Google Scholar]
- Qi, Y.; Denli, A.M.; Hannon, G.J. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 2005, 19, 421–428. [Google Scholar]
- Chen, X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar]
- Brodersen, P.; Sakvarelidze-Achard, L.; Bruun-Rasmussen, M.; Dunoyer, P.; Yamamoto, Y.Y.; Sieburth, L.; Voinnet, O. Widespread translational inhibition by plant miRNAs and siRNAs. Science 2008, 320, 1185–1190. [Google Scholar]
- Lanet, E.; Delannoy, E.; Sormani, R.; Floris, M.; Brodersen, P.; Crete, P.; Voinnet, O.; Robaglia, C. Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 2009, 21, 1762–1768. [Google Scholar]
- Matzke, M.A.; Matzke, A.J. Planting the seeds of a new paradigm. PLoS Biol 2004, 2, E133. [Google Scholar]
- Schramke, V.; Allshire, R. Those interfering little RNAs! Silencing and eliminating chromatin. Curr. Opin. Genet. Dev 2004, 14, 174–180. [Google Scholar]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004, 23, 4051–4060. [Google Scholar]
- Kim, V.N. MicroRNA precursors in motion: Exportin-5 mediates their nuclear export. Trends Cell Biol 2004, 14, 156–159. [Google Scholar]
- Park, M.Y.; Wu, G.; Gonzalez-Sulser, A.; Vaucheret, H.; Poethig, R.S. Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 3691–3696. [Google Scholar]
- Bao, N.; Lye, K.W.; Barton, M.K. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell 2004, 7, 653–662. [Google Scholar]
- Khraiwesh, B.; Zhu, J.K.; Zhu, J.H. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 2012, 1819, 137–148. [Google Scholar]
- Wu, L.; Zhou, H.; Zhang, Q.; Zhang, J.; Ni, F.; Liu, C.; Qi, Y. DNA methylation mediated by a microRNA pathway. Mol. Cell 2010, 38, 465–475. [Google Scholar]
- Addo-Quaye, C.; Snyder, J.A.; Park, Y.B.; Li, Y.F.; Sunkar, R.; Axtell, M.J. Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA 2009, 15, 2112–2121. [Google Scholar]
- Saleh, O.; Issman, N.; Seumel, G.I.; Stav, R.; Samach, A.; Reski, R.; Frank, W.; Arazi, T. MicroRNA534a control of BLADE-ON-PETIOLE 1 and 2 mediates juvenile-to-adult gametophyte transition in Physcomitrella patens. Plant J 2011, 65, 661–674. [Google Scholar]
- Xie, Z.; Allen, E.; Wilken, A.; Carrington, J.C. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2005, 102, 12984–12989. [Google Scholar]
- Gasciolli, V.; Mallory, A.C.; Bartel, D.P.; Vaucheret, H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol 2005, 15, 1494–1500. [Google Scholar]
- Adenot, X.; Elmayan, T.; Lauressergues, D.; Boutet, S.; Bouche, N.; Gasciolli, V.; Vaucheret, H. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol 2006, 16, 927–932. [Google Scholar]
- Yoshikawa, M.; Peragine, A.; Park, M.Y.; Poethig, R.S. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 2005, 19, 2164–2175. [Google Scholar]
- Montgomery, T.A.; Howell, M.D.; Cuperus, J.T.; Li, D.; Hansen, J.E.; Alexander, A.L.; Chapman, E.J.; Fahlgren, N.; Allen, E.; Carrington, J.C. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 2008, 133, 128–141. [Google Scholar]
- Li, F.; Orban, R.; Baker, B. SoMART: A web server for plant miRNA, tasiRNA and target gene analysis. Plant J 2012, 70, 891–901. [Google Scholar]
- Arif, M.A.; Fattash, I.; Ma, Z.; Cho, S.H.; Beike, A.K.; Reski, R.; Axtell, M.J.; Frank, W. DICER-LIKE3 activity in physcomitrella patens DICER-LIKE4 mutants causes severe developmental dysfunction and sterility. Mol. Plant 2012, 5, 1281–1294. [Google Scholar]
- Williams, L.; Carles, C.C.; Osmont, K.S.; Fletcher, J.C. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl. Acad. Sci. USA 2005, 102, 9703–9708. [Google Scholar]
- Garcia, D.; Collier, S.A.; Byrne, M.E.; Martienssen, R.A. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol 2006, 16, 933–938. [Google Scholar]
- Fahlgren, N.; Montgomery, T.A.; Howell, M.D.; Allen, E.; Dvorak, S.K.; Alexander, A.L.; Carrington, J.C. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol 2006, 16, 939–944. [Google Scholar]
- Dunoyer, P.; Brosnan, C.A.; Schott, G.; Wang, Y.; Jay, F.; Alioua, A.; Himber, C.; Voinnet, O. An endogenous, systemic RNAi pathway in plants. EMBO J 2010, 29, 1699–1712. [Google Scholar]
- Axtell, M.J. The small RNAs of Physcomitrella patens: Expression, function and evolution. Ann. Plant Rev 2009, 36, 113–142. [Google Scholar]
- MacRae, I.J.; Ma, E.; Zhou, M.; Robinson, C.V.; Doudna, J.A. In vitro reconstitution of the human RISC-loading complex. Proc. Natl. Acad. Sci. USA 2008, 105, 512–517. [Google Scholar]
- Nobuta, K.; McCormick, K.; Nakano, M.; Meyers, B.C. Bioinformatics analysis of small RNAs in plants using next generation sequencing technologies. Methods Mol. Biol 2010, 592, 89–106. [Google Scholar]
- Morin, R.D.; Aksay, G.; Dolgosheina, E.; Ebhardt, H.A.; Magrini, V.; Mardis, E.R.; Sahinalp, S.C.; Unrau, P.J. Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res 2008, 18, 571–584. [Google Scholar]
- Xie, Z.; Johansen, L.K.; Gustafson, A.M.; Kasschau, K.D.; Lellis, A.D.; Zilberman, D.; Jacobsen, S.E.; Carrington, J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2004, 2, E104. [Google Scholar]
- Pak, J.; Fire, A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 2007, 315, 241–244. [Google Scholar]
- Sijen, T.; Steiner, F.A.; Thijssen, K.L.; Plasterk, R.H. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 2007, 315, 244–247. [Google Scholar]
- Baulcombe, D.C. Molecular biology. Amplified silencing. Science 2007, 315, 199–200. [Google Scholar]
- Moissiard, G.; Parizotto, E.A.; Himber, C.; Voinnet, O. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA 2007, 13, 1268–1278. [Google Scholar]
- Mlotshwa, S.; Pruss, G.J.; Peragine, A.; Endres, M.W.; Li, J.; Chen, X.; Poethig, R.S.; Bowman, L.H.; Vance, V. DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS One 2008, 3, e1755. [Google Scholar]
- Howell, M.D.; Fahlgren, N.; Chapman, E.J.; Cumbie, J.S.; Sullivan, C.M.; Givan, S.A.; Kasschau, K.D.; Carrington, J.C. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 2007, 19, 926–942. [Google Scholar]
- Luo, Q.J.; Samanta, M. P.; Koksal, F.; Janda, J.; Galbraith, D.W.; Richardson, C.R.; Ou-Yang, F.; Rock, C.D. Evidence for antisense transcription associated with microRNA target mRNAs in Arabidopsis. PLoS Genet 2009, 5, e1000457. [Google Scholar]
- Alder, M.N.; Dames, S.; Gaudet, J.; Mango, S.E. Gene silencing in Caenorhabditis elegans by transitive RNA interference. RNA 2003, 9, 25–32. [Google Scholar]
- Nishikura, K. A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst. Cell 2001, 107, 415–418. [Google Scholar]
- Chen, H.M.; Chen, L.T.; Patel, K.; Li, Y.H.; Baulcombe, D.C.; Wu, S.H. From the cover: 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 2010, 107, 15269–15274. [Google Scholar]
- Vaucheret, H. Plant ARGONAUTES. Trends Plant Sci 2008, 13, 350–358. [Google Scholar]
- Asif, M.A.; Fattash, I.; Khraiwesh, B.; Frank, W. Physcomitrella patens Small RNA Pathways. In Non Coding RNAs in Plants; Erdmann, V.A., Barciszewski, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 139–173. [Google Scholar]
- Kurihara, Y.; Watanabe, Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 2004, 101, 12753–12758. [Google Scholar]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev 2002, 16, 1616–1626. [Google Scholar]
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 2005, 123, 1279–1291. [Google Scholar]
- Bouche, N.; Lauressergues, D.; Gasciolli, V.; Vaucheret, H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J 2006, 25, 3347–3356. [Google Scholar]
- Liu, B.; Chen, Z.; Song, X.; Liu, C.; Cui, X.; Zhao, X.; Fang, J.; Xu, W.; Zhang, H.; Wang, X.; et al. Oryza sativa dicer-like4 reveals a key role for small interfering RNA silencing in plant development. Plant Cell 2007, 19, 2705–2718. [Google Scholar]
- Dunoyer, P.; Himber, C.; Voinnet, O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet 2005, 37, 1356–1360. [Google Scholar]
- Vaucheret, H.; Mallory, A.C.; Bartel, D.P. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell 2006, 22, 129–136. [Google Scholar]
- Vaucheret, H.; Vazquez, F.; Crete, P.; Bartel, D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 2004, 18, 1187–1197. [Google Scholar]
- Harvey, J.J.; Lewsey, M.G.; Patel, K.; Westwood, J.; Heimstadt, S.; Carr, J.P.; Baulcombe, D.C. An antiviral defense role of AGO2 in plants. PLoS One 2011, 6, e14639. [Google Scholar]
- Rajeswaran, R.; Aregger, M.; Zvereva, A.S.; Borah, B.K.; Gubaeva, E.G.; Pooggin, M.M. Sequencing of RDR6-dependent double-stranded RNAs reveals novel features of plant siRNA biogenesis. Nucleic Acids Res 2012, 40, 6241–6254. [Google Scholar]
- Zilberman, D.; Cao, X.; Jacobsen, S.E. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 2003, 299, 716–719. [Google Scholar]
- Havecker, E.R.; Wallbridge, L.M.; Hardcastle, T.J.; Bush, M.S.; Kelly, K.A.; Dunn, R.M.; Schwach, F.; Doonan, J.H.; Baulcombe, D.C. The Arabidopsis RNA-directed DNA methylation ARGONAUTE functionally diverge based on their expression and interaction with target loci. Plant Cell 2010, 22, 321–334. [Google Scholar]
- Pontes, O.; Costa-Nunes, P.; Vithayathil, P.; Pikaard, C.S. RNA polymerase V functions in Arabidopsis interphase heterochromatin organization independently of the 24-nt siRNA-directed DNA methylation pathway. Mol. Plant 2009, 2, 700–710. [Google Scholar]
- Pontes, O.; Li, C.F.; Nunes, P.C.; Haag, J.; Ream, T.; Vitins, A.; Jacobsen, S.E.; Pikaard, C.S. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 2006, 126, 79–92. [Google Scholar]
- Wierzbicki, A.T.; Haag, J.R.; Pikaard, C.S. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 2008, 135, 635–648. [Google Scholar]
- Zilberman, D.; Cao, X.; Johansen, L.K.; Xie, Z.; Carrington, J.C.; Jacobsen, S.E. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol 2004, 14, 1214–1220. [Google Scholar]
- Zheng, X.; Zhu, J.; Kapoor, A.; Zhu, J.K. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J 2007, 26, 1691–1701. [Google Scholar]
- Olmedo-Monfil, V.; Duran-Figueroa, N.; Arteaga-Vazquez, M.; Demesa-Arevalo, E.; Autran, D.; Grimanelli, D.; Slotkin, R.K.; Martienssen, R.A.; Vielle-Calzada, J.P. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 2010, 464, 628–632. [Google Scholar]
- Liu, Q.; Yao, X.; Pi, L.; Wang, H.; Cui, X.; Huang, H. The ARGONAUTE10 gene modulates shoot apical meristem maintenance and leaf polarity establishment by repressing miR165/166 in Arabidopsis. Plant J 2009, 58, 27–40. [Google Scholar]
- Mallory, A.C.; Hinze, A.; Tucker, M.R.; Bouche, N.; Gasciolli, V.; Elmayan, T.; Lauressergues, D.; Jauvion, V.; Vaucheret, H.; Laux, T. Redundant and specific roles of the ARGONAUTE proteins AGO1 and ZLL in development and small RNA-directed gene silencing. PLoS Genet 2009, 5, e1000646. [Google Scholar]
- Sugiyama, T.; Cam, H.; Verdel, A.; Moazed, D.; Grewal, S.I. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc. Natl. Acad. Sci. USA 2005, 102, 152–157. [Google Scholar]
- Vaistij, F.E.; Jones, L.; Baulcombe, D.C. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 2002, 14, 857–867. [Google Scholar]
- Garcia-Ruiz, H.; Takeda, A.; Chapman, E.J.; Sullivan, C.M.; Fahlgren, N.; Brempelis, K.J.; Carrington, J.C. Arabidopsis RNA-Dependent RNA polymerases and Dicer-Like proteins in antiviral defense and small interfering RNA biogenesis during turnip mosaic virus infection. Plant Cell 2010, 22, 481–496. [Google Scholar]
- Chan, S.W.; Zilberman, D.; Xie, Z.; Johansen, L.K.; Carrington, J.C.; Jacobsen, S.E. RNA silencing genes control de novo DNA methylation. Science 2004, 303, 1336. [Google Scholar]
- Kasschau, K.D.; Fahlgren, N.; Chapman, E.J.; Sullivan, C.M.; Cumbie, J.S.; Givan, S.A.; Carrington, J.C. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 2007, 5, e57. [Google Scholar]
- Pandey, S.P.; Gaquerel, E.; Gase, K.; Baldwin, I.T. RNA-directed RNA polymerase3 from Nicotiana attenuata is required for competitive growth in natural environments. Plant Physiol 2008, 147, 1212–1224. [Google Scholar]
- Zong, J.; Yao, X.; Yin, J.; Zhang, D.; Ma, H. Evolution of the RNA-dependent RNA polymerase (RdRP) genes: Duplications and possible losses before and after the divergence of major eukaryotic groups. Gene 2009, 447, 29–39. [Google Scholar]
- Dalmay, T.; Hamilton, A.; Rudd, S.; Angell, S.; Baulcombe, D.C. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 2000, 101, 543–553. [Google Scholar]
- Elmayan, T.; Adenot, X.; Gissot, L.; Lauressergues, D.; Gy, I.; Vaucheret, H. A neomorphic sgs3 allele stabilizing miRNA cleavage products reveals that SGS3 acts as a homodimer. FEBS J 2009, 276, 835–844. [Google Scholar]
- Akbergenov, R.; Si-Ammour, A.; Blevins, T.; Amin, I.; Kutter, C.; Vanderschuren, H.; Zhang, P.; Gruissem, W.; Meins, F.; Hohn, T.; et al. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res 2006, 34, 462–471. [Google Scholar]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 2004, 18, 3016–3027. [Google Scholar]
- Fang, Y.; Spector, D.L. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol 2007, 17, 818–823. [Google Scholar]
- Qin, H.; Chen, F.; Huan, X.; Machida, S.; Song, J.; Yuan, Y.A. Structure of the Arabidopsis thaliana DCL4 DUF283 domain reveals a noncanonical double-stranded RNA-binding fold for protein-protein interaction. RNA 2010, 16, 474–481. [Google Scholar]
- Manavella, P.A.; Hagmann, J.; Ott, F.; Laubinger, S.; Franz, M.; Macek, B.; Weigel, D. Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 2012, 151, 859–870. [Google Scholar]
- Kim, S.; Yang, J.-Y.; Xu, J.; Jang, I.-C.; Prigge, M.J.; Chua, N.-H. Two cap-binding proteins CBP20 and CBP80 are involved in processing primary microRNAs. Plant Cell Physiol 2008, 49, 1634–1644. [Google Scholar]
- Laubinger, S.; Sachsenberg, T.; Zeller, G.; Busch, W.; Lohmann, J.U.; Rätsch, G.; Weigel, D. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing inArabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2008. [Google Scholar] [CrossRef]
- Smith, M.R.; Willmann, M.R.; Wu, G.; Berardini, T.Z.; Möller, B.; Weijers, D.; Poethig, R.S. Cyclophilin 40 is required for microRNA activity inArabidopsis. Proc. Natl. Acad. Sci. USA 2009. [Google Scholar] [CrossRef]
- Iki, T.; Yoshikawa, M.; Meshi, T.; Ishikawa, M. Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J 2012, 31, 267–278. [Google Scholar]
- Earley, K.W.; Poethig, R.S. Binding of the cyclophilin 40 Ortholog SQUINT to Hsp90 protein is required for SQUINT function in Arabidopsis. J. Biol. Chem 2011, 286, 38184–38189. [Google Scholar]
- Cao, X.; Aufsatz, W.; Zilberman, D.; Mette, M.F.; Huang, M.S.; Matzke, M.; Jacobsen, S.E. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol 2003, 13, 2212–2217. [Google Scholar]
- Cao, X.; Jacobsen, S.E. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. USA 2002, 99, 16491–16498. [Google Scholar]
- Law, J.A.; Ausin, I.; Johnson, L.M.; Vashisht, A.A.; Zhu, J.K.; Wohlschlegel, J.A.; Jacobsen, S.E. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr. Biol 2010, 20, 951–956. [Google Scholar]
- Huettel, B.; Kanno, T.; Daxinger, L.; Bucher, E.; van der Winden, J.; Matzke, A.J.; Matzke, M. RNA-directed DNA methylation mediated by DRD1 and Pol IVb: A versatile pathway for transcriptional gene silencing in plants. Biochim. Biophys. Acta 2007, 1769, 358–374. [Google Scholar]
- Chan, S.W.; Henderson, I.R.; Zhang, X.; Shah, G.; Chien, J.S.; Jacobsen, S.E. RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in arabidopsis. PLoS Genet 2006, 2, e83. [Google Scholar]
- Matzke, M.; Kanno, T.; Huettel, B.; Daxinger, L.; Matzke, A.J. RNA-directed DNA methylation and Pol IVb in Arabidopsis. Cold Spring Harbor Symp. Quant. Biol 2006, 71, 449–459. [Google Scholar]
- Huettel, B.; Kanno, T.; Daxinger, L.; Aufsatz, W.; Matzke, A.J.; Matzke, M. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J 2006, 25, 2828–2836. [Google Scholar]
- Jeddeloh, J.A.; Stokes, T.L.; Richards, E.J. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet 1999, 22, 94–97. [Google Scholar]
- Shaked, H.; Avivi-Ragolsky, N.; Levy, A.A. Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics 2006, 173, 985–994. [Google Scholar]
- Morel, J.B.; Mourrain, P.; Beclin, C.; Vaucheret, H. DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr. Biol 2000, 10, 1591–1594. [Google Scholar]
- Smith, L.M.; Pontes, O.; Searle, I.; Yelina, N.; Yousafzai, F.K.; Herr, A.J.; Pikaard, C.S.; Baulcombe, D.C. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell 2007, 19, 1507–1521. [Google Scholar]
- Ausin, I.; Mockler, T.C.; Chory, J.; Jacobsen, S.E. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat. Struct. Mol. Biol 2009, 16, 1325–1327. [Google Scholar]
- Zheng, Z.; Xing, Y.; He, X.J.; Li, W.; Hu, Y.; Yadav, S.K.; Oh, J.; Zhu, J.K. An SGS3-like protein functions in RNA-directed DNA methylation and transcriptional gene silencing in Arabidopsis. Plant J 2010, 62, 92–99. [Google Scholar]
- Golden, T.A.; Schauer, S.E.; Lang, J.D.; Pien, S.; Mushegian, A.R.; Grossniklaus, U.; Meinke, D.W.; Ray, A. Short integuments1/suspensor1/carpel factory, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol 2002, 130, 808–822. [Google Scholar]
- Satoh, N.; Itoh, J.; Nagato, Y. The SHOOTLESS2 and SHOOTLESS1 genes are involved in both initiation and maintenance of the shoot apical meristem through regulating the number of indeterminate cells. Genetics 2003, 164, 335–346. [Google Scholar]
- Nagasaki, H.; Itoh, J.; Hayashi, K.; Hibara, K.; Satoh-Nagasawa, N.; Nosaka, M.; Mukouhata, M.; Ashikari, M.; Kitano, H.; Matsuoka, M.; et al. The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl. Acad. Sci. USA 2007, 104, 14867–14871. [Google Scholar]
- Toriba, T.; Suzaki, T.; Yamaguchi, T.; Ohmori, Y.; Tsukaya, H.; Hirano, H.Y. Distinct regulation of adaxial-abaxial polarity in anther patterning in rice. Plant Cell 2010, 22, 1452–1462. [Google Scholar]
- Hamilton, A.; Voinnet, O.; Chappell, L.; Baulcombe, D. Two classes of short interfering RNA in RNA silencing. EMBO J 2002, 21, 4671–4679. [Google Scholar]
- Matzke, M.A.; Birchler, J.A. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet 2005, 6, 24–35. [Google Scholar]
- Melquist, S.; Bender, J. Transcription from an upstream promoter controls methylation signaling from an inverted repeat of endogenous genes in Arabidopsis. Genes Dev 2003, 17, 2036–2047. [Google Scholar]
- Aufsatz, W.; Mette, M.F.; van der Winden, J.; Matzke, A.J.; Matzke, M. RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 16499–16506. [Google Scholar]
- Pelissier, T.; Thalmeir, S.; Kempe, D.; Sanger, H.L.; Wassenegger, M. Heavy de novo methylation at symmetrical and non-symmetrical sites is a hallmark of RNA-directed DNA methylation. Nucleic Acids Res 1999, 27, 1625–1634. [Google Scholar]
- Hall, I.M.; Shankaranarayana, G.D.; Noma, K.; Ayoub, N.; Cohen, A.; Grewal, S.I. Establishment and maintenance of a heterochromatin domain. Science 2002, 297, 2232–2237. [Google Scholar]
- Aufsatz, W.; Mette, M.F.; Matzke, A.J.; Matzke, M. The role of MET1 in RNA-directed de novo and maintenance methylation of CG dinucleotides. Plant Mol. Biol 2004, 54, 793–804. [Google Scholar]
- Verdel, A.; Jia, S.; Gerber, S.; Sugiyama, T.; Gygi, S.; Grewal, S.I.; Moazed, D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 2004, 303, 672–676. [Google Scholar]
- Liu, Q.; Rand, T.A.; Kalidas, S.; Du, F.; Kim, H.E.; Smith, D.P.; Wang, X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 2003, 301, 1921–1925. [Google Scholar]
- Tabara, H.; Yigit, E.; Siomi, H.; Mello, C.C. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 2002, 109, 861–871. [Google Scholar]
- Tomari, Y.; Zamore, P.D. Perspective: Machines for RNAi. Genes Dev 2005, 19, 517–529. [Google Scholar]
- Xie, Z.; Kasschau, K.D.; Carrington, J.C. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol 2003, 13, 784–789. [Google Scholar]
- Alvarez, J.P.; Pekker, I.; Goldshmidt, A.; Blum, E.; Amsellem, Z.; Eshed, Y. Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 2006, 18, 1134–1151. [Google Scholar]
- Niu, Q.W.; Lin, S.S.; Reyes, J.L.; Chen, K.C.; Wu, H.W.; Yeh, S.D.; Chua, N.H. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol 2006, 24, 1420–1428. [Google Scholar]
- Ossowski, S.; Schwab, R.; Weigel, D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 2008, 53, 674–690. [Google Scholar]
- Parizotto, E.A.; Dunoyer, P.; Rahm, N.; Himber, C.; Voinnet, O. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev 2004, 18, 2237–2242. [Google Scholar]
- Qu, J.; Ye, J.; Fang, R. Artificial microRNA-mediated virus resistance in plants. J. Virol 2007, 81, 6690–6699. [Google Scholar]
- Schwab, R.; Ossowski, S.; Riester, M.; Warthmann, N.; Weigel, D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 2006, 18, 1121–1133. [Google Scholar]
- Warthmann, N.; Chen, H.; Ossowski, S.; Weigel, D.; Herve, P. Highly specific gene silencing by artificial miRNAs in rice. PLoS One 2008, 3, e1829. [Google Scholar]
- Strepp, R.; Scholz, S.; Kruse, S.; Speth, V.; Reski, R. Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc. Natl. Acad. Sci. USA 1998, 95, 4368–4373. [Google Scholar]
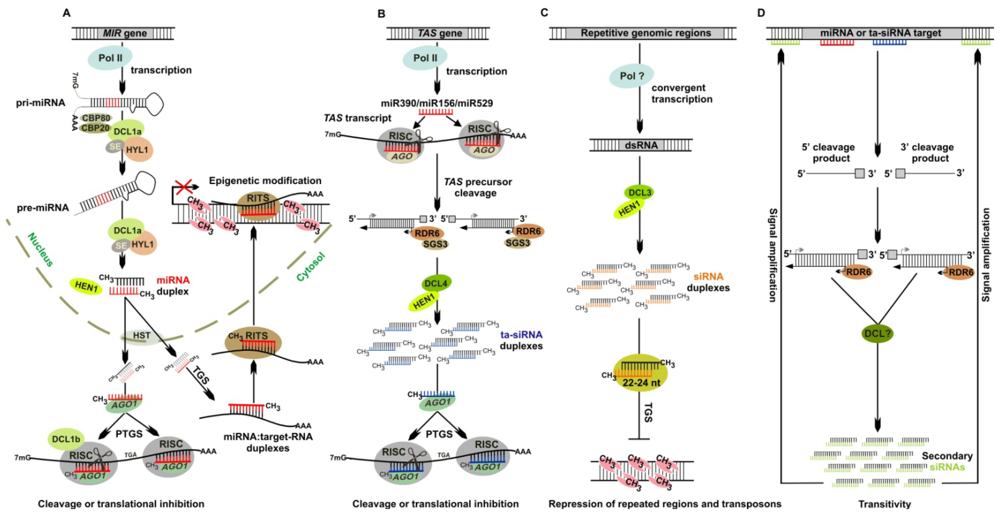
| Protein family | P. patens homologues | NCBI/Gene model number | E-value, % Identity | Molecular function | References |
|---|---|---|---|---|---|
| AtDCL1 | PpDCL1a | EF670436 | 0.0, 68% | miRNA biogenesis Indispensable for target cleavage | [5,29,82,83] |
| PpDCL1b | DQ675601 | 0.0, 65% | |||
| AtDCL2 | n.i 1 | - | - | Generates endogenous siRNAs from a convergently transcribed and overlapping gene pairs Transitive silencing of transgenes Produces viral siRNAs | [74,84,85] |
| AtDCL3 | PpDCL3 | EF670437 | 1e−116, 32% | Generates siRNAs that guide chromatin modification in P. patens and A. thaliana | [21,69] |
| AtDCL4 | PpDCL4 | EF670438 | 1e−124, 33% | Generates trans-acting siRNAs (ta-siRNAs) | [54,55,57,60,86,87] |
| AtAGO1 | PpAGO1a 2 | Phypa_205541 | 0.0, 78% | Associates with the majority of miRNAs to guide the cleavage of their targets | [2,39,40,88,89] |
| PpAGO1b 2 | Phypa_158832 | 0.0, 77% | |||
| PpAGO1c 2 | Phypa_141045 | 0.0, 75% | |||
| AtAGO2 | n.i | - | -- | Known to be function in antiviral defense and ta-siRNAs biogenesis | [90,91] |
| AtAGO3 | n.i | - | - | Not analyzed | |
| AtAGO4 | PpAGO42 | Phypa_200513 | 1e−164, 38% | Involved in 24nt siRNA mediated gene silencing | [92–97] |
| AtAGO5 | n.i | - | - | Not analyzed | |
| AtAGO6 | PpAGO62 | Phypa_117253 | 1e−152, 39% | Involved in 24nt siRNA mediated DNA methylation | [93,98] |
| AtAGO7 | n.i | - | - | Associates specifically with miR390 and directs cleavage of the AtTAS3 precursor | [13–16,56,58] |
| AtAGO8 | n.i | - | - | Not analyzed | |
| AtAGO9 | PpAGO92 | Phypa_134255 | 6.1e−160, 40% | Preferentially interacts with 24nt siRNAs derived from transposable elements (TEs), required to silence TEs in female gametes and their accessory cells. Cell fate determination in the ovule. | [93,99] |
| AtAGO10 | n.i | - | - | Implicated in miRNA-directed translational inhibition and repression of miR165/166 levels | [42,100,101] |
| AtRDR1 | PpRDR12 | Phypa_219654 | 1.3e−204, 48% | Synthesis of long dsRNA from transgenes that can initiate different RNAi pathways Biogenesis of secondary siRNAs from RNA viruses | [102–104] |
| AtRDR2 | n.i | - | - | Biogenesis of 24nt siRNAs from repeat loci involved in DNA methylation | [105,106] |
| AtRDR3a | n.i. | - | - | Not analyzed | [107,108] |
| AtRDR3b | PpRDR3b 2 | Phypa_169723 | 1.8e−96, 33% | ||
| AtRDR3c | PpRDR3c 2 | Phypa_172848 | 6.1e−89, 31% | ||
| AtRDR6 | PpRDR6 | Phypa_379 | 2.8e−226, 42% | Initiation and maintenance of dsRNA-induced RNAi in A. thaliana Conversion of TAS precursors into dsRNA in P. patens and A. thaliana | [12,14,109,110] |
| AtHEN1 | PpHEN12 | Phypa_148777 | 3e−56, 33% | Methylates miRNA and siRNA duplexes at the 3′ end | [29–31,111] |
| AtHYL | PpHYL12 | Phypa_34761 | 7e−31, 50% | Interacts with AtDCL1 and confers stability to miRNA precursors | [69,112–114] |
| AtHASTY | PpHASTY12 | Phypa_137344 | 1.3e−228, 40% | Exports miRNA-miRNA * duplex to the cytoplasm | [14,48] |
| PpHASTY22 | Phypa_151199 | 2.1e−173, 41% | |||
| AtSE | PpSE12 | Phypa_133793 | 1.7e−92, 41% | Interacts with AtDCL1 and confers stability to miRNA precursors | [69,82,112,113] |
| PpSE22 | Phypa_124567 | 1.8e−70, 41% | |||
| PpSE32 | Phypa_99415 | 3.3e−53, 35% | |||
| AtCPL1 | PpCPL12 | Phypa_432395 | 1e−126, 49% | Required for HYL1 dephosphorylation, which in turn is essential for accurate miRNA processing and strand selection. | [115] |
| PpCPL22 | Phypa_429817 | 1e−126, 51% | |||
| AtCBP20 | PpCBP20a2 | Phypa_442048 | 5e−42, 53% | Involved in pre-miRNA splicing and miRNA processing | [116,117] |
| PpCBP20b2 | Phypa_442049 | 7e−71, 58% | |||
| PpCBP20c2 | Phypa_442050 | 7e−69,76% | |||
| AtCBP80 | PpCBP80.12 | Phypa_425787 | 0.0, 47% | Involved in pre-miRNA splicing and miRNA processing | [116,117] |
| PpCBP80.22 | Phypa_432264 | 0.0, 47% | |||
| AtSQN/CYP40 | PpSQNa2 | Phypa_433182 | 1e−136, 66% | Required for miRNA activity by promoting the activity of AGO1. Plays a unique and important role in plant RISC assembly | [118–120] |
| PpSQNb2 | Phypa_433181 | 1e−136, 66% | |||
| AtHSP90 | PpHsp90.12 | Phypa_456075 | 0.0, 80% | Plays a unique and important role in plant RISC assembly | [119,120] |
| PpHsp90.22 | Phypa_454408 | 0.0, 80% | |||
| PpHsp90.32 | Phypa_452062 | 0.0, 79% | |||
| PpHsp90.42 | Phypa_452093 | 0.0, 80% | |||
| AtSGS3 | PpSGS32 | Phypa_448213 | 3.0e−71, 37% | Involved in the production of ta-siRNAs, through direct or indirect stabilisation of TAS cleavage products | [14,110] |
| AtPol IV | PpPol IV2 | Phypa_132119 | 1.3e−72, 49% | Required for the biogenesis of 24nt siRNAs (with RDR2 and DCL3) that associate with AGO4 and direct DNA and histone modifications | [94–96] |
| AtPol V | PpPol V2 | Phypa_129844 | 1e−132, 70% | Generates transcripts from heterochromatic regions (with DRD1) that are discussed to bind siRNA-AGO4 complexes directing DNA and histone modifications | [94–96] |
| AtDRM1 | PpDRM12 | Phypa_148057 | 5e−92, 51% | Involved in the siRNA-directed de novo DNA methylation and maintenance of DNA methylation at CHH sites | [105,121,122] |
| AtDRM2 | PpDRM22 | Phypa_133529 | 6.3e−87, 47% | ||
| AtDRD1 | PpDRD12 | Phypa_113504 | 1e−109, 35% | Cooperates with Pol V | [94,96,122–127] |
| AtSNF2 | PpSNF22 | Phypa_211797 | 1.3e−187, 46% | Involved in the spreading of transgene silencing (with AtRDR2 and AtPol IV) and in the production of endogenous 24 nt siRNAs | [128–131] |
| AtRDM12 | PpRDM122 | Phypa_98999 | 1e−46, 26% | Involved in the de novo DNA methylation and siRNA-mediated maintenance of DNA methylation | [132,133] |
© 2013 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Arif, M.A.; Frank, W.; Khraiwesh, B. Role of RNA Interference (RNAi) in the Moss Physcomitrella patens. Int. J. Mol. Sci. 2013, 14, 1516-1540. https://doi.org/10.3390/ijms14011516
Arif MA, Frank W, Khraiwesh B. Role of RNA Interference (RNAi) in the Moss Physcomitrella patens. International Journal of Molecular Sciences. 2013; 14(1):1516-1540. https://doi.org/10.3390/ijms14011516
Chicago/Turabian StyleArif, Muhammad Asif, Wolfgang Frank, and Basel Khraiwesh. 2013. "Role of RNA Interference (RNAi) in the Moss Physcomitrella patens" International Journal of Molecular Sciences 14, no. 1: 1516-1540. https://doi.org/10.3390/ijms14011516




