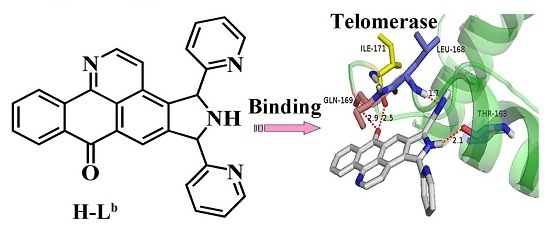
Oxoisoaporphine as Potent Telomerase Inhibitor
Abstract
:1. Introduction
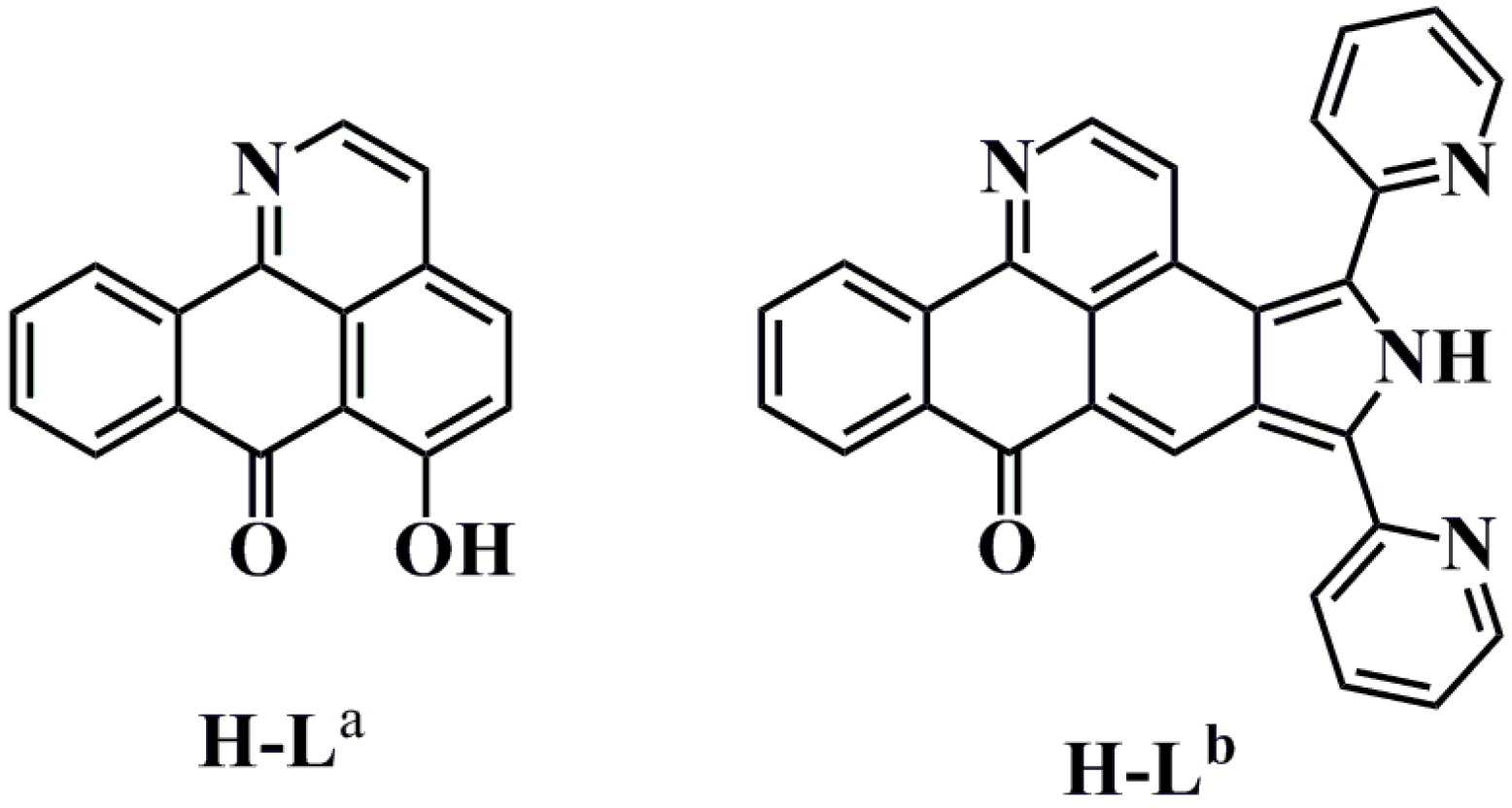
2. Results and Discussion
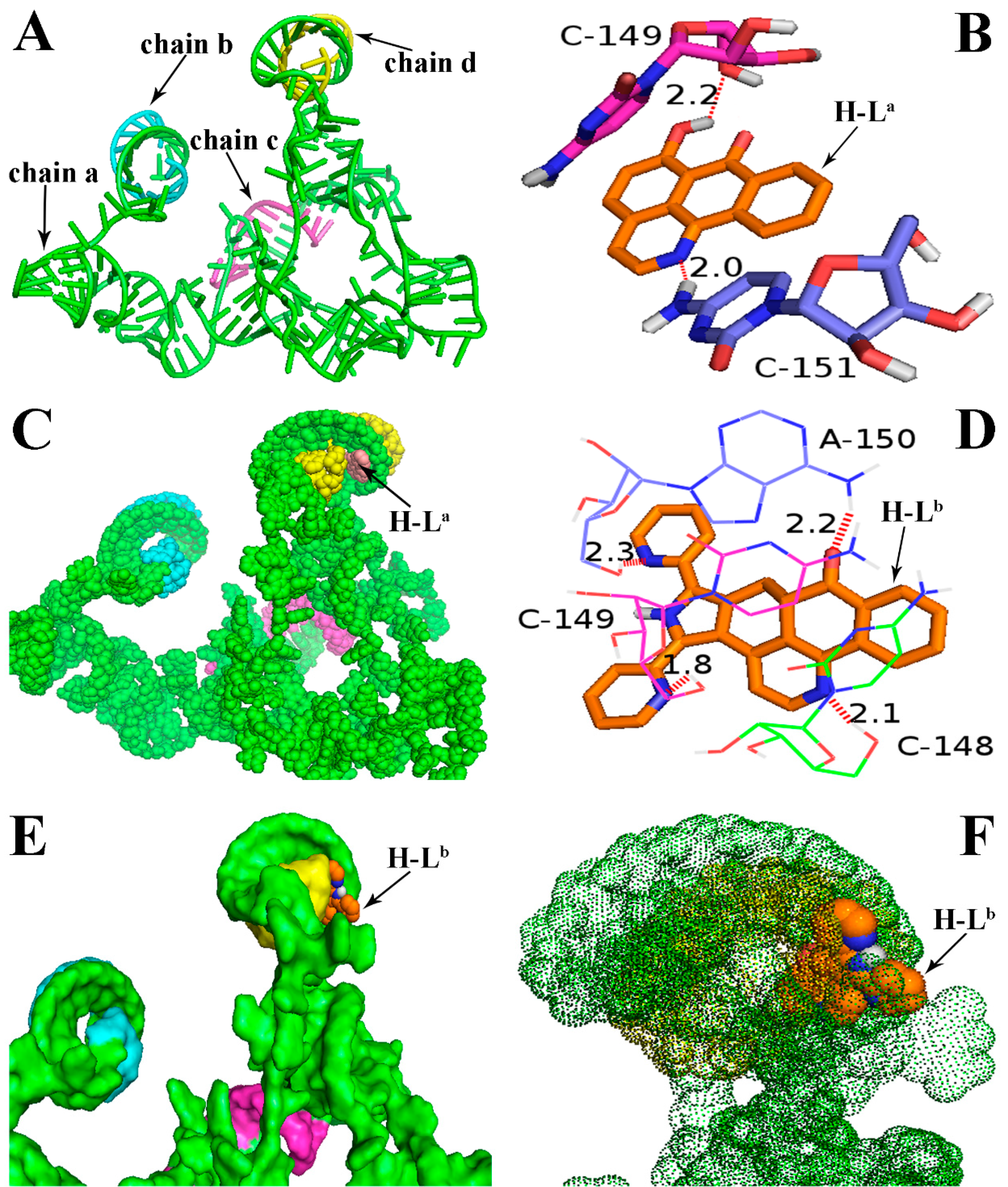
2.1. Molecular Docking
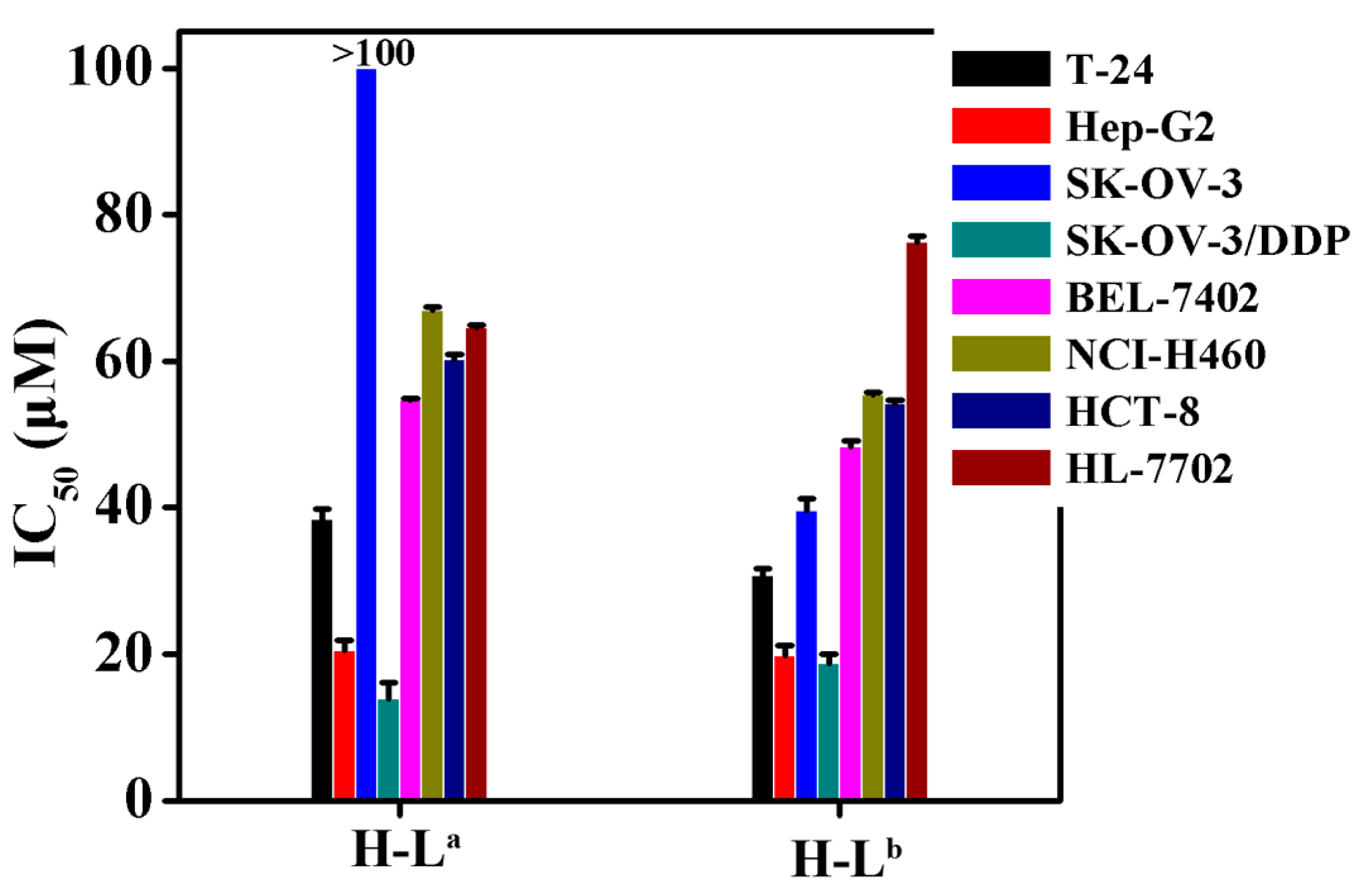
2.2. In Vitro Cytotoxicity
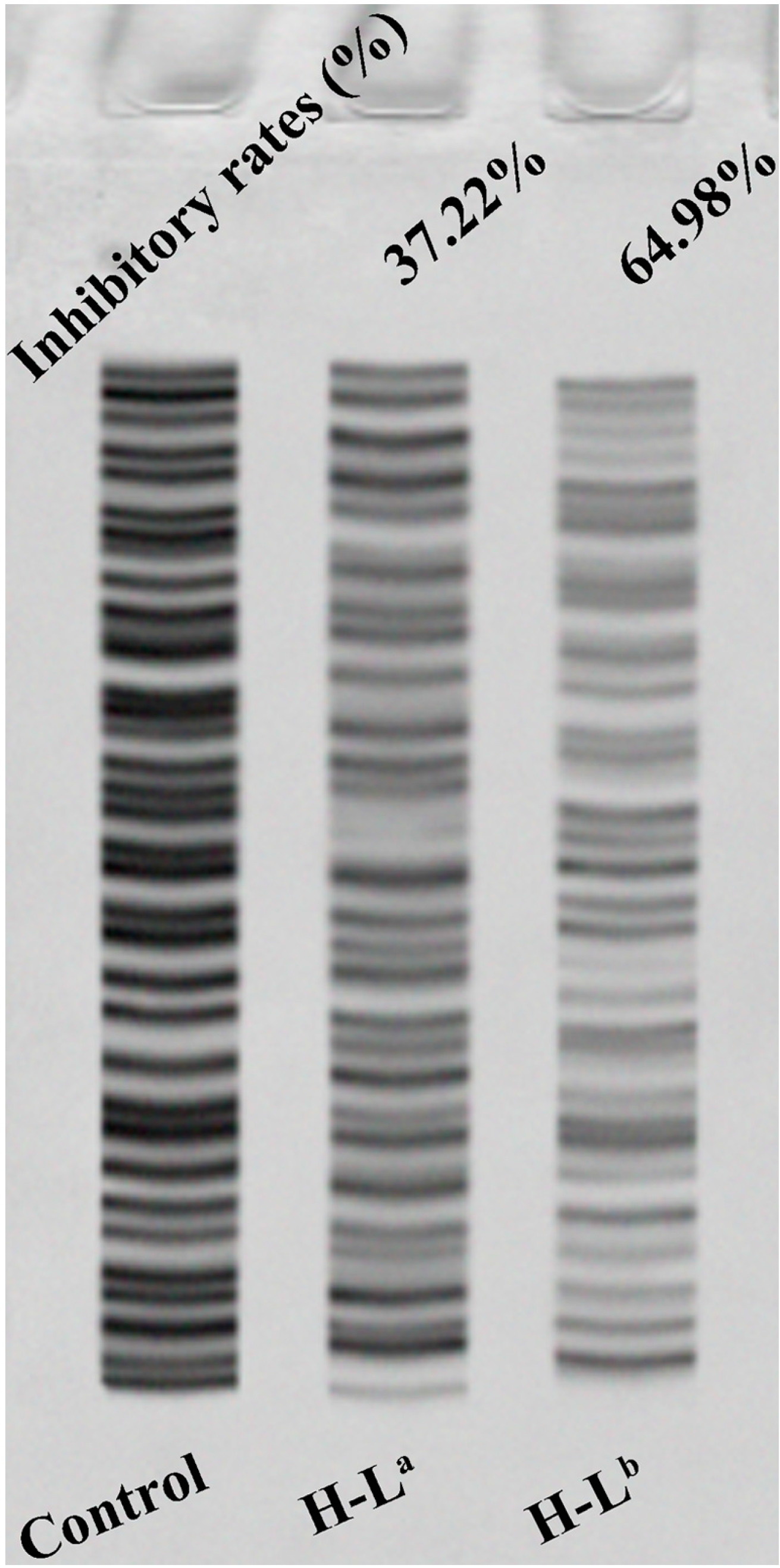
2.3. Telomerase Activity Inhibition Studies by TRAP-Silver Staining Assay
3. Experimental Methods
3.1. Materials and Instrumentation
3.2. Synthesis of Oxoisoaporphine Ligands
3.3. MTT Assay and Telomerase Activity Assay
3.4. Docking Study
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rezler, E.M.; Bearss, D.J.; Hurley, L.H. Telomeres and telomerases as drug targets. Curr. Opin. Pharmacol. 2002, 2, 415–423. [Google Scholar] [CrossRef]
- Balasubramanian, S.H.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y. Chemistry in human telomere biology: Structure, function and targeting of telomere DNA/RNA. Chem. Soc. Rev. 2011, 40, 2719–2740. [Google Scholar] [CrossRef] [PubMed]
- Sekaran, V.; Soares, J.; Jarstfer, M.B. Telomere maintenance as a target for drug discovery. J. Med. Chem. 2014, 57, 521–538. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.L.; Riou, J.F.; Mailliet, P.; Teulade-Fichou, M.P.; Gilson, E. Natural and pharmacological regulation of telomerase. Nucleic Acids Res. 2002, 30, 839–865. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Patel, D.J. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: Distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003, 125, 15021–15027. [Google Scholar] [CrossRef] [PubMed]
- Ying, L.; Green, J.J.; Li, H.; Klenerman, D.; Balasubramanian, S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 2003, 100, 14629–14634. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, G.N.; Lee, M.P.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S.; Parkinson, G.N. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003, 13, 275–283. [Google Scholar] [CrossRef]
- Baran, N.; Pucshansky, L.; Marco, Y.; Benjamin, S.; Manor, H. The SV40 large T-antigen helicase can unwind four stranded DNA structures linked by G-quartets. Nucleic Acids Res. 1997, 25, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Yabuki, A.; Maizels, N. A human nuclease specific for G4 DNA. Proc. Natl. Acad. Sci. USA 2001, 98, 12444–12449. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.C.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Amrane, S.; Kerkour, A.; Bedrat, A.; Vialet, B.; Andreola, M.L.; Mergny, J.L. Topology of a DNA G-qudruplex structure formed in the HIV-1 promoter: A potential target for anti-HIV drug development. J. Am. Chem. Soc. 2014, 136, 5249–5252. [Google Scholar] [CrossRef] [PubMed]
- Paritala, H.; Firestine, S.M. Characterization of insulin ILPR sequences for their ability to adopt a G-quadruplex structure. Nucleosides Nucleotides Nucleic Acids 2010, 29, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.-X.; Huang, Z.-S.; Tan, J.-H. Targeting G-quadruplex nucleic acids with heterocyclic alkaloids and their derivatives. Eur. J. Med. Chem. 2015, 97, 538–551. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-L.; Qin, Q.-P.; Liu, Y.-C.; Chen, Z.-F.; Liang, H. A platinum(II) complex of liriodenine from traditional Chinese medicine (TCM): Cell cycle arrest, cell apoptosis induction and telomerase inhibition activity via G-quadruplex DNA stabilization. J. Inorg. Biochem. 2014, 137, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.-F.; Qin, Q.-P.; Qin, J.-L.; Zhou, J.; Li, Y.-L.; Li, N.; Liu, Y.-C.; Liang, H. Water-soluble ruthenium(II) complexes with chiral 4-(2,3-Dihydroxypropyl)-formamide oxoaporphine (FOA): In vitro and in vivo anticancer activity by stabilization of G-quadruplex DNA, inhibition of telomerase activity, and induction of tumor cell apoptosis. J. Med. Chem. 2015, 58, 4771–4789. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.-F.; Qin, Q.-P.; Qin, J.-L.; Liu, Y.-C.; Huang, K.-B.; Li, Y.-L.; Meng, T.; Zhang, G.-H.; Peng, Y.; Luo, X.-J.; et al. Stabilization of G-quadruplex DNA, inhibition of telomerase activity, and tumor cell apoptosis by organ platinum(II) complexes with oxoisoaporphine. J. Med. Chem. 2015, 58, 2159–2179. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.-L.; Qin, Q.-P.; Wei, Z.-Z.; Yu, C.-C.; Meng, T.; Wu, C.-X.; Liang, Y.-L.; Liang, H.; Chen, Z.-F. Stabilization of c-myc G-quadruplex DNA, inhibition of telomerase activity, disruption of mitochondrial functions and tumor cell apoptosis by platinum(II) complex with 9-amino-oxoisoaporphine. Eur. J. Med. Chem. 2016, 124, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.-P.; Qin, J.-L.; Meng, T.; Lin, W.-H.; Zhang, C.-H.; Wei, Z.-Z.; Chen, J.-N.; Liu, Y.-C.; Liang, H.; Chen, Z.-F. High in vivo antitumor activity of cobalt oxoisoaporphine complexes by targeting G-quadruplex DNA, telomerase and disrupting mitochondrial functions. Eur. J. Med. Chem. 2016, 124, 380–392. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Kim, N.K.; Feigon, J. Architecture of human telomerase RNA. Proc. Natl. Acad. Sci. USA 2011, 108, 20325–20332. [Google Scholar] [CrossRef] [PubMed]
- Jansson, L.I.; Akiyama, B.M.; Ooms, A.; Lu, C.; Rubin, S.M.; Stone, M.D. Structural basis of template-boundary definition in Tetrahymena telomerase. Nat. Struct. Mol. Biol. 2015, 22, 883–888. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Wang, Z.; Koo, B.K.; Patel, A.; Cascio, D.; Collins, K.; Feigon, J. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol. Cell 2012, 47, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Bryan, C.; Rice, C.; Hoffman, H.; Harkisheimer, M.; Sweeney, M.; Skordalakes, E. Structural basis of telomerase inhibition by the highly specific BIBR1532. Structure 2015, 23, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gavory, G.; Symmons, M.F.; Krishnan Ghosh, Y.; Klenerman, D.; Balasubramanian, S. Structural analysis of the catalytic core of human telomerase RNA by FRET and molecular modeling. Biochemistry 2006, 45, 13304–13311. [Google Scholar] [CrossRef] [PubMed]
- Holt, S.E.; Wright, W.E.; Shay, J.W. Regulation of telomerase activity in immortal cell lines. Mol. Cell. Biol. 1996, 16, 2932–2939. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Kumar, R.; Mandal, M.; Sharma, N.; Sharma, H.W.; Dhingra, U.; Sokoloski, J.A.; Hsiao, R.; Narayanan, R. Cell cycle-dependent modulation of telomerase activity in tumor cells. Proc. Natl. Acad. Sci. USA 1996, 93, 6091–6095. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.E.; Arnal, A.A.; Neidle, S.; Vilar, R. Stabilization of G-quadruplex DNA and inhibition of telomerase activity by square-planar nickel(II) complexes. J. Am. Chem. Soc. 2006, 128, 5592–5993. [Google Scholar] [CrossRef] [PubMed]
- Meng, T.; Tang, S.-F.; Qin, Q.-P.; Liang, Y.-L.; Wu, C.-X.; Wang, C.-Y.; Yan, H.-T.; Dong, J.-X.; Liu, Y.-C. Evaluation of the effect of iodine substitution of 8-hydroxyquinoline on its platinum(II) complex: Cytotoxicity, cell apoptosis and telomerase inhibition. Med. Chem. Commun. 2016, 7, 1802–1811. [Google Scholar] [CrossRef]
- Cole, D.K.; Rizkallah, P.J.; Gao, F.; Watson, N.I.; Boulter, J.M.; Bell, J.I.; Sami, M.; Gao, G.F.; Jakobsen, B.K. Crystal structure of HLA-A* 2402 complexed with a telomerase peptide. Eur. J. Immunol. 2006, 36, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-N.; Wang, X.-F.; Li, T.; Wu, D.-W.; Fu, X.-B.; Zhang, G.-J.; Shen, X.-C.; Wang, H.-S. Design, synthesis, and biological evaluation of novel quinazolinyl-diaryl urea derivatives as potential anticancer agent. Eur. J. Med. Chem. 2016, 107, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds H-La and H-Lb are available from the authors.
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Z.-Z.; Qin, Q.-P.; Chen, J.-N.; Chen, Z.-F. Oxoisoaporphine as Potent Telomerase Inhibitor. Molecules 2016, 21, 1534. https://doi.org/10.3390/molecules21111534
Wei Z-Z, Qin Q-P, Chen J-N, Chen Z-F. Oxoisoaporphine as Potent Telomerase Inhibitor. Molecules. 2016; 21(11):1534. https://doi.org/10.3390/molecules21111534
Chicago/Turabian StyleWei, Zu-Zhuang, Qi-Pin Qin, Jia-Nian Chen, and Zhen-Feng Chen. 2016. "Oxoisoaporphine as Potent Telomerase Inhibitor" Molecules 21, no. 11: 1534. https://doi.org/10.3390/molecules21111534