The Plant Decapeptide OSIP108 Can Alleviate Mitochondrial Dysfunction Induced by Cisplatin in Human Cells
Abstract
:1. Introduction
2. Results and Discussion
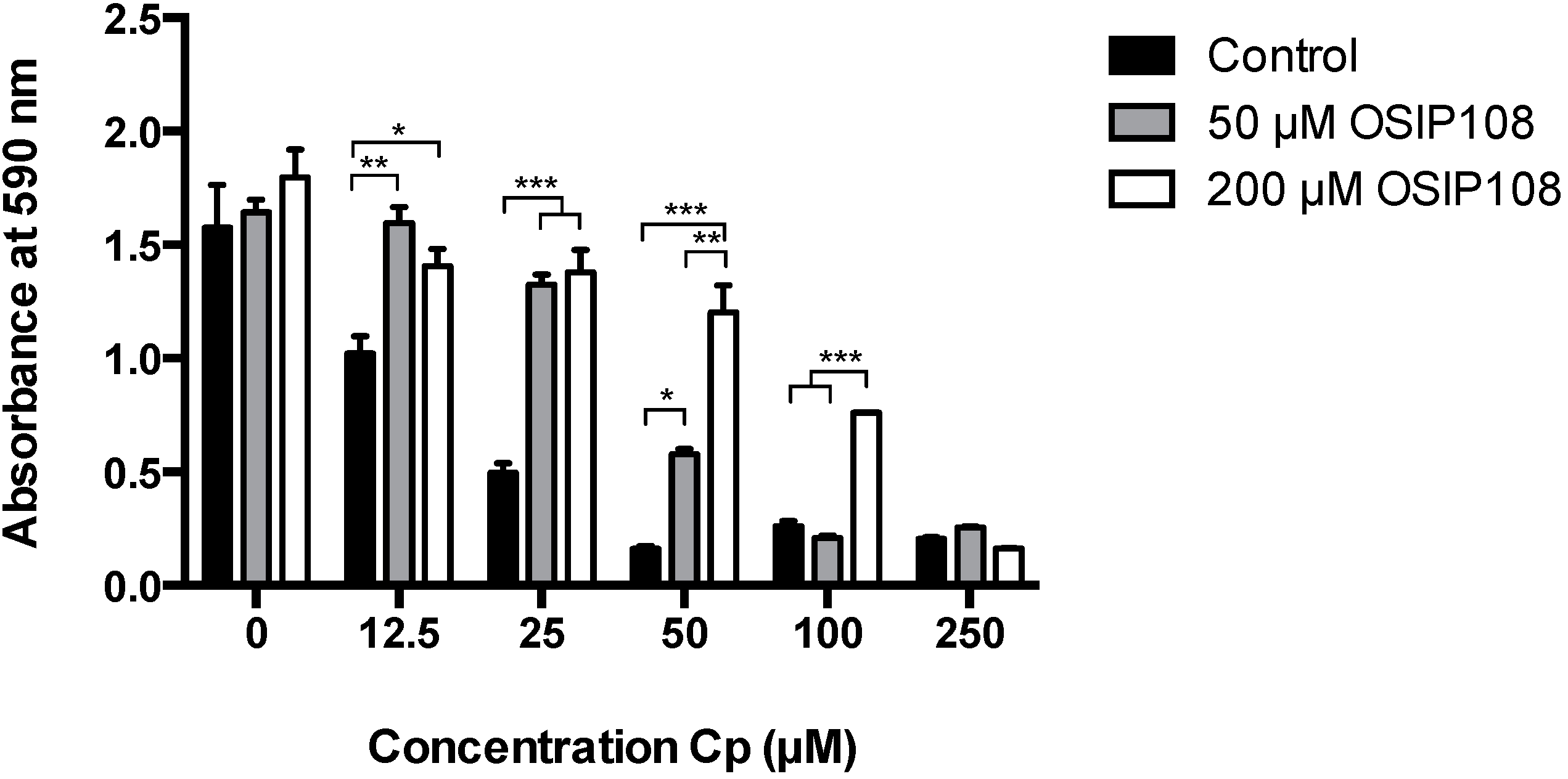
2.1. OSIP108 Increases Tolerance of the Human Hepatocyte HepG2 Model Cell Line to Cp
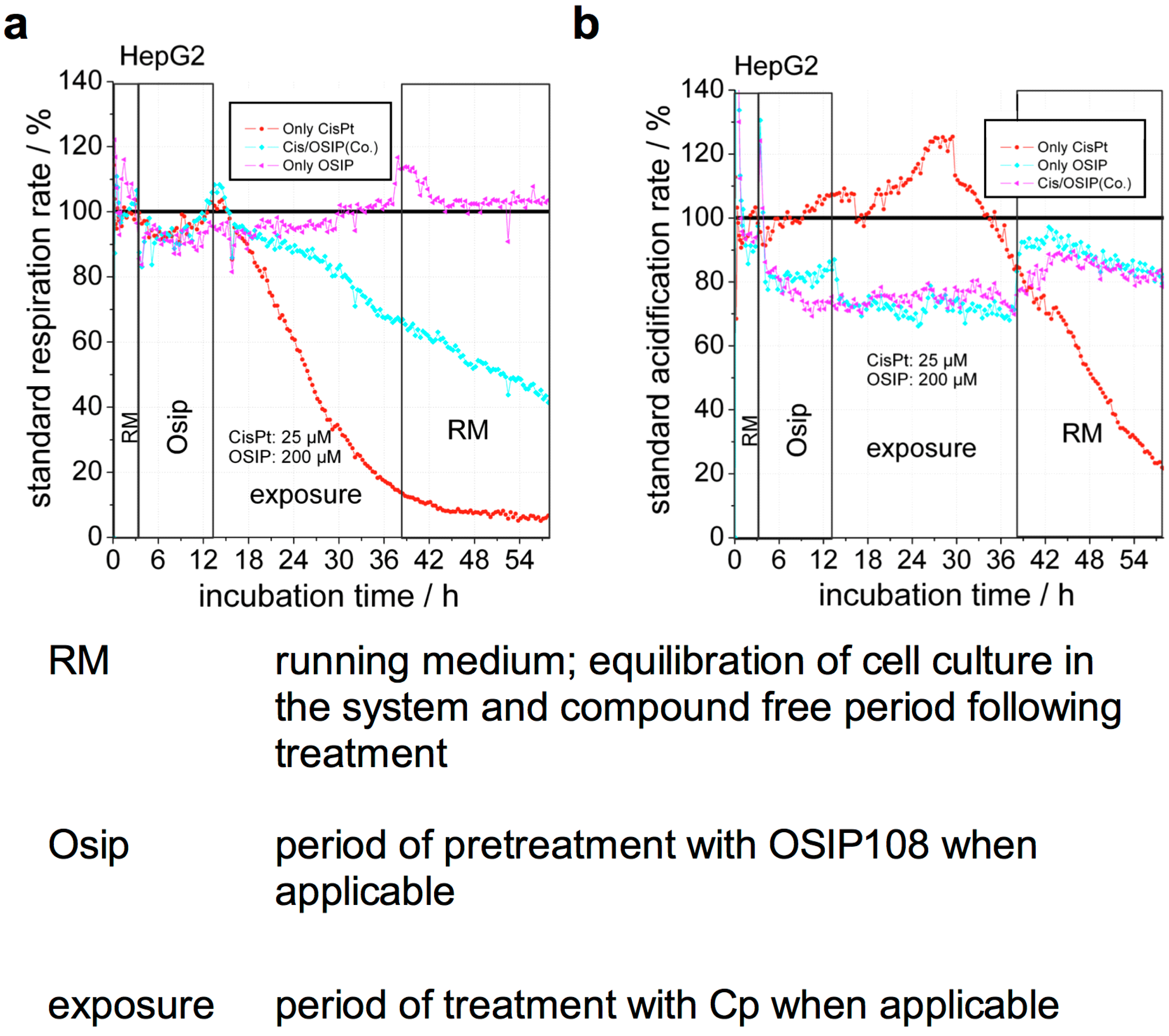
2.2. OSIP108 Reduces Cp-Induced Inhibition of Respiration
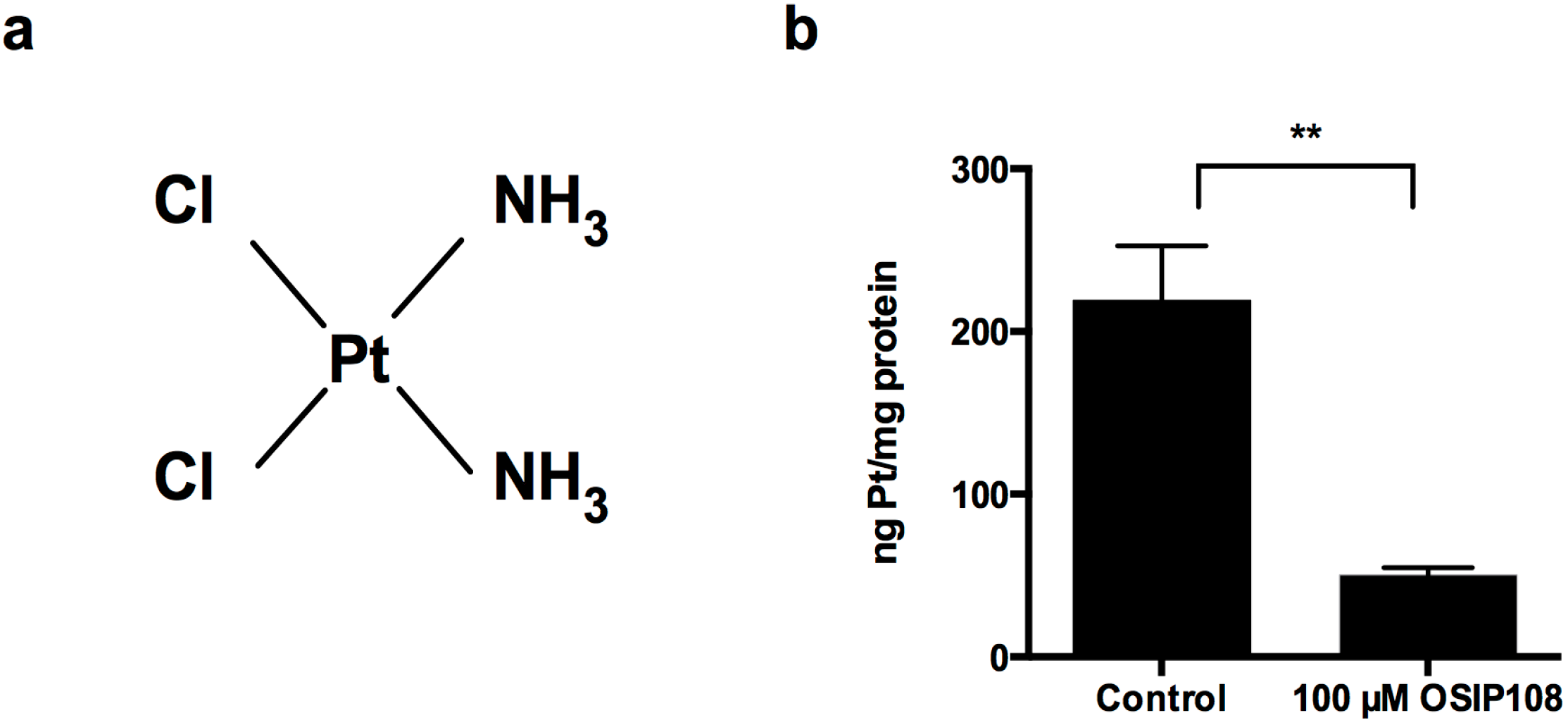
2.3. OSIP108 Affects Cp Uptake in HepG2 Cells
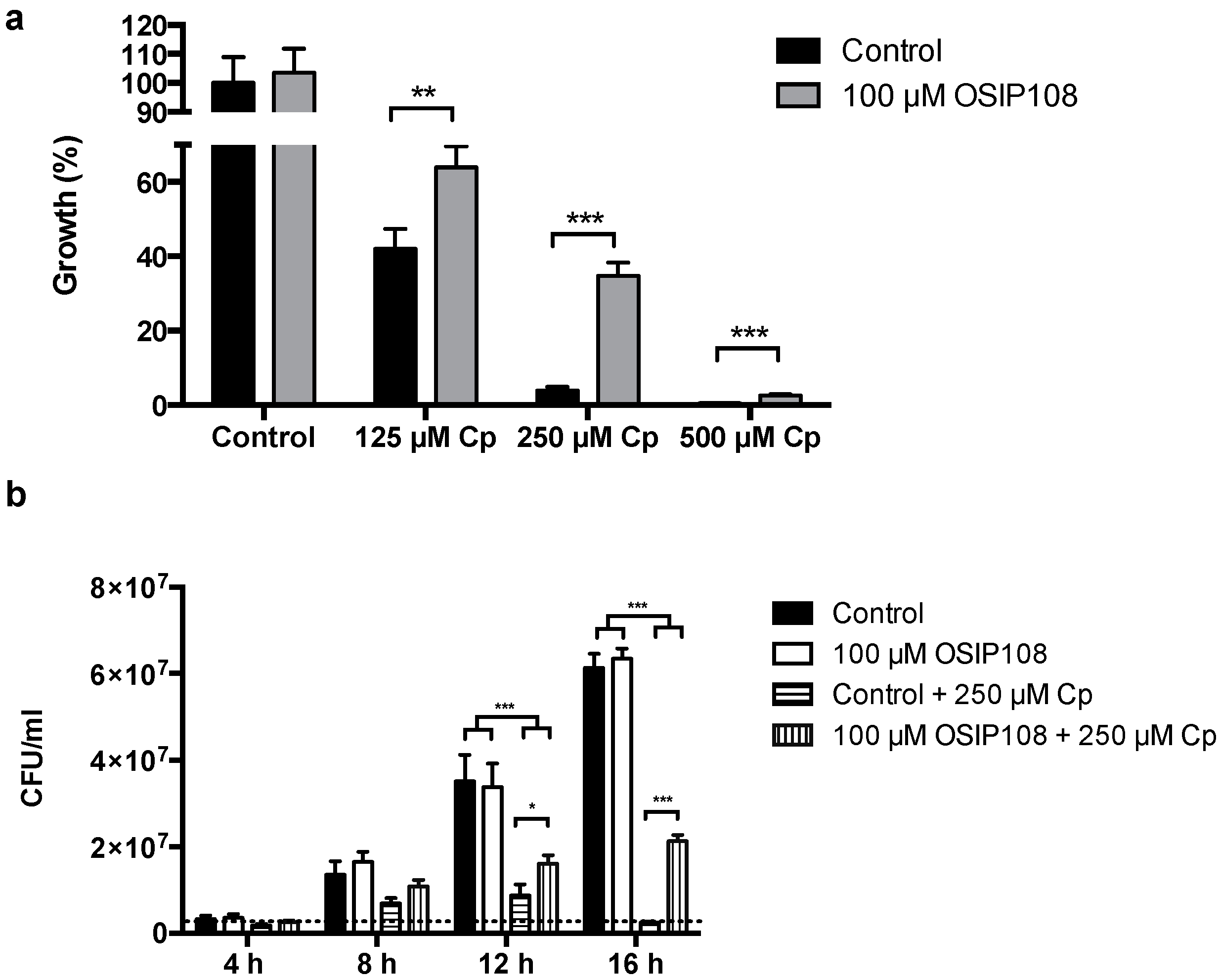
2.4. Activity of OSIP108 Analogs on Cp-Stressed Yeast
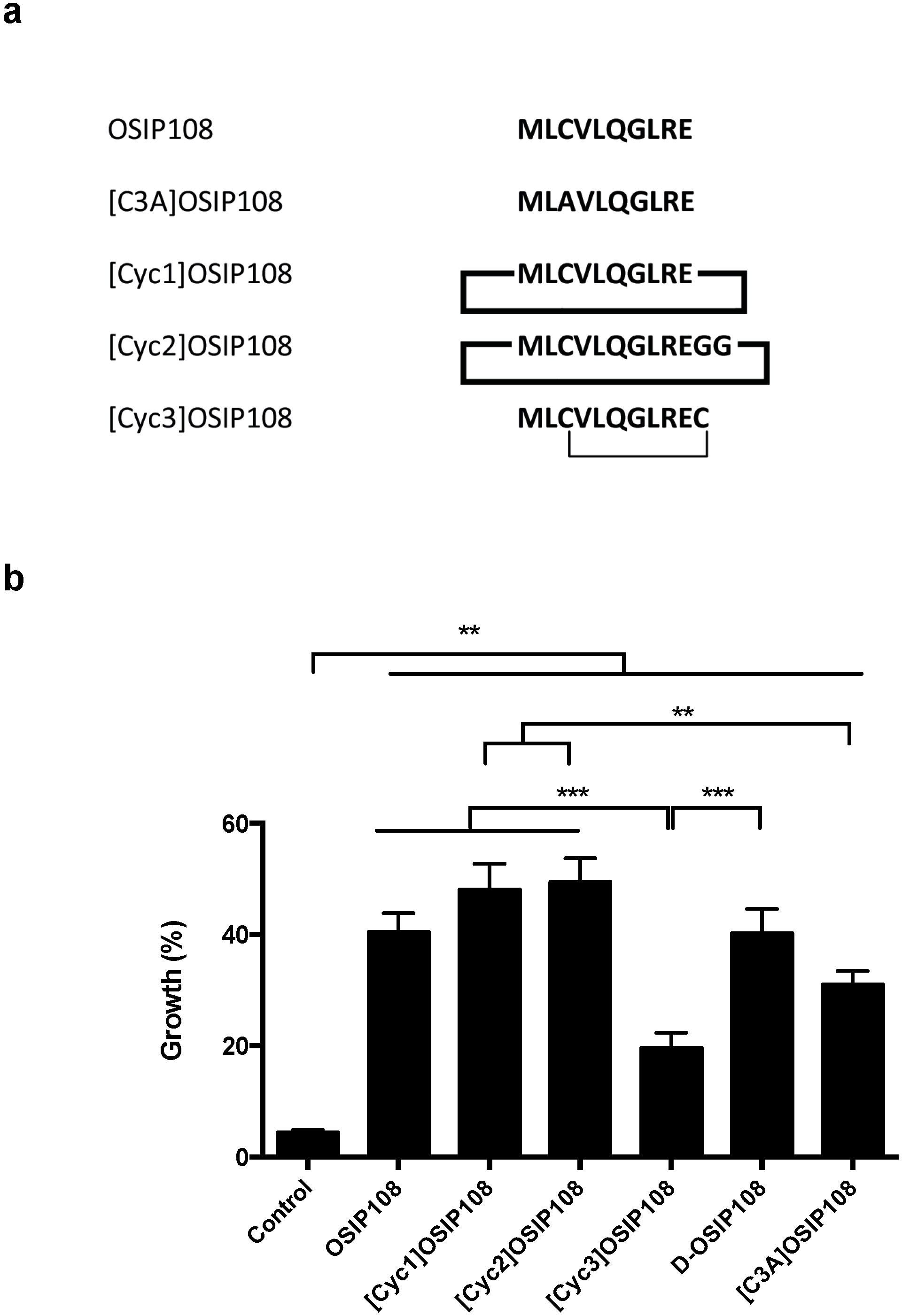
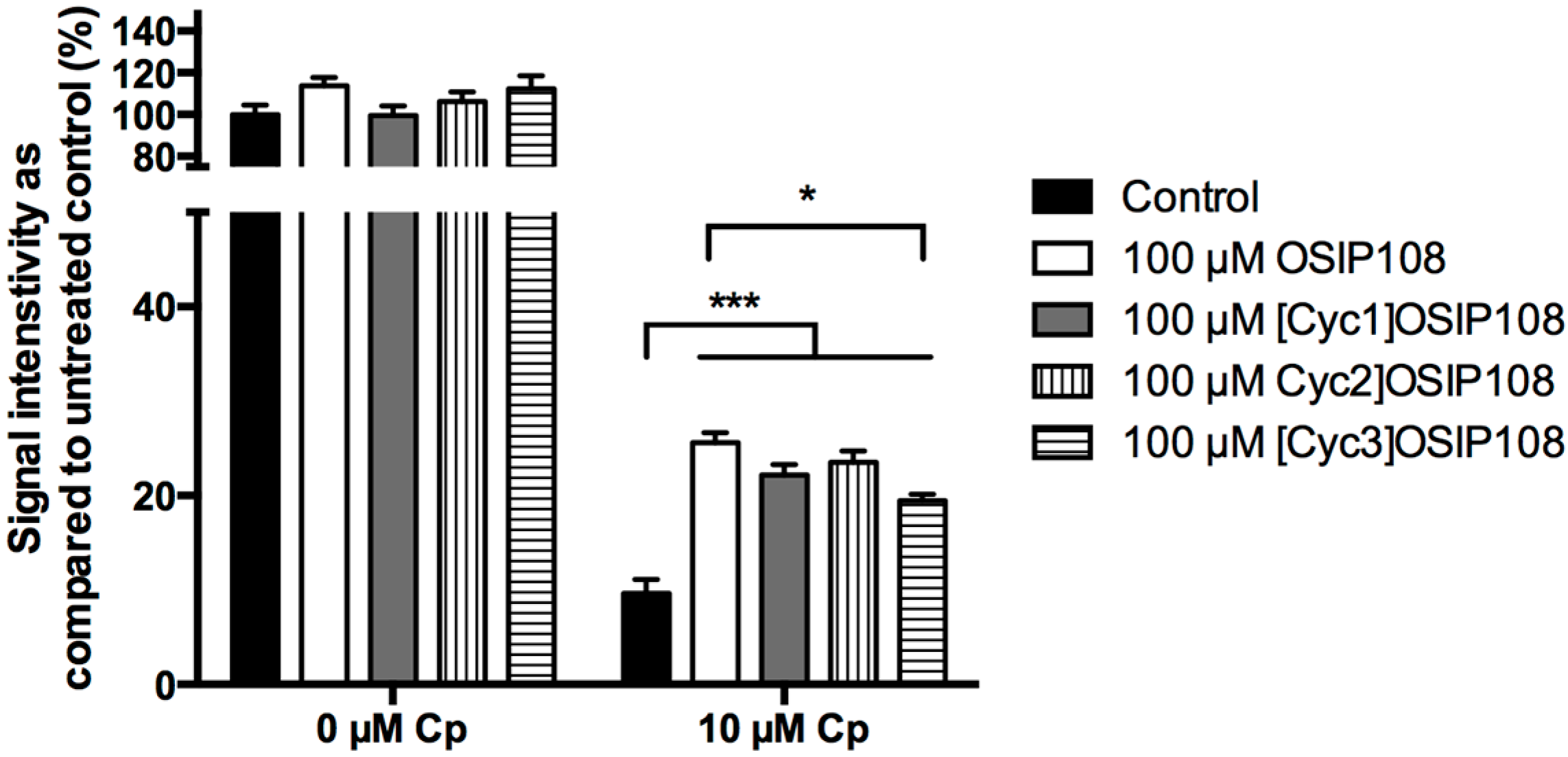
3. Experimental Section
3.1. Materials and Microorganisms
3.2. Cp Toxicity in HepG2 Cells
3.3. BrdU Assay in Cp-Treated HepG2 Cells
3.4. Real-Time Monitoring of HepG2 Metabolism
3.5. Cellular Cp Uptake in HepG2 Cells
3.6. Yeast Growth Assays
3.7. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Einhorn, E.H. Testicular cancer: An oncological success story. Clin. Cancer Res. 1997, 3, 2630–2632. [Google Scholar]
- Hanigan, M.H.; Devarajan, P. Cisplatin nephrotoxicity: Molecular mechanisms. Cancer Ther. 2003, 1, 47–61. [Google Scholar]
- Florea, A.M.; Büsselberg, D. Cisplatin as an Anti-Tumor Drug: Cellular Mechanisms of Activity, Drug Resistance and Induced Side Effects. Cancers 2011, 3, 1351–1371. [Google Scholar] [CrossRef]
- Koberle, B.; Tomicic, M.T.; Usanova, S.; Kaina, B. Cisplatin resistance: Preclinical findings and clinical implications. Biochim. Biophys. Acta 2010, 1806, 172–182. [Google Scholar]
- Jordan, P.; Carmo-Fonseca, M. Molecular mechanisms involved in cisplatin cytotoxicity. Cell. Mol. Life Sci. 2000, 57, 1229–1235. [Google Scholar] [CrossRef]
- Rebillard, A.; Rioux-Leclercq, N.; Muller, C.; Bellaud, P.; Jouan, F.; Meurette, O.; Jouan, E.; Vernhet, L.; Le Quement, C.; Carpinteiro, A.; et al. Acid sphingomyelinase deficiency protects from cisplatin-induced gastrointestinal damage. Oncogene 2008, 27, 6590–6595. [Google Scholar] [CrossRef]
- Thevissen, K.; Francois, I.E.; Winderickx, J.; Pannecouque, C.; Cammue, B.P. Ceramide involvement in apoptosis and apoptotic diseases. Mini Rev. Med. Chem. 2006, 6, 699–709. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, H.; Liu, F.; Dong, Z. Mitochondrial dysregulation and protection in cisplatin nephrotoxicity. Arch. Toxicol. 2014, 88, 1249–1256. [Google Scholar] [CrossRef]
- Kruidering, M.; van de Water, B.; de Heer, E.; Mulder, G.J.; Nagelkerke, J.F. Cisplatin-induced nephrotoxicity in porcine proximal tubular cells: Mitochondrial dysfunction by inhibition of complexes I to IV of the respiratory chain. J. Pharmacol. Exp. Ther. 1997, 280, 638–649. [Google Scholar]
- Alborzinia, H.; Can, S.; Holenya, P.; Scholl, C.; Lederer, E.; Kitanovic, I.; Wolfl, S. Real-time monitoring of cisplatin-induced cell death. PLoS One 2011, 6, e19714. [Google Scholar] [CrossRef]
- Mukhopadhyay, P.; Horvath, B.; Zsengeller, Z.; Zielonka, J.; Tanchian, G.; Holovac, E.; Kechrid, M.; Patel, V.; Stillman, I.E.; Parikh, S.M.; et al. Mitochondrial-targeted antioxidants represent a promising approach for prevention of cisplatin-induced nephropathy. Free Radic. Biol. Med. 2012, 52, 497–506. [Google Scholar] [CrossRef]
- Marullo, R.; Werner, E.; Degtyareva, N.; Moore, B.; Altavilla, G.; Ramalingam, S.S.; Doetsch, P.W. Cisplatin Induces a Mitochondrial-ROS Response That Contributes to Cytotoxicity Depending on Mitochondrial Redox Status and Bioenergetic Functions. PLoS One 2013, 8, e81162. [Google Scholar] [CrossRef]
- De Coninck, B.; Carron, D.; Tavormina, P.; Willem, L.; Craik, D.J.; Vos, C.; Thevissen, K.; Mathys, J.; Cammue, B.P. Mining the genome of Arabidopsis thaliana as a basis for the identification of novel bioactive peptides involved in oxidative stress tolerance. J. Exp. Bot. 2013, 64, 5297–5307. [Google Scholar] [CrossRef]
- Farrington, J.A.; Ebert, M.; Land, E.J.; Fletcher, K. Bipyridylium quaternary salts and related compounds. V. Pulse radiolysis studies of the reaction of paraquat radical with oxygen. Implications for the mode of action of bipyridyl herbicides. Biochim. Biophys. Acta 1973, 314, 372–381. [Google Scholar]
- Spincemaille, P.; Chandhok, G.; Newcomb, B.; Verbeek, J.; Vriens, K.; Zibert, A.; Schmidt, H.; Hannun, Y.A.; van Pelt, J.; Cassiman, D.; et al. The plant decapeptide OSIP108 prevents copper-induced apoptosis in yeast and human cells. Biochim. Biophys. Acta 2014, 1843. [Google Scholar]
- Lang, P.A.; Schenck, M.; Nicolay, J.P.; Becker, J.U.; Kempe, D.S.; Lupescu, A.; Koka, S.; Eisele, K.; Klarl, B.A.; Rubben, H.; et al. Liver cell death and anemia in Wilson disease involve acid sphingomyelinase and ceramide. Nat. Med. 2007, 13, 164–170. [Google Scholar] [CrossRef]
- Seth, R.; Yang, S.; Choi, S.; Sabean, M.; Roberts, E.A. In vitro assessment of copper-induced toxicity in the human hepatoma line, Hep G2. Toxicol. In Vitro 2004, 18, 501–509. [Google Scholar] [CrossRef]
- Sokol, R.J.; Twedt, D.; McKim, J.M., Jr.; Devereaux, M.W.; Karrer, F.M.; Kam, I.; von Steigman, G.; Narkewicz, M.R.; Bacon, B.R.; Britton, R.S.; et al. Oxidant injury to hepatic mitochondria in patients with Wilson's disease and Bedlington terriers with copper toxicosis. Gastroenterology 1994, 107, 1788–1798. [Google Scholar]
- Gaetke, L.M.; Chow, C.K. Copper toxicity, oxidative stress, and antioxidant nutrients. Toxicology 2003, 189, 147–163. [Google Scholar] [CrossRef]
- Zischka, H.; Lichtmannegger, J. Pathological mitochondrial copper overload in livers of Wilson’s disease patients and related animal models. Ann. N. Y. Acad. Sci. 2014, 1315, 6–15. [Google Scholar]
- Ferenci, P. Regional distribution of mutations of the ATP7B gene in patients with Wilson disease: Impact on genetic testing. Hum. Genet. 2006, 120, 151–159. [Google Scholar] [CrossRef]
- Loudianos, G.; Gitlin, J.D. Wilson’s disease. Semin. Liver Dis. 2000, 20, 353–364. [Google Scholar] [CrossRef]
- Huster, D. Wilson disease. Best Pract. Res. Clin. Gastroenterol. 2010, 24, 531–539. [Google Scholar] [CrossRef]
- Ala, A.; Walker, A.P.; Ashkan, K.; Dooley, J.S.; Schilsky, M.L. Wilson’s disease. Lancet 2007, 369, 397–408. [Google Scholar] [CrossRef]
- Spincemaille, P.; Pham, D.H.; Chandhok, G.; Verbeek, J.; Zibert, A.; Libbrecht, L.; Schmidt, H.; Esguerra, C.V.; de Witte, P.A.; Cammue, B.P.; et al. The plant decapeptide OSIP108 prevents copper-induced toxicity in various models for Wilson disease. Toxicol. Appl. Pharmacol. 2014, in press. [Google Scholar]
- Roberts, E.A.; Robinson, B.H.; Yang, S. Mitochondrial structure and function in the untreated Jackson toxic milk (tx-j) mouse, a model for Wilson disease. Mol. Genet. Metab. 2008, 93, 54–65. [Google Scholar] [CrossRef]
- Thedinga, E.; Kob, A.; Holst, H.; Keuer, A.; Drechsler, S.; Niendorf, R.; Baumann, W.; Freund, I.; Lehmann, M.; Ehret, R. Online monitoring of cell metabolism for studying pharmacodynamic effects. Toxicol. Appl. Pharmacol. 2007, 220, 33–44. [Google Scholar] [CrossRef]
- Bayley, J.P.; Devilee, P. The Warburg effect in 2012. Curr. Opin. Oncol. 2012, 24, 62–67. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of Tumors in the Body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef]
- Seeland, S.; Torok, M.; Kettiger, H.; Treiber, A.; Hafner, M.; Huwyler, J. A cell-based, multiparametric sensor approach characterises drug-induced cytotoxicity in human liver HepG2 cells. Toxicol. In Vitro 2013, 27, 1109–1120. [Google Scholar] [CrossRef]
- Felser, A.; Blum, K.; Lindinger, P.W.; Bouitbir, J.; Krahenbuhl, S. Mechanisms of hepatocellular toxicity associated with dronedarone—A comparison to amiodarone. Toxicol. Sci. 2013, 131, 480–490. [Google Scholar] [CrossRef]
- Tolosa, L.; Pinto, S.; Donato, M.T.; Lahoz, A.; Castell, J.V.; O’Connor, J.E.; Gomez-Lechon, M.J. Development of a multiparametric cell-based protocol to screen and classify the hepatotoxicity potential of drugs. Toxicol. Sci. 2012, 127, 187–198. [Google Scholar] [CrossRef]
- Jennen, D.; Ruiz-Aracama, A.; Magkoufopoulou, C.; Peijnenburg, A.; Lommen, A.; van Delft, J.; Kleinjans, J. Integrating transcriptomics and metabonomics to unravel modes-of-action of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in HepG2 cells. BMC Syst. Biol. 2011, 5, 139. [Google Scholar] [CrossRef]
- Raza, H.; John, A. Implications of altered glutathione metabolism in aspirin-induced oxidative stress and mitochondrial dysfunction in HepG2 cells. PLoS One 2012, 7, e36325. [Google Scholar] [CrossRef]
- Kitagaki, H.; Cowart, L.A.; Matmati, N.; Montefusco, D.; Gandy, J.; de Avalos, S.V.; Novgorodov, S.A.; Zheng, J.; Obeid, L.M.; Hannun, Y.A. ISC1-dependent metabolic adaptation reveals an indispensable role for mitochondria in induction of nuclear genes during the diauxic shift in Saccharomyces cerevisiae. J. Biol. Chem. 2009, 284, 10818–10830. [Google Scholar] [CrossRef]
- Bartke, N.; Hannun, Y.A. Bioactive sphingolipids: Metabolism and function. J. Lipid Res. 2009, 50, S91–S96. [Google Scholar]
- Montefusco, D.J.; Chen, L.; Matmati, N.; Lu, S.; Newcomb, B.; Cooper, G.F.; Hannun, Y.A.; Lu, X. Distinct signaling roles of ceramide species in yeast revealed through systematic perturbation and systems biology analyses. Sci. Signal. 2013, 6, rs14. [Google Scholar]
- Kogot-Levin, A.; Saada, A. Ceramide and the mitochondrial respiratory chain. Biochimie 2014, 100, 88–94. [Google Scholar] [CrossRef]
- Spincemaille, P.; Matmati, N.; Hannun, Y.A.; Cammue, B.P.A.; Thevissen, K. Sphingolipids and Mitochondrial Function in Budding Yeast. Biochim. Biophys. Acta 2014, in press. [Google Scholar]
- Spincemaille, P.; Cammue, B.P.A.; Thevissen, K. Sphingolipids and Mitochondrial Function, Lessons Learned from Yeast. Microb. Cell 2014, 1, 210–224. [Google Scholar]
- Wensing, K.U.; Ciarimboli, G. Saving ears and kidneys from cisplatin. Anticancer Res. 2013, 33, 4183–4188. [Google Scholar]
- Frohlich, K.U.; Fussi, H.; Ruckenstuhl, C. Yeast apoptosis—From genes to pathways. Semin. Cancer Biol. 2007, 17, 112–121. [Google Scholar] [CrossRef]
- Rea, S.L.; Graham, B.H.; Nakamaru-Ogiso, E.; Kar, A.; Falk, M.J. Bacteria, yeast, worms, and flies: Exploiting simple model organisms to investigate human mitochondrial diseases. Dev. Disabil. Res. Rev. 2010, 16, 200–218. [Google Scholar] [CrossRef]
- Craik, D.J.; Daly, N.L.; Mulvenna, J.; Plan, M.R.; Trabi, M. Discovery, structure and biological activities of the cyclotides. Curr. Protein Pept. Sci. 2004, 5, 297–315. [Google Scholar] [CrossRef]
- Gould, A.; Ji, Y.; Aboye, T.L.; Camarero, J.A. Cyclotides, a novel ultrastable polypeptide scaffold for drug discovery. Curr. Pharm. Des. 2011, 17, 4294–4307. [Google Scholar] [CrossRef]
- Sando, L.; Henriques, S.T.; Foley, F.; Simonsen, S.M.; Daly, N.L.; Hall, K.N.; Gustafson, K.R.; Aguilar, M.I.; Craik, D.J. A synthetic mirror image of kalata B1 reveals that cyclotide activity is independent of a protein receptor. ChemBioChem 2011, 12, 2456–2462. [Google Scholar] [CrossRef]
- Hamamoto, K.; Kida, Y.; Zhang, Y.; Shimizu, T.; Kuwano, K. Antimicrobial activity and stability to proteolysis of small linear cationic peptides with d-amino acid substitutions. Microbiol. Immunol. 2002, 46, 741–749. [Google Scholar] [CrossRef]
- Fischer, P.M. The design, synthesis and application of stereochemical and directional peptide isomers: A critical review. Curr. Protein Pept. Sci. 2003, 4, 339–356. [Google Scholar] [CrossRef]
- Delattin, N.; de Brucker, K.; Craik, D.J.; Cheneval, O.; Frohlich, M.; Veber, M.; Girandon, L.; Davis, T.R.; Weeks, A.E.; Kumamoto, C.A.; et al. Plant-derived decapeptide OSIP108 interferes with Candida albicans biofilm formation without affecting cell viability. Antimicrob. Agents Chemother. 2014, 58, 2647–2656. [Google Scholar] [CrossRef]
- Custodio, J.B.; Cardoso, C.M.; Santos, M.S.; Almeida, L.M.; Vicente, J.A.; Fernandes, M.A. Cisplatin impairs rat liver mitochondrial functions by inducing changes on membrane ion permeability: Prevention by thiol group protecting agents. Toxicology 2009, 259, 18–24. [Google Scholar] [CrossRef]
- Cunha, D.; Cunha, R.; Corte-Real, M.; Chaves, S.R. Cisplatin-induced cell death in Saccharomyces cerevisiae is programmed and rescued by proteasome inhibition. DNA Repair 2013, 12, 444–449. [Google Scholar] [CrossRef]
- Sample Availability: Not available.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Spincemaille, P.; Alborzinia, H.; Dekervel, J.; Windmolders, P.; Van Pelt, J.; Cassiman, D.; Cheneval, O.; Craik, D.J.; Schur, J.; Ott, I.; et al. The Plant Decapeptide OSIP108 Can Alleviate Mitochondrial Dysfunction Induced by Cisplatin in Human Cells. Molecules 2014, 19, 15088-15102. https://doi.org/10.3390/molecules190915088
Spincemaille P, Alborzinia H, Dekervel J, Windmolders P, Van Pelt J, Cassiman D, Cheneval O, Craik DJ, Schur J, Ott I, et al. The Plant Decapeptide OSIP108 Can Alleviate Mitochondrial Dysfunction Induced by Cisplatin in Human Cells. Molecules. 2014; 19(9):15088-15102. https://doi.org/10.3390/molecules190915088
Chicago/Turabian StyleSpincemaille, Pieter, Hamed Alborzinia, Jeroen Dekervel, Petra Windmolders, Jos Van Pelt, David Cassiman, Olivier Cheneval, David J. Craik, Julia Schur, Ingo Ott, and et al. 2014. "The Plant Decapeptide OSIP108 Can Alleviate Mitochondrial Dysfunction Induced by Cisplatin in Human Cells" Molecules 19, no. 9: 15088-15102. https://doi.org/10.3390/molecules190915088
APA StyleSpincemaille, P., Alborzinia, H., Dekervel, J., Windmolders, P., Van Pelt, J., Cassiman, D., Cheneval, O., Craik, D. J., Schur, J., Ott, I., Wölfl, S., Cammue, B. P. A., & Thevissen, K. (2014). The Plant Decapeptide OSIP108 Can Alleviate Mitochondrial Dysfunction Induced by Cisplatin in Human Cells. Molecules, 19(9), 15088-15102. https://doi.org/10.3390/molecules190915088




