Beads-Based Electrochemical Assay for the Detection of Influenza Hemagglutinin Labeled with CdTe Quantum Dots
Abstract
:1. Introduction
2. Results and Discussion
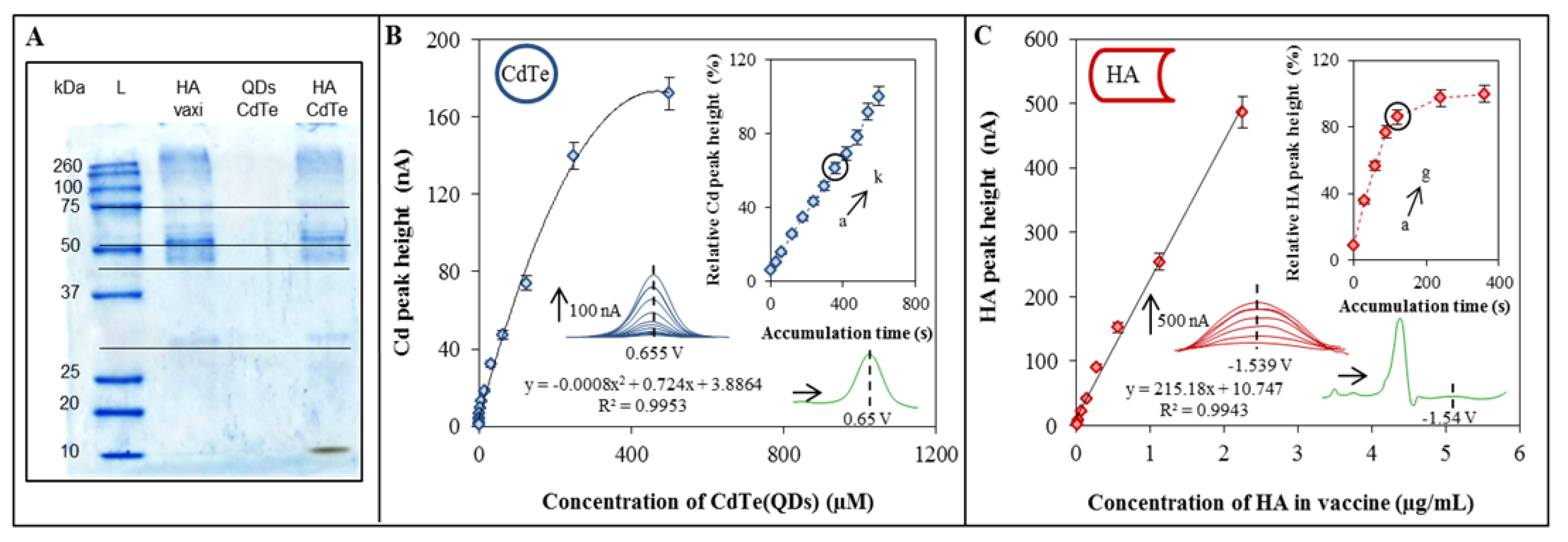
2.1. Characterization of vaxi HA, CdTe and HA-CdTe Complex by Gel Electrophoresis
2.2. Characterization of CdTe by Electrochemical Analysis
2.3. Characterization of Influenza Vaccine by the Brdicka Reaction
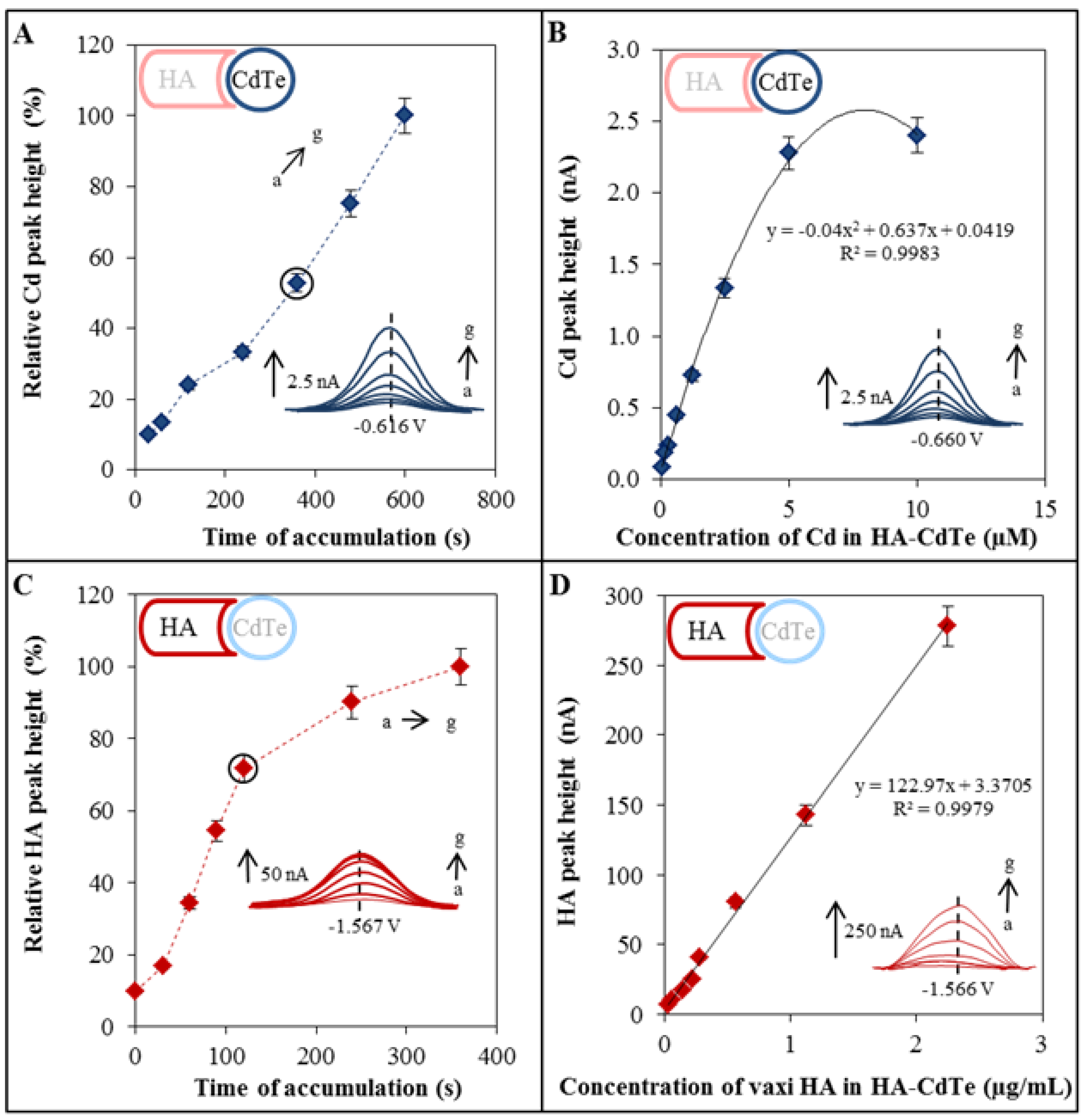
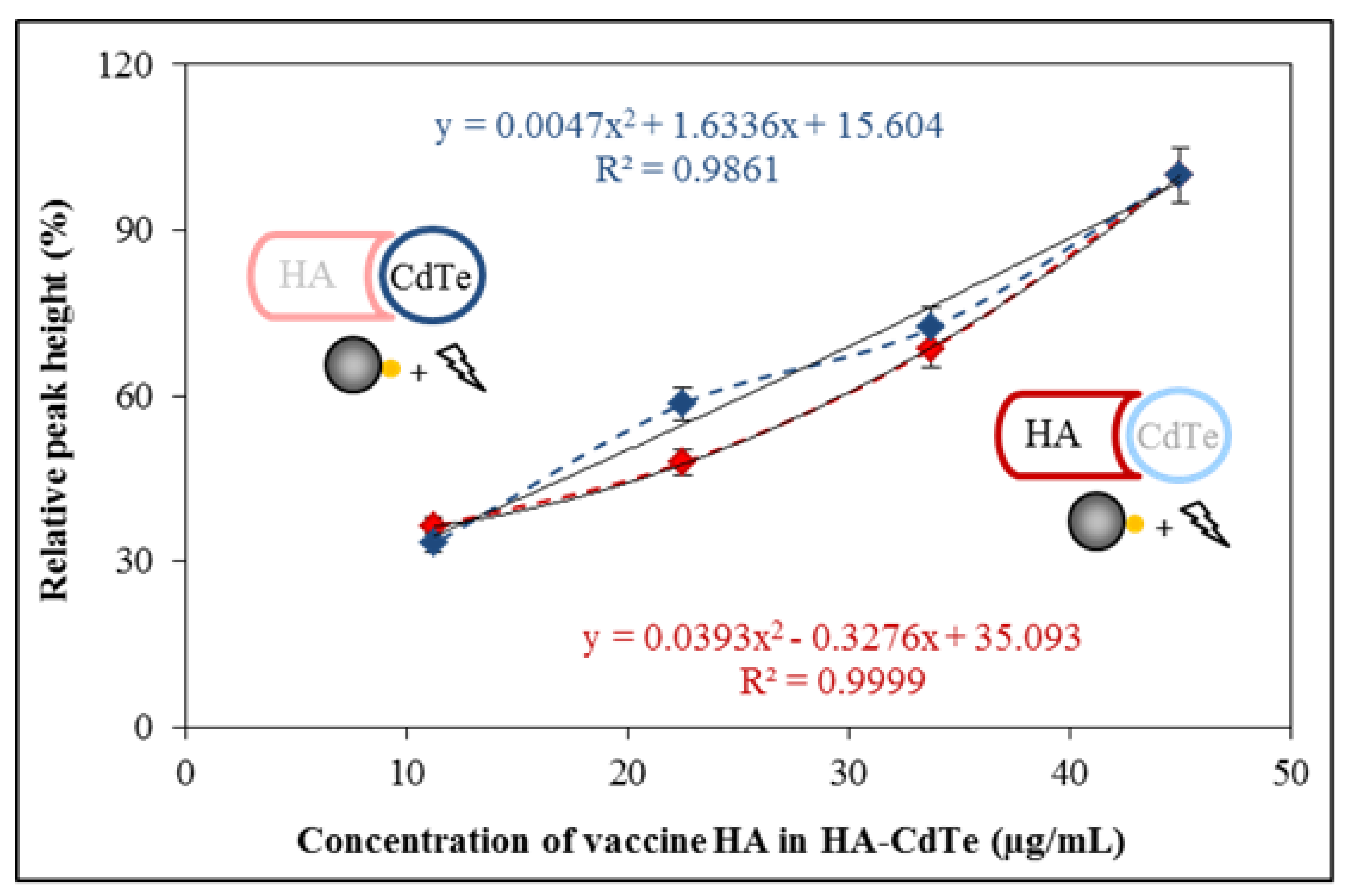
2.4. Electrochemical Characterization of HA-CdTe Complex
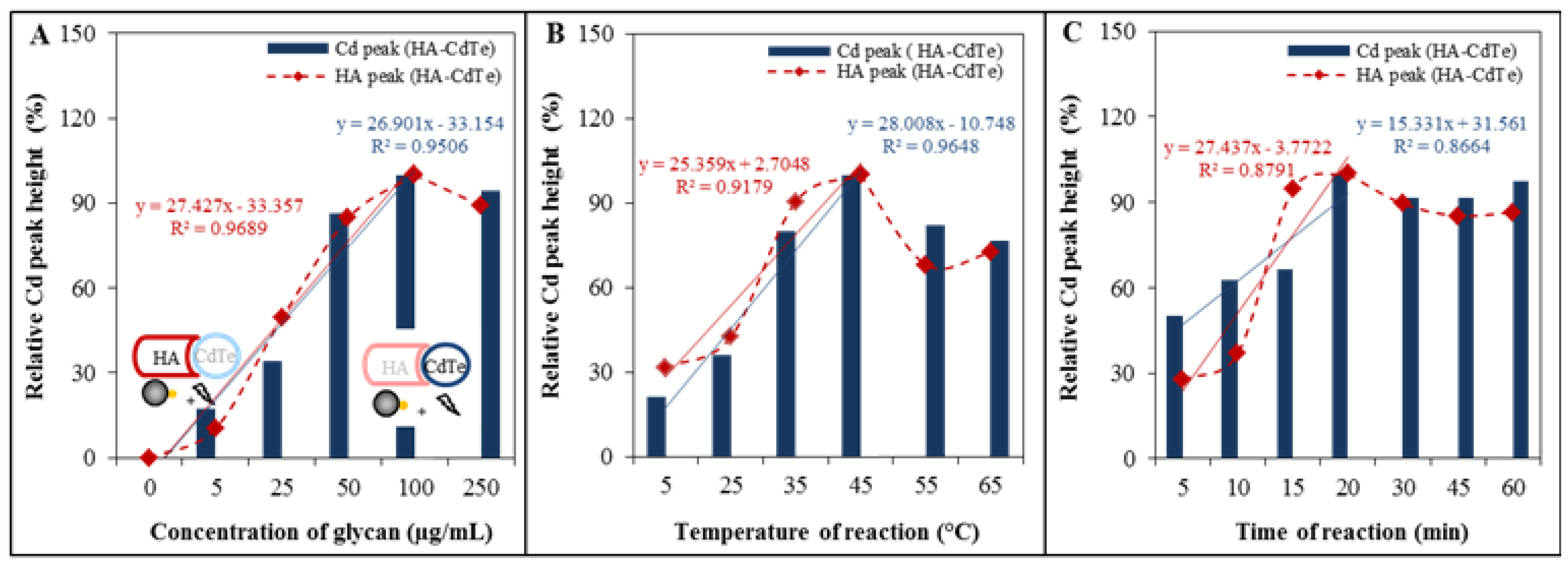
2.5. Optimization of the Automated Isolation Procedure
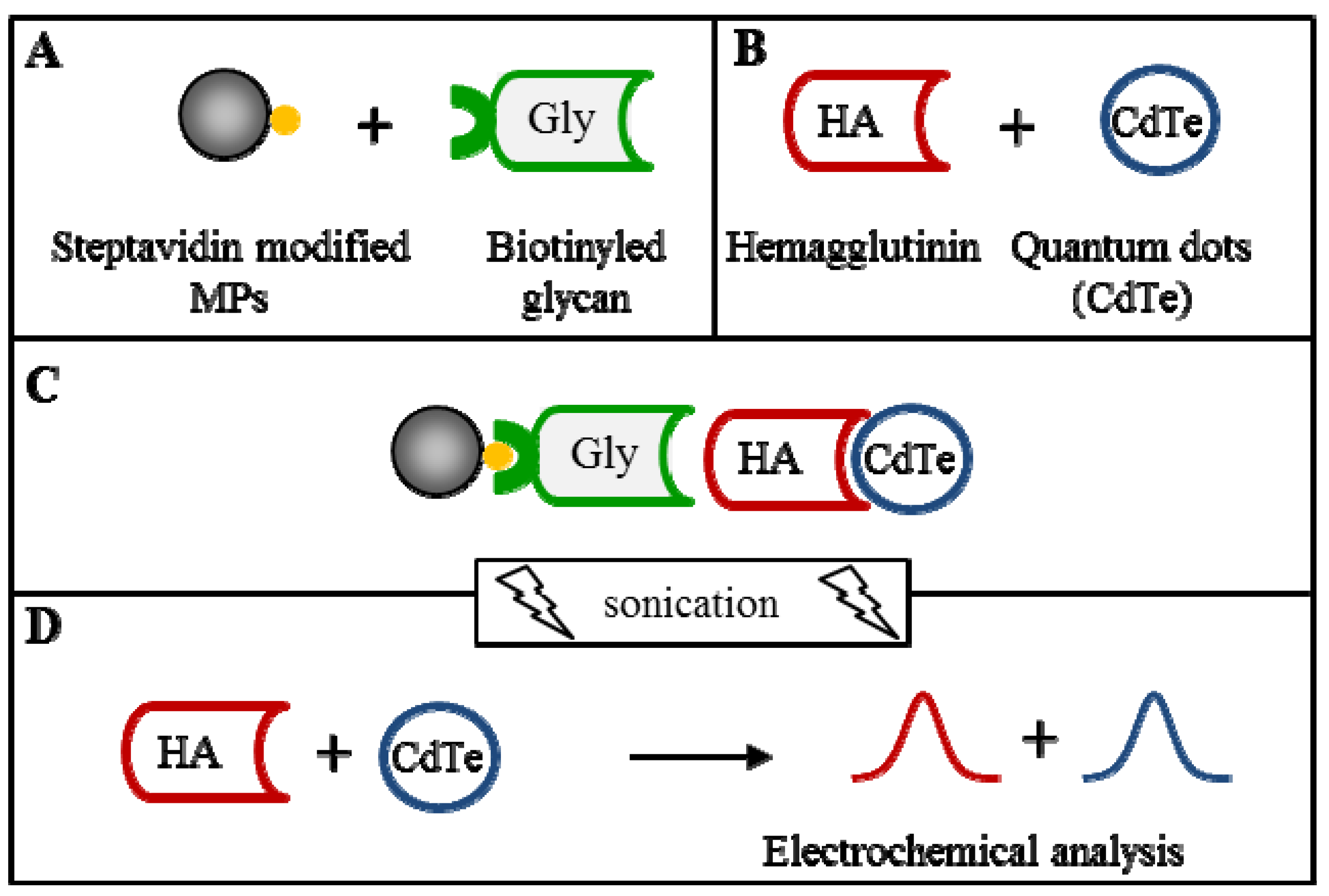
2.6. Hemagglutinin Detection from the Influenza Vaccine
3. Experimental
3.1. Chemicals
3.2. Hemagglutinin
3.3. Preparation of QDs (CdTe)
3.4. Labelling of Vaxigrip® HA by CdTe QDs
3.5. Characterization of vaxi HA and HA-CdTe Complex by Gel Electrophoresis
3.6. Isolation of HA-QDs by Glycan Modified MPs Using a Robotic Pipetting Station
3.6.1. Robotic Pipetting Station
3.6.2. The Automated Isolation Procedure
3.7. Electrochemical Detection of CdTe and HA-CdTe Complex
3.8. Electrochemical Detection of vaxi HA and HA-CdTe Complex
3.9. Descriptive Statistics
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Shoji, Y.; Farrance, C.E.; Bautista, J.; Bi, H.; Musiychuk, K.; Horsey, A.; Park, H.; Jaje, J.; Green, B.J.; Shamloul, M.; et al. A plant-based system for rapid production of influenza vaccine antigens. Influenza Other Respir. Viruses 2012, 6, 204–210. [Google Scholar] [CrossRef]
- Ding, X.F.; Jiang, L.F.; Ke, C.W.; Yang, Z.; Lei, C.L.; Cao, K.Y.; Xu, J.; Xu, L.; Yang, X.F.; Zhang, Y.H.; et al. Amino acid sequence analysis and identification of mutations under positive selection in hemagglutinin of 2009 influenza A (H1N1) isolates. Virus Genes 2010, 41, 329–340. [Google Scholar] [CrossRef]
- Chen, Q.J.; Huang, S.P.; Chen, J.J.; Zhang, S.Q.; Chen, Z. NA proteins of influenza A viruses H1N1/2009, H5N1, and H9N2 show differential effects on infection initiation, virus release, and cell-cell fusion. PLoS One 2013, 8, e54334. [Google Scholar]
- Samson, M.; Pizzorno, A.; Abed, Y.; Boivin, G. Influenza virus resistance to neuraminidase inhibitors. Antivir. Res. 2013, 98, 174–185. [Google Scholar] [CrossRef]
- Rudrawar, S.; Pascolutti, M.; Bhatt, B.; Thomson, R.J.; von Itzstein, M. An efficient synthesis of C3 C-alkylated Neu5Ac2en derivatives. Tetrahedron Lett. 2013, 54, 1198–1201. [Google Scholar] [CrossRef]
- Cho, S.; Lee, B.R.; Cho, B.K.; Kim, J.H.; Kim, B.G. In vitro selection of sialic acid specific RNA aptamer and its application to the rapid sensing of sialic acid modified sugars. Biotechnol. Bioeng. 2013, 110, 905–913. [Google Scholar] [CrossRef]
- Feng, Z.L.; Towers, S.; Yang, Y.D. Modeling the effects of vaccination and treatment on pandemic influenza. AAPS J. 2011, 13, 427–437. [Google Scholar] [CrossRef]
- Couch, R.B. Seasonal inactivated influenza virus vaccines. Vaccine 2008, 26, D5–D9. [Google Scholar]
- Lanthier, P.A.; Huston, G.E.; Moquin, A.; Eaton, S.M.; Szaba, F.M.; Kummer, L.W.; Tighe, M.P.; Kohlmeier, J.E.; Blair, P.J.; Broderick, M.; et al. Live attenuated influenza vaccine (LAIV) impacts innate and adaptive immune responses. Vaccine 2011, 29, 7849–7856. [Google Scholar] [CrossRef]
- Thompson, C.M.; Petiot, E.; Lennaertz, A.; Henry, O.; Kamen, A.A. Analytical technologies for influenza virus-like particle candidate vaccines: Challenges and emerging approaches. Virol. J. 2013, 10, 1–14. [Google Scholar] [CrossRef]
- Adam, V.; Sileny, A.; Hubalek, J.; Beklova, M.; Zehnalek, J.; Havel, L.; Kizek, R. Microsensors as a tool to detect heavy metals. Toxicol. Lett. 2008, 180, S227–S228. [Google Scholar]
- Huska, D.; Zitka, O.; Adam, V.; Beklova, M.; Krizkova, S.; Zeman, L.; Horna, A.; Havel, L.; Zehnalek, J.; Kizek, R. A sensor for investigating the interaction between biologically important heavy metals and glutathione. Czech J. Anim. Sci. 2007, 52, 37–43. [Google Scholar]
- Liu, G.D.; Lin, Y.H. Nanomaterial labels in electrochemical immunosensors and immunoassays. Talanta 2007, 74, 308–317. [Google Scholar] [CrossRef]
- Majzlik, P.; Prasek, J.; Trnkova, L.; Zehnalek, J.; Adam, V.; Havel, L.; Hubalek, J.; Kizek, R. Biosensors for detection of heavy metals. Listy Cukrov. Reparske 2010, 126, 413–414. [Google Scholar]
- Amano, Y.; Cheng, Q. Detection of influenza virus: Traditional approaches and development of biosensors. Anal. Bioanal. Chem. 2005, 381, 156–164. [Google Scholar] [CrossRef]
- Hubalek, J.; Adam, V.; Kizek, R. New Approach in Rapid Viruses Detection and Its Implementation on a Chip. In Proceedings of the 2009 International Conference on eHealthTelemedicineand Social Medicine, Los Alamitos, CA, USA, 1–7 February 2009; IEEE Computer Society: Washington, DC, USA, 2009; pp. 108–112. [Google Scholar]
- Miller, S.A.; Hiatt, L.A.; Keil, R.G.; Wright, D.W.; Cliffel, D.E. Multifunctional nanoparticles as simulants for a gravimetric immunoassay. Anal. Bioanal. Chem. 2011, 399, 1021–1029. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, W.P.; Zhang, Z.L.; He, R.L.; Lin, Y.; Xie, M.; Wang, H.Z.; Pang, D.W. Robust and highly sensitive fluorescence approach for Point-of-Care virus detection based on immunomagnetic separation. Anal. Chem. 2012, 84, 2358–2365. [Google Scholar]
- Chandrasekaran, A.; Srinivasan, A.; Raman, R.; Viswanathan, K.; Raguram, S.; Tumpey, T.M.; Sasisekharan, V.; Sasisekharan, R. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat. Biotechnol. 2008, 26, 107–113. [Google Scholar]
- Stevens, J.; Blixt, O.; Paulson, J.C.; Wilson, I.A. Glycan microarray technologies: Tools to survey host specificity of influenza viruses. Nat. Rev. Microbiol. 2006, 4, 857–864. [Google Scholar] [CrossRef]
- Stevens, J.; Corper, A.L.; Basler, C.F.; Taubenberger, J.K.; Palese, P.; Wilson, I.A. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 2004, 303, 1866–1870. [Google Scholar] [CrossRef]
- Stevens, J.; Blixt, O.; Tumpey, T.M.; Taubenberger, J.K.; Paulson, J.C.; Wilson, I.A. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 2006, 312, 404–410. [Google Scholar] [CrossRef]
- Takahashi, Y.; Hirano, Y.; Yasukawa, T.; Shiku, H.; Yamada, H.; Matsue, T. Topographic, electrochemical, and optical images captured using standing approach mode scanning electrochemical/optical microscopy. Langmuir 2006, 22, 10299–10306. [Google Scholar] [CrossRef]
- Bewley, C.A. Illuminating the switch in influenza viruses. Nat. Biotechnol. 2008, 26, 60–62. [Google Scholar] [CrossRef]
- Suenaga, E.; Mizuno, H.; Penmetcha, K.K.R. Monitoring influenza hemagglutinin and glycan interactions using surface plasmon resonance. Biosens. Bioelectron. 2012, 32, 195–201. [Google Scholar] [CrossRef]
- Krejcova, L.; Dospivova, D.; Ryvolova, M.; Kopel, P.; Hynek, D.; Krizkova, S.; Hubalek, J.; Adam, V.; kizek, R. Paramagnetic particles coupled with an automated flow injection analysis as a tool for influenza viral protein detection. Electrophoresis 2012, 33, 3195–3204. [Google Scholar] [CrossRef]
- Garcia-Canas, V.; Lorbetskie, B.; Cyr, T.D.; Hefford, M.A.; Smith, S.; Girard, M. Approach to the profiling and characterization of influenza vaccine constituents by the combined use of size-exclusion chromatography, gel electrophoresis and mass spectrometry. Biologicals 2010, 38, 294–302. [Google Scholar] [CrossRef]
- Epand, R.M.; Epand, R.F. Thermal denaturation of influenza virus and its relationship to membrane fusion. Biochem. J. 2002, 365, 841–848. [Google Scholar]
- Schwarzer, J.; Rapp, E.; Hennig, R.; Genzel, Y.; Jordan, I.; Sandig, V.; Reichl, U. Glycan analysis in cell culture-based influenza vaccine production: Influence of host cell line and virus strain on the glycosylation pattern of viral hemagglutinin. Vaccine 2009, 27, 4325–4336. [Google Scholar]
- Chou, T.C.; Hsu, W.; Wang, C.H.; Chen, Y.J.; Fang, J.M. Rapid and specific influenza virus detection by functionalized magnetic nanoparticles and mass spectrometry. J. Nanobiotechnol. 2011, 9, 1–13. [Google Scholar] [CrossRef]
- Hoffman, L.R.; Kuntz, I.D.; White, J.M. Structure-based identification of an inducer of the low-pH conformational change in the influenza virus hemagglutinin: Irreversible inhibition of infectivity. J. Virol. 1997, 71, 8808–8820. [Google Scholar]
- Doneley, B. Avian Medicine and Surgery in Practice: Companion and Aviary Birds; Manson Publishing Ltd.: London, UK, 2010; p. 330. [Google Scholar]
- Korte, T.; Ludwig, K.; Booy, F.P.; Blumenthal, R.; Herrmann, A. Conformational intermediates and fusion activity of influenza virus hemagglutinin. J. Virol. 1999, 73, 4567–4574. [Google Scholar]
- Ruigrok, R.W.H.; Martin, S.R.; Wharton, S.A.; Skehel, J.J.; Bayley, P.M.; Wiley, D.C. Conformational-changes in the hemagglutinin of influenza-virus which accompany heat-induced fusion of virus with liposomes. Virology 1986, 155, 484–497. [Google Scholar] [CrossRef]
- Kamikawa, T.L.; Mikolajczyk, M.G.; Kennedy, M.; Zhang, P.; Wang, W.; Scott, D.E.; Alocilja, E.C. Nanoparticle-based biosensor for the detection of emerging pandemic influenza strains. Biosens. Bioelectron. 2010, 26, 1346–1352. [Google Scholar] [CrossRef]
- Egashira, N.; Morita, S.; Hifumi, E.; Mitoma, Y.; Uda, T. Attomole detection of hemagglutinin molecule of influenza virus by combining an electrochemiluminescence sensor with an immunoliposome that encapsulates a Ru complex. Anal. Chem. 2008, 80, 4020–4025. [Google Scholar] [CrossRef]
- Wagner, R.; Matrosovich, M.; Klenk, H.D. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev. Med. Virol. 2002, 12, 159–166. [Google Scholar] [CrossRef]
- Legastelois, I.; Chevalier, M.; Bernard, M.C.; de Montfort, A.; Fouque, M.; Pilloud, A.; Serraille, C.; Devard, N.; Engel, O.; Sodoyer, R.; et al. Avian glycan-specific IgM monoclonal antibodies for the detection and quantitation of type A and B haemagglutinins in egg-derived influenza vaccines. J. Virol. Methods 2011, 178, 129–136. [Google Scholar] [CrossRef]
- Bousse, T.; Shore, D.A.; Goldsmith, C.S.; Hossain, M.J.; Jang, Y.; Davis, C.T.; Donis, R.O.; Stevens, J. Quantitation of influenza virus using field flow fractionation and multi-angle light scattering for quantifying influenza A particles. J. Virol. Methods 2013, 193, 589–596. [Google Scholar] [CrossRef]
- Duan, J.L.; Song, L.X.; Zhan, J.H. One-pot synthesis of highly luminescent CdTe quantum dots by microwave irradiation reduction and their Hg2+-sensitive properties. Nano Res. 2009, 2, 61–68. [Google Scholar] [CrossRef]
- Wong, C.; Sridhara, S.; Bardwell, J.C.A.; Jakob, U. Heating greatly speeds Coomassie blue staining and destaining. Biotechniques 2000, 28, 426–428. [Google Scholar]
- Skalickova, S.; Zitka, O.; Nejdl, L.; Krizkova, S.; Sochor, J.; Janu, L.; Ryvolova, M.; Hynek, D.; Zidkova, J.; Zidek, V.; et al. Study of interaction between metallothionein and CdTe quantum dots. Chromatographia 2013, 76, 345–353. [Google Scholar] [CrossRef]
- Long, G.L.; Winefordner, J.D. Limit of detection. Anal. Chem. 1983, 55, A712–A724. [Google Scholar]
- Samples Availability: Samples of the quantum dots are available from the authors.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Krejcova, L.; Nejdl, L.; Hynek, D.; Krizkova, S.; Kopel, P.; Adam, V.; Kizek, R. Beads-Based Electrochemical Assay for the Detection of Influenza Hemagglutinin Labeled with CdTe Quantum Dots. Molecules 2013, 18, 15573-15586. https://doi.org/10.3390/molecules181215573
Krejcova L, Nejdl L, Hynek D, Krizkova S, Kopel P, Adam V, Kizek R. Beads-Based Electrochemical Assay for the Detection of Influenza Hemagglutinin Labeled with CdTe Quantum Dots. Molecules. 2013; 18(12):15573-15586. https://doi.org/10.3390/molecules181215573
Chicago/Turabian StyleKrejcova, Ludmila, Lukas Nejdl, David Hynek, Sona Krizkova, Pavel Kopel, Vojtech Adam, and Rene Kizek. 2013. "Beads-Based Electrochemical Assay for the Detection of Influenza Hemagglutinin Labeled with CdTe Quantum Dots" Molecules 18, no. 12: 15573-15586. https://doi.org/10.3390/molecules181215573




