Microorganisms 2023, 11(2), 359; https://doi.org/10.3390/microorganisms11020359 - 31 Jan 2023
Cited by 8 | Viewed by 3220
Abstract
Patients with cancer have a higher risk of severe bacterial infections. This study aims to determine the frequency, susceptibility profiles, and resistance genes of bacterial species involved in bacteremia, as well as risk factors associated with mortality in cancer patients in Colombia. In
[...] Read more.
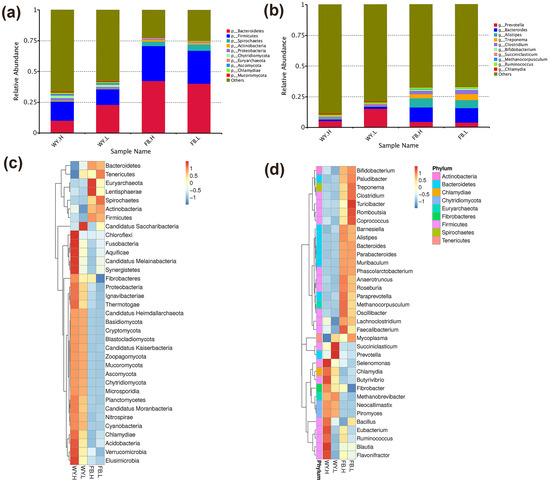
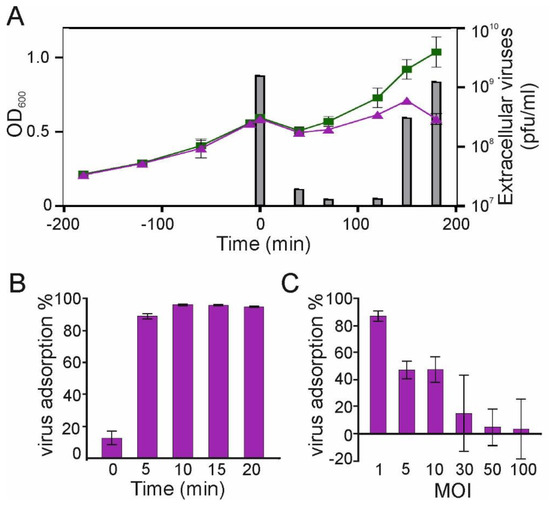
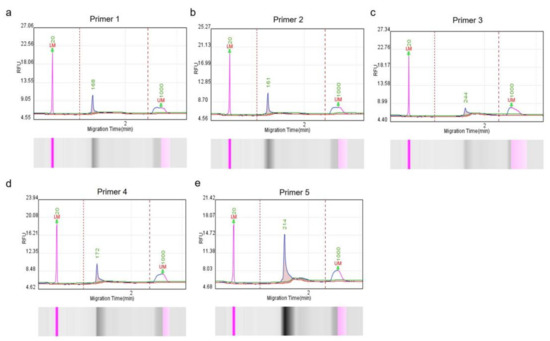
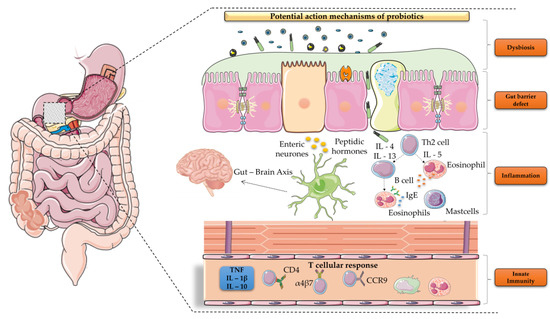
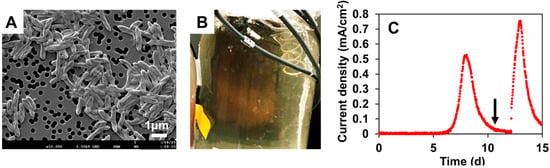
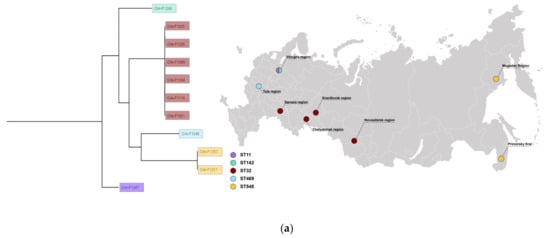
Patients with cancer have a higher risk of severe bacterial infections. This study aims to determine the frequency, susceptibility profiles, and resistance genes of bacterial species involved in bacteremia, as well as risk factors associated with mortality in cancer patients in Colombia. In this prospective multicenter cohort study of adult patients with cancer and bacteremia, susceptibility testing was performed and selected resistance genes were identified. A multivariate regression analysis was carried out for the identification of risk factors for mortality. In 195 patients, 206 microorganisms were isolated. Gram-negative bacteria were more frequently found, in 142 cases (68.9%): 67 Escherichia coli (32.5%), 36 Klebsiella pneumoniae (17.4%), and 21 Pseudomonas aeruginosa (10.1%), and 18 other Gram-negative isolates (8.7%). Staphylococcus aureus represented 12.4% (n = 25). Among the isolates, resistance to at least one antibiotic was identified in 63% of them. Genes coding for extended-spectrum beta-lactamases and carbapenemases, blaCTX-M and blaKPC, respectively, were commonly found. Mortality rate was 25.6% and it was lower in those with adequate empirical antibiotic treatment (22.0% vs. 45.2%, OR: 0.26, 95% CI: 0.1–0.63, in the multivariate model). In Colombia, in patients with cancer and bacteremia, bacteria have a high resistance profile to beta-lactams, with a high incidence of extended-spectrum beta-lactamases and carbapenemases. Adequate empirical treatment diminishes mortality, and empirical selection of treatment in this environment of high resistance is of key importance.
Full article
(This article belongs to the Section Medical Microbiology)