Current Advances and Methodologies in Gene Editing
A topical collection in Methods and Protocols (ISSN 2409-9279).
Viewed by 70588Editors
Interests: mouse models; recombineering technology; CRISPR/Cas9 technology; Cas9-screens; dCas9-imaging; general regulation of transcription and epigenesis
Special Issues, Collections and Topics in MDPI journals
2. Yale-NUS College, 10 College Avenue West, Singapore 138609, Singapore
Interests: butterfly wing patterns; evo-devo; functional genetics; plasticity; eyespots; evolution; sexual and natural selection; behaviour
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear colleagues,
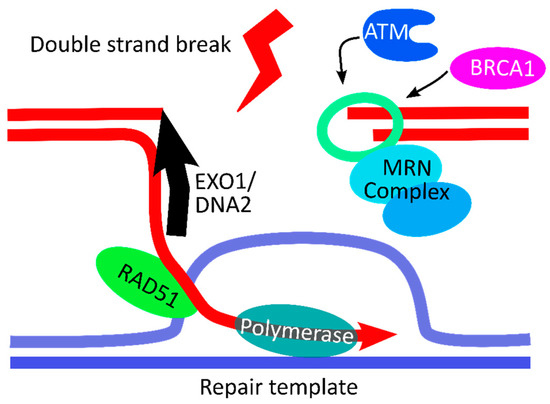
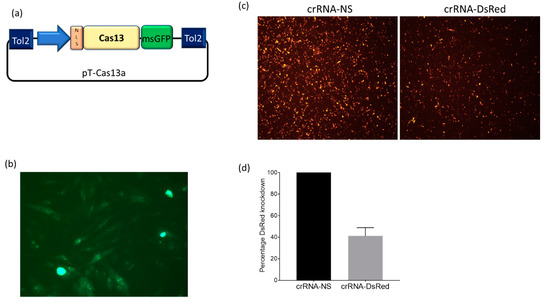
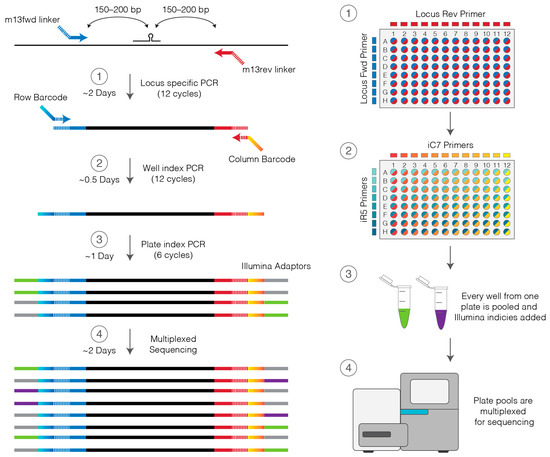
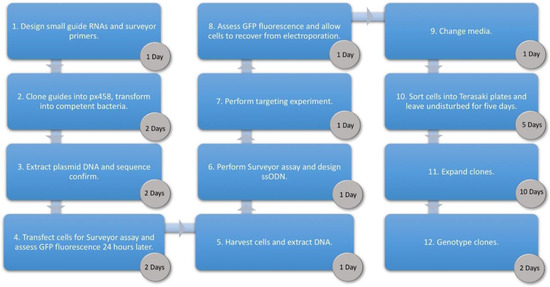
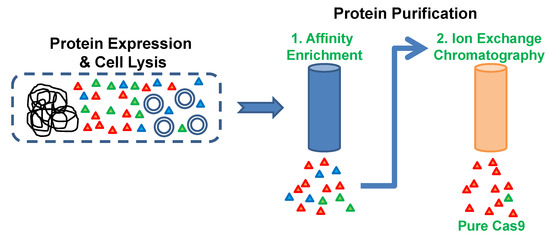
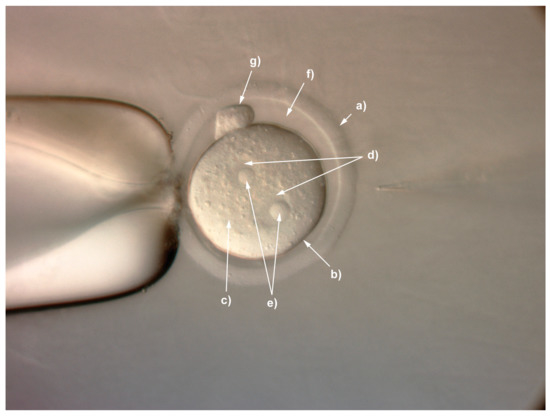
With the emergence of CRISPR/Cas9, an unprecedented variety of novel approaches and possibilities for genomic engineering are unveiled on a regular basis. These developments are accelerating the pace of research in model systems, as well as creating a plethora of emerging model systems invaluable for comparative functional approaches across the tree of life. The CRISPR/Cas9 toolbox has already allowed the manipulation of gene function in basal land plants, butterflies, crickets and Atlantic salmon, and many more are sure to follow.
This topical collection on “Gene Editing” aims to provide a forum for discussions on the latest technical developments in the fields of general genome engineering technologies, including (i) the establishment of cell culture systems, and (ii) the development of established and emerging organismal models by CRISPR/Cas9 or similar genome engineering tools. A constant flow of reports demonstrates the continuous refinement of the CRISPR/Cas9 revolutionary tool. Invariably, however, new challenges or missing details in the implementation of this technology become apparent when applied to specific model systems, which need to be addressed. Our topical collection aims to keep up with the most recent developments, refinements, and latest achievements in gene editing available today. We encourage contributions from laboratories working at the forefront of the development of novel CRISPR/Cas9 approaches, with the goal of sharing these details and accelerating the speed of functional genetic discovery across the tree of life, in well-established as well as in emerging model systems.
Dr. Philip Hublitz
Dr. Antónia Monteiro
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Methods and Protocols is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 1800 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- CRISPR/Cas9
- Genome engineering
- Gene editing
- Established and emerging model systems
- Model generation
- Functional genetics using KI and KO
- Spatiotemporal regulation
- Arrayed and pooled sgRNA-Screening
- Cas9 efficiency and fidelity
- New tools in the CRISPR/Cas9 toolbox