Gut Microbiota-Derived Tryptophan Metabolites Alleviate Allergic Asthma Inflammation in Ovalbumin-Induced Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Tryptophan Metabolite Injections
2.2. Experimental Design
2.3. Weight Measurement and Evaluation of Asthmatic Mice
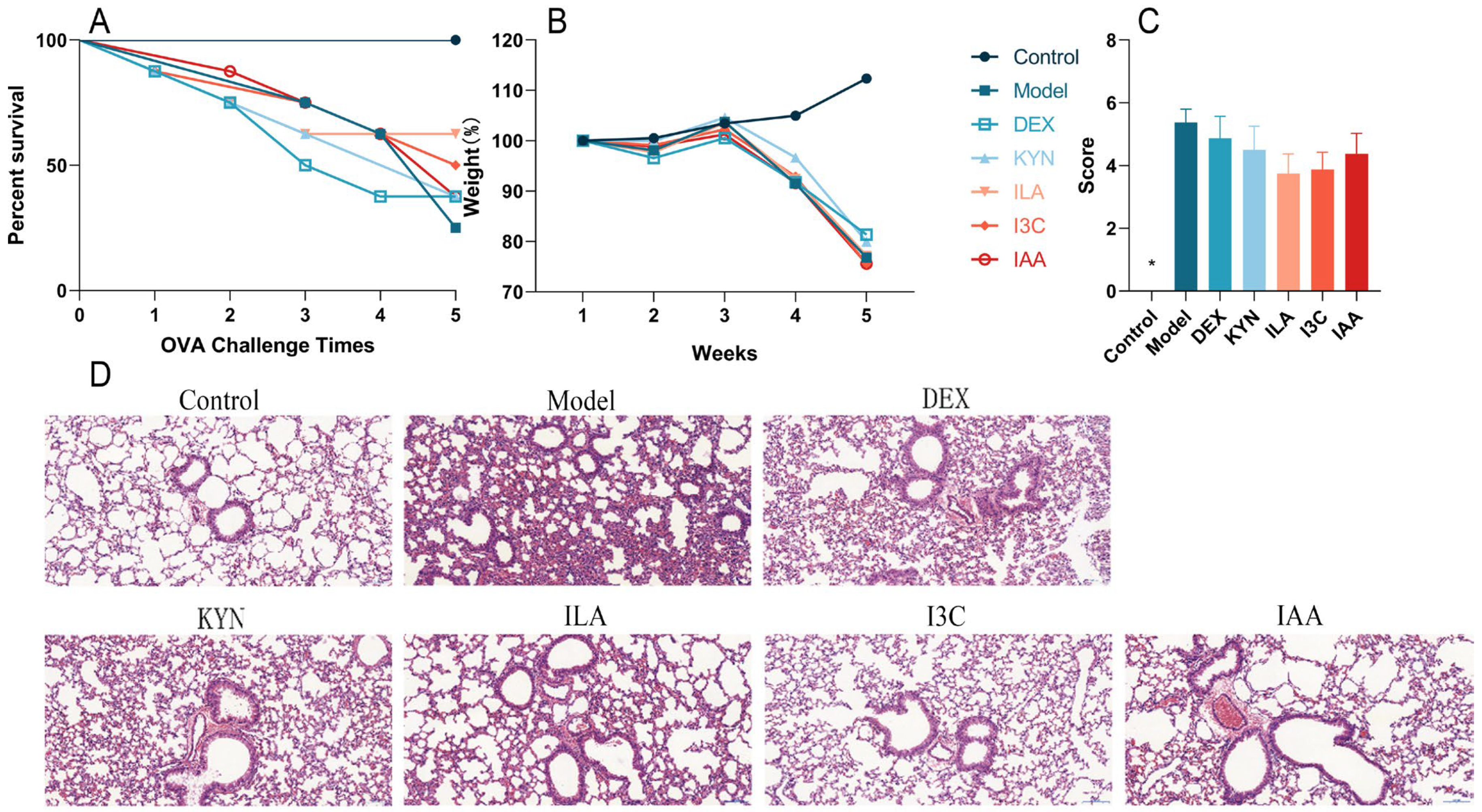
2.4. Histopathological Changes in Mouse Lung Tissues
2.5. Serum Immunoglobulin Assay
2.6. Measurement of Inflammatory Cytokines in BALF
2.7. Sequencing the 16S Ribosomal RNA Gene
2.8. Detection of Tryptophan-Targeted Metabolism in Mouse Feces
2.9. Statistical Analysis
3. Results
3.1. Tryptophan Metabolites Alleviate Changes in Body Weight and Lung Tissues in Allergic Asthma Mice
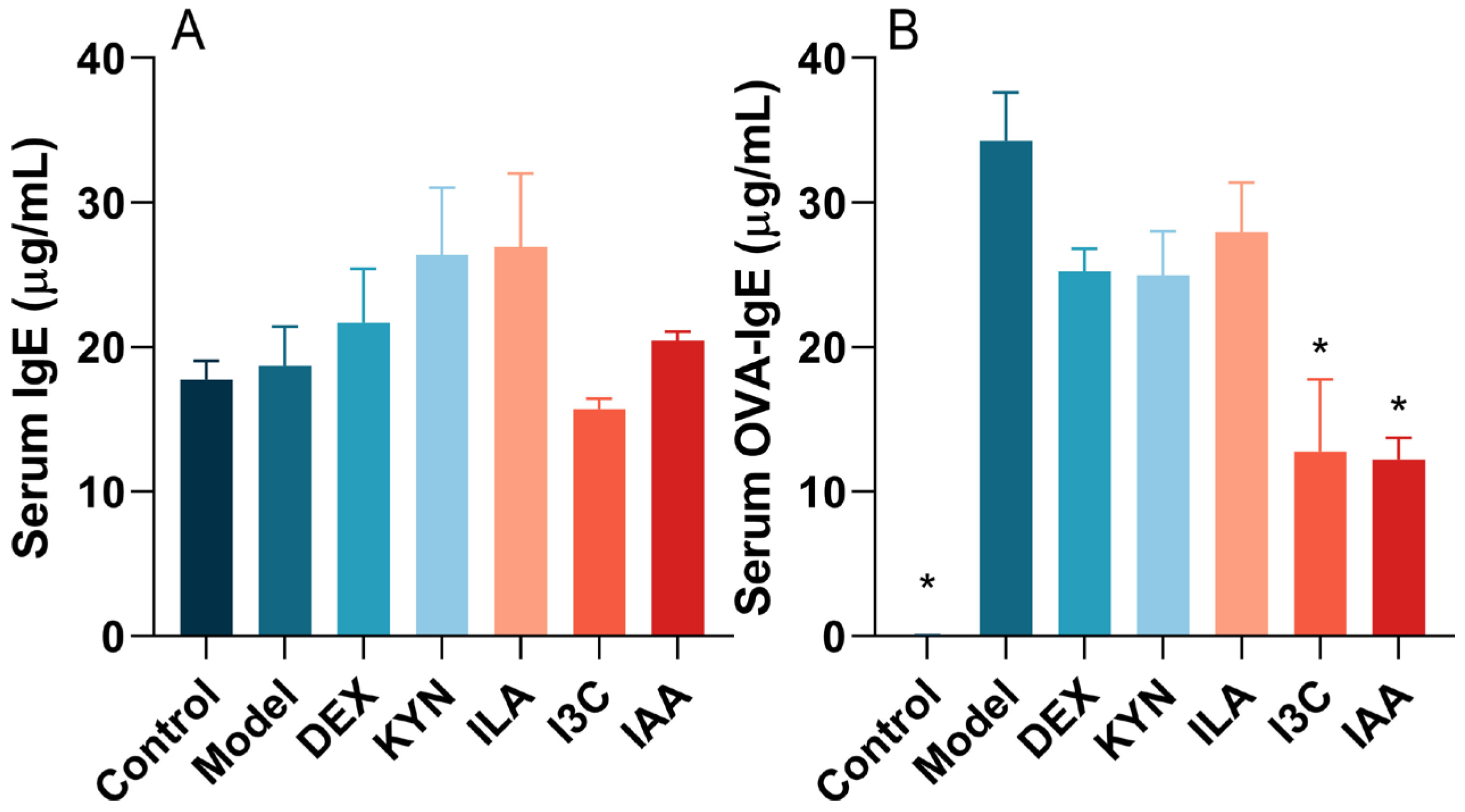
3.2. Tryptophan Metabolites Regulate IgE and OVA-sIgE Levels in Allergic Asthma Mice
3.3. Tryptophan Metabolites Influence Inflammatory Cytokine Levels in Allergic Asthma Mice
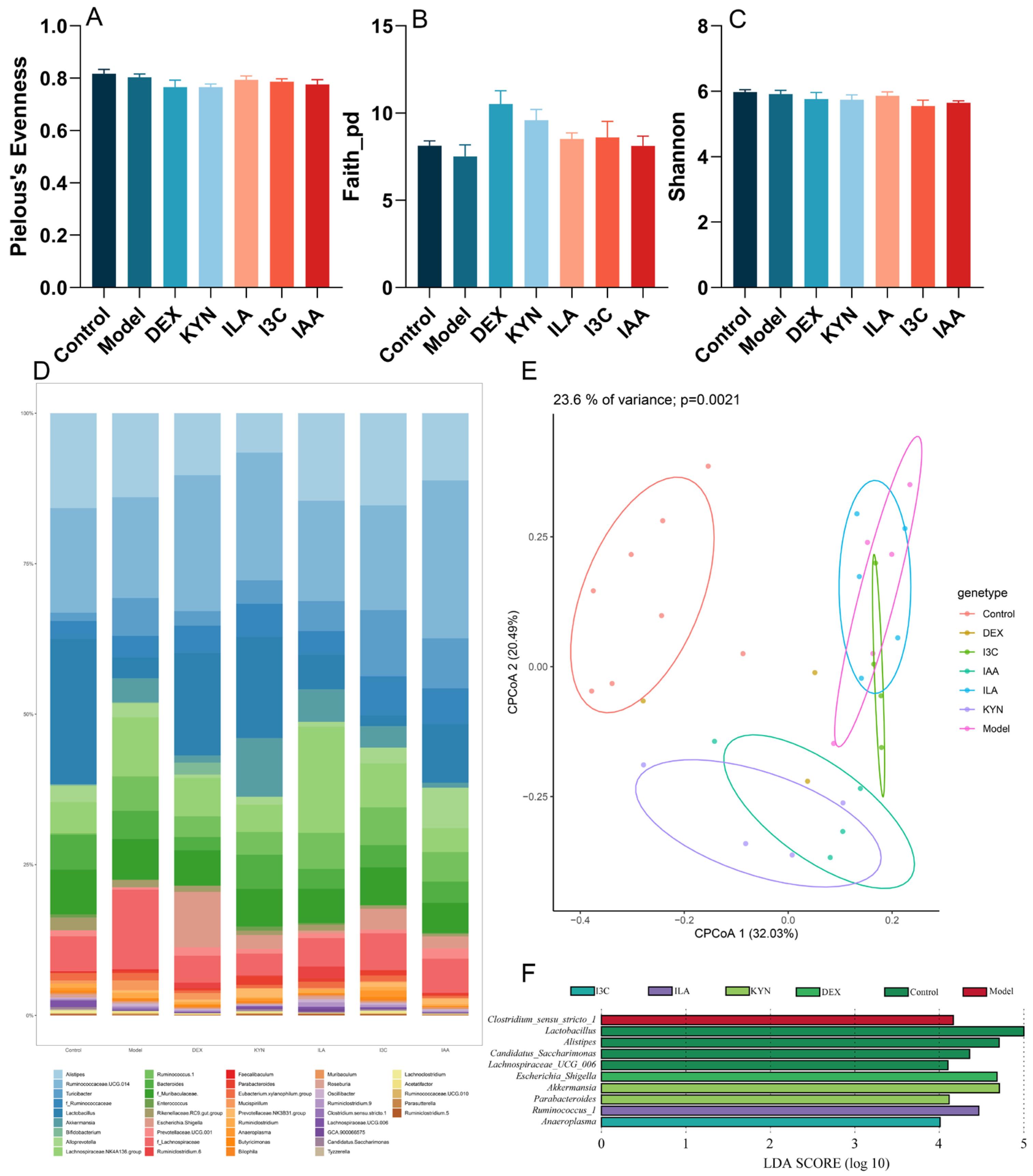
3.4. Tryptophan Metabolites Modulate the Gut Microbiota Composition in Allergic Asthma Mice
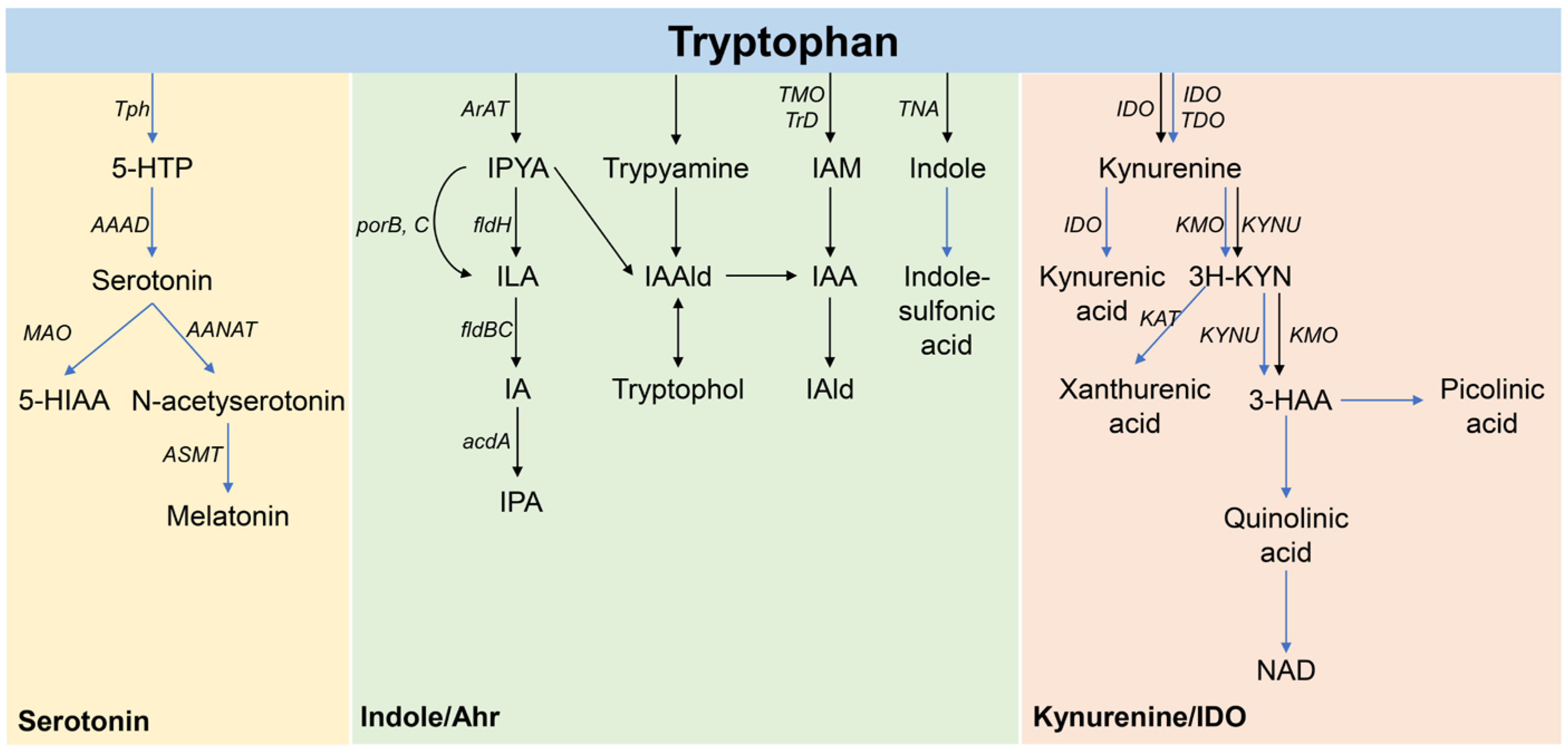
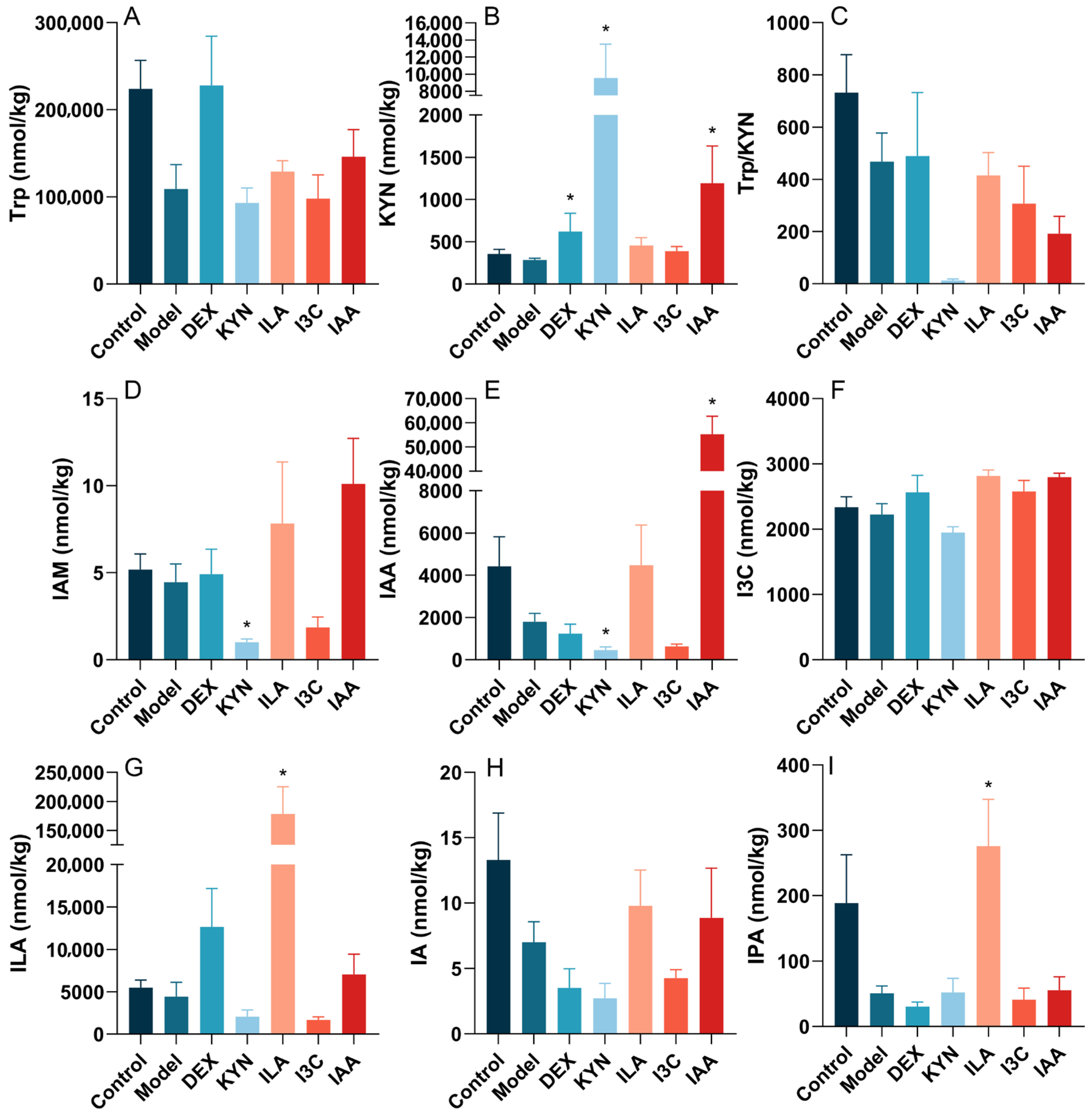
3.5. Tryptophan Metabolites Influence the Tryptophan Metabolite Content in the Gut Microbiota of Allergic Asthma Mice
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Chapman, D.G.; Irvin, C.G. Mechanisms of airway hyper-responsiveness in asthma: The past, present and yet to come. Clin. Exp. Allergy 2015, 45, 706–719. [Google Scholar] [CrossRef] [PubMed]
- Pate, C.A.; Zahran, H.S.; Qin, X.; Johnson, C.; Hummelman, E.; Malilay, J. Asthma Surveillance-United States, 2006–2018. MMWR Surveill. Summ. 2021, 70, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Amin, F.; Sadeeqa, S. Prevalence of asthma and its management: A review. J. Pak. Med. Assoc. 2018, 68, 1823–1827. [Google Scholar] [PubMed]
- Athari, S.S.; Athari, S.M. The importance of eosinophil, platelet and dendritic cell in asthma. Asian Pac. J. Trop. Dis. 2014, 4, S41–S47. [Google Scholar] [CrossRef]
- Patel, V.H.; Thannir, S.; Dhanani, M.; Augustine, I.; Sandeep, S.L.; Mehadi, A.; Avanthika, C.; Jhaveri, S. Current Limitations and Recent Advances in the Management of Asthma. Disease-a-Month 2023, 69, 101483. [Google Scholar] [CrossRef] [PubMed]
- Seys, S.F.; Quirce, S.; Agache, I.; Akdis, C.A.; Alvaro-Lozano, M.; Antolín-Amérigo, D.; Bjermer, L.; Bobolea, I.; Bonini, M.; Bossios, A.; et al. Severe asthma: Entering an era of new concepts and emerging therapies: Highlights of the 4th international severe asthma forum, Madrid, 2018. Allergy 2019, 74, 2244–2248. [Google Scholar] [CrossRef] [PubMed]
- Roth-Walter, F.; Adcock, I.M.; Benito-Villalvilla, C.; Bianchini, R.; Bjermer, L.; Boyman, O.; Caramori, G.; Cari, L.; Fan Chung, K.; Diamant, Z.; et al. Immune modulation via T regulatory cell enhancement: Disease-modifying therapies for autoimmunity and their potential for chronic allergic and inflammatory diseases-An EAACI position paper of the Task Force on Immunopharmacology (TIPCO). Allergy 2021, 76, 90–113. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Shang, Y.X. Epigallocatechin gallate ameliorates airway inflammation by regulating Treg/Th17 imbalance in an asthmatic mouse model. Int. Immunopharmacol. 2019, 72, 422–428. [Google Scholar] [CrossRef]
- Zheng, R.; Wang, F.; Huang, Y.; Xiang, Q.; Dai, H.; Zhang, W. Elevated Th17 cell frequencies and Th17/Treg ratio are associated with airway hyperresponsiveness in asthmatic children. J. Asthma 2021, 58, 707–716. [Google Scholar] [CrossRef]
- Jiang, H.; Wu, X.; Zhu, H.; Xie, Y.; Tang, S.; Jiang, Y. FOXP3(+)Treg/Th17 cell imbalance in lung tissues of mice with asthma. Int. J. Clin. Exp. Med. 2015, 8, 4158–4163. [Google Scholar]
- Chang, Y.D.; Li, C.H.; Tsai, C.H.; Cheng, Y.W.; Kang, J.J.; Lee, C.C. Aryl hydrocarbon receptor deficiency enhanced airway inflammation and remodeling in a murine chronic asthma model. FASEB J. 2020, 34, 15300–15313. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Y.; Li, N.; Wang, Y.; Li, Q.; Ge, D.; Peng, G.; Zhou, M. Dachengqi Decoction alleviates intestinal inflammation in ovalbumin-induced asthma by reducing group 2 innate lymphoid cells in a microbiota-dependent manner. J. Tradit. Complement. Med. 2023, 13, 183–192. [Google Scholar] [CrossRef]
- Begley, L.; Madapoosi, S.; Opron, K.; Ndum, O.; Baptist, A.; Rysso, K.; Erb-Downward, J.R.; Huang, Y.J. Gut microbiota relationships to lung function and adult asthma phenotype: A pilot study. BMJ Open Respir. Res. 2018, 5, e000324. [Google Scholar] [CrossRef]
- Nagai, H. Recent research and developmental strategy of anti-asthma drugs. Pharmacol. Ther. 2012, 133, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Cui, Z.; Wang, J.; Feng, Y.; Xie, R.; Li, D.; Peng, J.; Huang, R.; Li, T. Aryl hydrocarbon receptor modulates airway inflammation in mice with cockroach allergen-induced asthma by regulating Th17/Treg differentiation. Nan Fang. Yi Ke Da Xue Xue Bao 2021, 41, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Dunican, E.M.; Fahy, J.V. Asthma and corticosteroids: Time for a more precise approach to treatment. Eur. Respir. J. 2017, 49, 1701167. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Yang, J.; Kong, W.; Liu, Y.; Liu, S.; Su, L. Cordyceps militaris polysaccharide alleviates ovalbumin-induced allergic asthma through the Nrf2/HO-1 and NF-κB signaling pathways and regulates the gut microbiota. Int. J. Biol. Macromol. 2023, 238, 124333. [Google Scholar] [CrossRef]
- Platten, M.; Nollen, E.A.A.; Röhrig, U.F.; Fallarino, F.; Opitz, C.A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 2019, 18, 379–401. [Google Scholar] [CrossRef]
- Dehhaghi, M.; Kazemi Shariat Panahi, H.; Guillemin, G.J. Microorganisms, Tryptophan Metabolism, and Kynurenine Pathway: A Complex Interconnected Loop Influencing Human Health Status. Int. J. Tryptophan Res. 2019, 12, 1178646919852996. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef]
- Al-Rashidi, H.E. Gut microbiota and immunity relevance in eubiosis and dysbiosis. Saudi J. Biol. Sci. 2022, 29, 1628–1643. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Chu, J.; Feng, S.; Guo, C.; Xue, B.; He, K.; Li, L. Immunological mechanisms of inflammatory diseases caused by gut microbiota dysbiosis: A review. Biomed. Pharmacother. 2023, 164, 114985. [Google Scholar] [CrossRef] [PubMed]
- Wypych, T.P.; Wickramasinghe, L.C.; Marsland, B.J. The influence of the microbiome on respiratory health. Nat. Immunol. 2019, 20, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Turturice, B.A.; McGee, H.S.; Oliver, B.; Baraket, M.; Nguyen, B.T.; Ascoli, C.; Ranjan, R.; Rani, A.; Perkins, D.L.; Finn, P.W. Atopic asthmatic immune phenotypes associated with airway microbiota and airway obstruction. PLoS ONE 2017, 12, e0184566. [Google Scholar] [CrossRef] [PubMed]
- Buendía, E.; Zakzuk, J.; San-Juan-Vergara, H.; Zurek, E.; Ajami, N.J.; Caraballo, L. Gut microbiota components are associated with fixed airway obstruction in asthmatic patients living in the tropics. Sci. Rep. 2018, 8, 9582. [Google Scholar] [CrossRef] [PubMed]
- Michalovich, D.; Rodriguez-Perez, N.; Smolinska, S.; Pirozynski, M.; Mayhew, D.; Uddin, S.; Van Horn, S.; Sokolowska, M.; Altunbulakli, C.; Eljaszewicz, A.; et al. Obesity and disease severity magnify disturbed microbiome-immune interactions in asthma patients. Nat. Commun. 2019, 10, 5711. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.L.; Wu, J.J.; Ye, H.X.; Feng, D.Y.; Meng, P.; Yang, H.L.; Wu, W.B.; Li, H.T.; He, Z.; Zhang, T.T. Associations Between Gut Microbiota and Asthma Endotypes: A Cross-Sectional Study in South China Based on Patients with Newly Diagnosed Asthma. J. Asthma Allergy 2021, 14, 981–992. [Google Scholar] [CrossRef]
- Vijay, A.; Valdes, A.M. Role of the gut microbiome in chronic diseases: A narrative review. Eur. J. Clin. Nutr. 2022, 76, 489–501. [Google Scholar] [CrossRef]
- Lee-Sarwar, K.A.; Lasky-Su, J.; Kelly, R.S.; Litonjua, A.A.; Weiss, S.T. Gut Microbial-Derived Metabolomics of Asthma. Metabolites 2020, 10, 97. [Google Scholar] [CrossRef]
- Tagé, B.S.S.; Gonzatti, M.B.; Vieira, R.P.; Keller, A.C.; Bortoluci, K.R.; Aimbire, F. Three Main SCFAs Mitigate Lung Inflammation and Tissue Remodeling Nlrp3-Dependent in Murine HDM-Induced Neutrophilic Asthma. Inflammation 2024. Epub ahead of printing. [Google Scholar] [CrossRef]
- Alsharairi, N.A. The Role of Short-Chain Fatty Acids in the Interplay between a Very Low-Calorie Ketogenic Diet and the Infant Gut Microbiota and Its Therapeutic Implications for Reducing Asthma. Int. J. Mol. Sci. 2020, 21, 9580. [Google Scholar] [CrossRef] [PubMed]
- Larabi, A.B.; Masson, H.L.P.; Bäumler, A.J. Bile acids as modulators of gut microbiota composition and function. Gut Microbes 2023, 15, 2172671. [Google Scholar] [CrossRef] [PubMed]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef] [PubMed]
- Esser, C.; Rannug, A.; Stockinger, B. The aryl hydrocarbon receptor in immunity. Trends Immunol. 2009, 30, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Pan, T.; Li, L.; Wang, H.; Zhu, J.; Zhang, H.; Zhao, J.; Chen, W.; Lu, W. Bifidobacterium longum mediated tryptophan metabolism to improve atopic dermatitis via the gut-skin axis. Gut Microbes 2022, 14, 2044723. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Qian, L.; Fang, Z.; Wang, H.; Zhu, J.; Lee, Y.K.; Zhao, J.; Zhang, H.; Chen, W. Probiotic strains alleviated OVA-induced food allergy in mice by regulating the gut microbiota and improving the level of indoleacrylic acid in fecal samples. Food Funct. 2022, 13, 3704–3719. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Wu, Y.; Chen, B.; Yao, Y.; Wang, Y.; Bai, H.; Li, C.; Yang, Y.; Chen, Y. Cyanidin-3-O-β-glucoside attenuates allergic airway inflammation by modulating the IL-4Rα-STAT6 signaling pathway in a murine asthma model. Int. Immunopharmacol. 2019, 69, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Spacova, I.; Petrova, M.I.; Fremau, A.; Pollaris, L.; Vanoirbeek, J.; Ceuppens, J.L.; Seys, S.; Lebeer, S. Intranasal administration of probiotic Lactobacillus rhamnosus GG prevents birch pollen-induced allergic asthma in a murine model. Allergy 2019, 74, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Maeda, S.; Horiguchi, K.; Maehara, T.; Aritake, K.; Choi, B.I.; Iwakura, Y.; Urade, Y.; Murata, T. PGD2 deficiency exacerbates food antigen-induced mast cell hyperplasia. Nat. Commun. 2015, 6, 7514. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.-H.; Hu, T.-Y.; Mo, L.-H.; Ma, L.; Hu, W.-H.; Li, Y.-S.; Liu, Z.-Q.; Qiu, S.-Q. Yan-Hou-Qing formula attenuates allergic airway inflammation via up-regulation of Treg and suppressing Th2 responses in Ovalbumin-induced asthmatic mice. J. Ethnopharmacol. 2019, 231, 275–282. [Google Scholar] [CrossRef]
- Yang, Z.; Li, X.; Fu, R.; Hu, M.; Wei, Y.; Hu, X.; Tan, W.; Tong, X.; Huang, F. Therapeutic Effect of Renifolin F on Airway Allergy in an Ovalbumin-Induced Asthma Mouse Model In Vivo. Molecules 2022, 27, 3789. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Yang, B.; Ross, R.P.; Stanton, C.; Zhao, J.; Zhang, H.; Chen, W. Crosstalk between sIgA-Coated Bacteria in Infant Gut and Early-Life Health. Trends Microbiol. 2021, 29, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Hata, T.; Asano, Y.; Yoshihara, K.; Kimura-Todani, T.; Miyata, N.; Zhang, X.T.; Takakura, S.; Aiba, Y.; Koga, Y.; Sudo, N. Regulation of gut luminal serotonin by commensal microbiota in mice. PLoS ONE 2017, 12, e0180745. [Google Scholar] [CrossRef] [PubMed]
- Moskvin, S.V.; Khadartsev, A.A. Methods of effective low-level laser therapy in the treatment of patients with bronchial asthma (literature review). Biomed. Pharmacother. 2020, 10, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Moeller, A.; Carlsen, K.H.; Sly, P.D.; Baraldi, E.; Piacentini, G.; Pavord, I.; Lex, C.; Saglani, S. Monitoring asthma in childhood: Lung function, bronchial responsiveness and inflammation. Eur. Respir. Rev. 2015, 24, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Yeh, S.Y.; Schwartzstein, R. Asthma: Pathophysiology and Diagnosis. In Asthma, Health and Society; Springer: Berlin/Heidelberg, Germany, 2009; pp. 19–42. [Google Scholar] [CrossRef]
- Moon, E.Y.; Ryu, S.K. TACI:Fc scavenging B cell activating factor (BAFF) alleviates ovalbumin-induced bronchial asthma in mice. Exp. Mol. Med. 2007, 39, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Harker, J.A.; Lloyd, C.M. T helper 2 cells in asthma. J. Exp. Med. 2023, 220, 1094. [Google Scholar] [CrossRef] [PubMed]
- Gasiuniene, E.; Lavinskiene, S.; Sakalauskas, R.; Sitkauskiene, B. Levels of IL-32 in Serum, Induced Sputum Supernatant, and Bronchial Lavage Fluid of Patients with Chronic Obstructive Pulmonary Disease. COPD 2016, 13, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zeng, B.; Liu, Z.; Liao, Z.; Li, W.; Wei, H.; Fang, X. Comparative Diversity Analysis of Gut Microbiota in Two Different Human Flora-Associated Mouse Strains. Curr. Microbiol. 2014, 69, 365–373. [Google Scholar] [CrossRef]
- Redhu, N.S.; Saleh, A.; Lee, H.C.; Halayko, A.J.; Ziegler, S.F.; Gounni, A.S. IgE induces transcriptional regulation of thymic stromal lymphopoietin in human airway smooth muscle cells. J. Allergy Clin. Immunol. 2011, 128, 892–896.e2. [Google Scholar] [CrossRef]
- Justiz Vaillant, A.A.; Vashisht, R.; Zito, P.M. Immediate Hypersensitivity Reactions. In StatPearls; StatPearls Publishing: St. Petersburg, FL, USA, 2023. [Google Scholar]
- Takeda, M.; Tanabe, M.; Ito, W.; Ueki, S.; Konnno, Y.; Chihara, M.; Itoga, M.; Kobayashi, Y.; Moritoki, Y.; Kayaba, H.; et al. Gender difference in allergic airway remodelling and immunoglobulin production in mouse model of asthma. Respirology 2013, 18, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.H.; Shi, G.C.; Wan, H.Y.; Jiang, L.H.; Ai, X.Y.; Zhu, H.X.; Tang, W.; Ma, J.Y.; Jin, X.Y.; Zhang, B.Y. Coexistence of Th1/Th2 and Th17/Treg imbalances in patients with allergic asthma. Chin. Med. J. (Engl.) 2011, 124, 1951–1956. [Google Scholar] [PubMed]
- Kyler, K.E.; Jones, B.L. The chicken or the egg: The role of IL-6 in pediatric obese and allergen-exposed asthma. J. Allergy Clin. Immunol. 2023, 152, 1420–1422. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.S. Tumour necrosis factor-alpha: The role of this multifunctional cytokine in asthma. Immunol. Cell Biol. 2001, 79, 132–140. [Google Scholar] [CrossRef]
- Calderón, M.A.; Devalia, J.L.; Prior, A.J.; Sapsford, R.J.; Davies, R.J. A comparison of cytokine release from epithelial cells cultured from nasal biopsy specimens of atopic patients with and without rhinitis and nonatopic subjects without rhinitis. J. Allergy Clin. Immunol. 1997, 99, 65–76. [Google Scholar] [CrossRef]
- Odamaki, T.; Xiao, J.Z.; Iwabuchi, N.; Sakamoto, M.; Takahashi, N.; Kondo, S.; Iwatsuki, K.; Kokubo, S.; Togashi, H.; Enomoto, T.; et al. Fluctuation of fecal microbiota in individuals with Japanese cedar pollinosis during the pollen season and influence of probiotic intake. J. Investig. Allergol. Clin. Immunol. 2007, 17, 92–100. [Google Scholar]
- Tesmer, L.A.; Lundy, S.K.; Sarkar, S.; Fox, D.A. Th17 cells in human disease. Immunol. Rev. 2008, 223, 87–113. [Google Scholar] [CrossRef]
- Lajoie, S.; Lewkowich, I.P.; Suzuki, Y.; Clark, J.R.; Sproles, A.A.; Dienger, K.; Budelsky, A.L.; Wills-Karp, M. Complement-mediated regulation of the IL-17A axis is a central genetic determinant of the severity of experimental allergic asthma. Nat. Immunol. 2010, 11, 928–935. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Lopes, J.E.; Chong, M.M.; Ivanov, I.I.; Min, R.; Victora, G.D.; Shen, Y.; Du, J.; Rubtsov, Y.P.; Rudensky, A.Y.; et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature 2008, 453, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sheng, A.; Jia, X.; Zeng, Z.; Zhang, X.; Zhao, W.; Zhang, W. Th17/Treg dysregulation in allergic asthmatic children is associated with elevated notch expression. J. Asthma 2018, 55, 1–7. [Google Scholar] [CrossRef]
- Zhang, D.; Li, S.; Wang, N.; Tan, H.Y.; Zhang, Z.; Feng, Y. The Cross-Talk Between Gut Microbiota and Lungs in Common Lung Diseases. Front. Microbiol. 2020, 11, 301. [Google Scholar] [CrossRef] [PubMed]
- Hufnagl, K.; Pali-Schöll, I.; Roth-Walter, F.; Jensen-Jarolim, E. Dysbiosis of the gut and lung microbiome has a role in asthma. Semin. Immunopathol. 2020, 42, 75–93. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.L.; Gold, M.J.; Hartmann, M.; Willing, B.P.; Thorson, L.; Wlodarska, M.; Gill, N.; Blanchet, M.R.; Mohn, W.W.; McNagny, K.M.; et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012, 13, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Duan, W.; Guan, T.; Zhou, Q.; Yan, W.; Geng, Y. Water extract of Pingchuan formula ameliorated murine asthma through modulating metabolites and gut microbiota. J. Pharm. Biomed. Anal. 2023, 236, 115728. [Google Scholar] [CrossRef]
- Salamon, D.; Gosiewski, T.; Krawczyk, A.; Sroka-Oleksiak, A.; Duplaga, M.; Fyderek, K.; Kowalska-Duplaga, K. Quantitative changes in selected bacteria in the stool during the treatment of Crohn’s disease. Adv. Med. Sci. 2020, 65, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.J.; Zhang, Q.Y.; Wang, L.H.; Zhang, C.R.; Li, Y.; Zhang, Y.G. Growth performance, digestibility, blood metabolites, ruminal fermentation, and bacterial communities in response to the inclusion of gallic acid in the starter feed of preweaning dairy calves. J. Dairy Sci. 2022, 105, 3078–3089. [Google Scholar] [CrossRef] [PubMed]
- Oka, A.; Sartor, R.B. Microbial-Based and Microbial-Targeted Therapies for Inflammatory Bowel Diseases. Dig. Dis. Sci. 2020, 65, 757–788. [Google Scholar] [CrossRef] [PubMed]
- Durack, J.; Kimes, N.E.; Lin, D.L.; Rauch, M.; McKean, M.; McCauley, K.; Panzer, A.R.; Mar, J.S.; Cabana, M.D.; Lynch, S.V. Delayed gut microbiota development in high-risk for asthma infants is temporarily modifiable by Lactobacillus supplementation. Nat. Commun. 2018, 9, 707. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Zhang, B.; Yu, Z.; Hu, Y.; Lv, H.; Ji, X.; Wang, J.; Peng, B.; Wang, S. Ameliorative Effect of Dietary Tryptophan on Neurodegeneration and Inflammation in d-Galactose-Induced Aging Mice with the Potential Mechanism Relying on AMPK/SIRT1/PGC-1α Pathway and Gut Microbiota. J. Agric. Food Chem. 2021, 69, 4732–4744. [Google Scholar] [CrossRef]
- Demirci, M.; Tokman, H.B.; Uysal, H.K.; Demiryas, S.; Karakullukcu, A.; Saribas, S.; Cokugras, H.; Kocazeybek, B.S. Reduced Akkermansia muciniphila and Faecalibacterium prausnitzii levels in the gut microbiota of children with allergic asthma. Allergol. Immunopathol. 2019, 47, 365–371. [Google Scholar] [CrossRef]
- Su, X.; Gao, Y.; Yang, R. Gut Microbiota-Derived Tryptophan Metabolites Maintain Gut and Systemic Homeostasis. Cells 2022, 11, 2296. [Google Scholar] [CrossRef] [PubMed]
- Rankin, L.C.; Kaiser, K.A.; de Los Santos-Alexis, K.; Park, H.; Uhlemann, A.C.; Gray, D.H.D.; Arpaia, N. Dietary tryptophan deficiency promotes gut RORγt(+) Treg cells at the expense of Gata3(+) Treg cells and alters commensal microbiota metabolism. Cell Rep. 2023, 42, 112135. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Jin, L.; Zeng, J.; Wang, J.; Zhong, S.; Fan, W.; Liao, W. Tryptophan metabolite-regulated Treg responses contribute to attenuation of airway inflammation during specific immunotherapy in a mouse asthma model. Hum. Vaccin. Immunother. 2020, 16, 1891–1899. [Google Scholar] [CrossRef]
- Losol, P.; Sokolowska, M.; Chang, Y.S. Interactions between microbiome and underlying mechanisms in asthma. Respir. Med. 2023, 208, 107118. [Google Scholar] [CrossRef] [PubMed]
- Goudot, C.; Coillard, A.; Villani, A.C.; Gueguen, P.; Cros, A.; Sarkizova, S.; Tang-Huau, T.L.; Bohec, M.; Baulande, S.; Hacohen, N.; et al. Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages. Immunity 2017, 47, 582–596.e6. [Google Scholar] [CrossRef] [PubMed]
- Sadik, A.; Somarribas Patterson, L.F.; Öztürk, S.; Mohapatra, S.R.; Panitz, V.; Secker, P.F.; Pfänder, P.; Loth, S.; Salem, H.; Prentzell, M.T.; et al. IL4I1 Is a Metabolic Immune Checkpoint that Activates the AHR and Promotes Tumor Progression. Cell 2020, 182, 1252–1270.e34. [Google Scholar] [CrossRef]
- Poulain-Godefroy, O.; Bouté, M.; Carrard, J.; Alvarez-Simon, D.; Tsicopoulos, A.; de Nadai, P. The Aryl Hydrocarbon Receptor in Asthma: Friend or Foe? Int. J. Mol. Sci. 2020, 21, 8797. [Google Scholar] [CrossRef]
- Akkoc, T.; O’Mahony, L.; Ferstl, R.; Akdis, C.; Akkoc, T. Mouse Models of Asthma: Characteristics, Limitations and Future Perspectives on Clinical Translation. In Cell Biology and Translational Medicine, Volume 15: Stem Cells in Tissue Differentiation, Regulation and Disease; Turksen, K., Ed.; Springer International Publishing: Cham, Switzerland, 2022; pp. 119–133. [Google Scholar]

| Score | 1 (Mild) | 2 (Moderate) | 3 (Severe) |
|---|---|---|---|
| Nasal itching | Slight scratching of the nose with the front paws (<10 times) | Scratching the nose (between slight scratching and rubbing around) | Severe scratching of the nose (rubbing around) |
| Sneeze number | 1–3 | 4–10 | ≥11 |
| Runny nose | Flowing to the front nostril | Flowing beyond the front nostrils | Flowing all over the face |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; He, Y.; Dang, D.; Zhao, Y.; Zhao, J.; Lu, W. Gut Microbiota-Derived Tryptophan Metabolites Alleviate Allergic Asthma Inflammation in Ovalbumin-Induced Mice. Foods 2024, 13, 1336. https://doi.org/10.3390/foods13091336
Wang H, He Y, Dang D, Zhao Y, Zhao J, Lu W. Gut Microbiota-Derived Tryptophan Metabolites Alleviate Allergic Asthma Inflammation in Ovalbumin-Induced Mice. Foods. 2024; 13(9):1336. https://doi.org/10.3390/foods13091336
Chicago/Turabian StyleWang, Hongchao, Yuan He, Danting Dang, Yurong Zhao, Jianxin Zhao, and Wenwei Lu. 2024. "Gut Microbiota-Derived Tryptophan Metabolites Alleviate Allergic Asthma Inflammation in Ovalbumin-Induced Mice" Foods 13, no. 9: 1336. https://doi.org/10.3390/foods13091336





