Isolation and Characterisation of Streptococcus spp. with Human Milk Oligosaccharides Utilization Capacity from Human Milk
Abstract
:1. Introduction
2. Materials and Methods
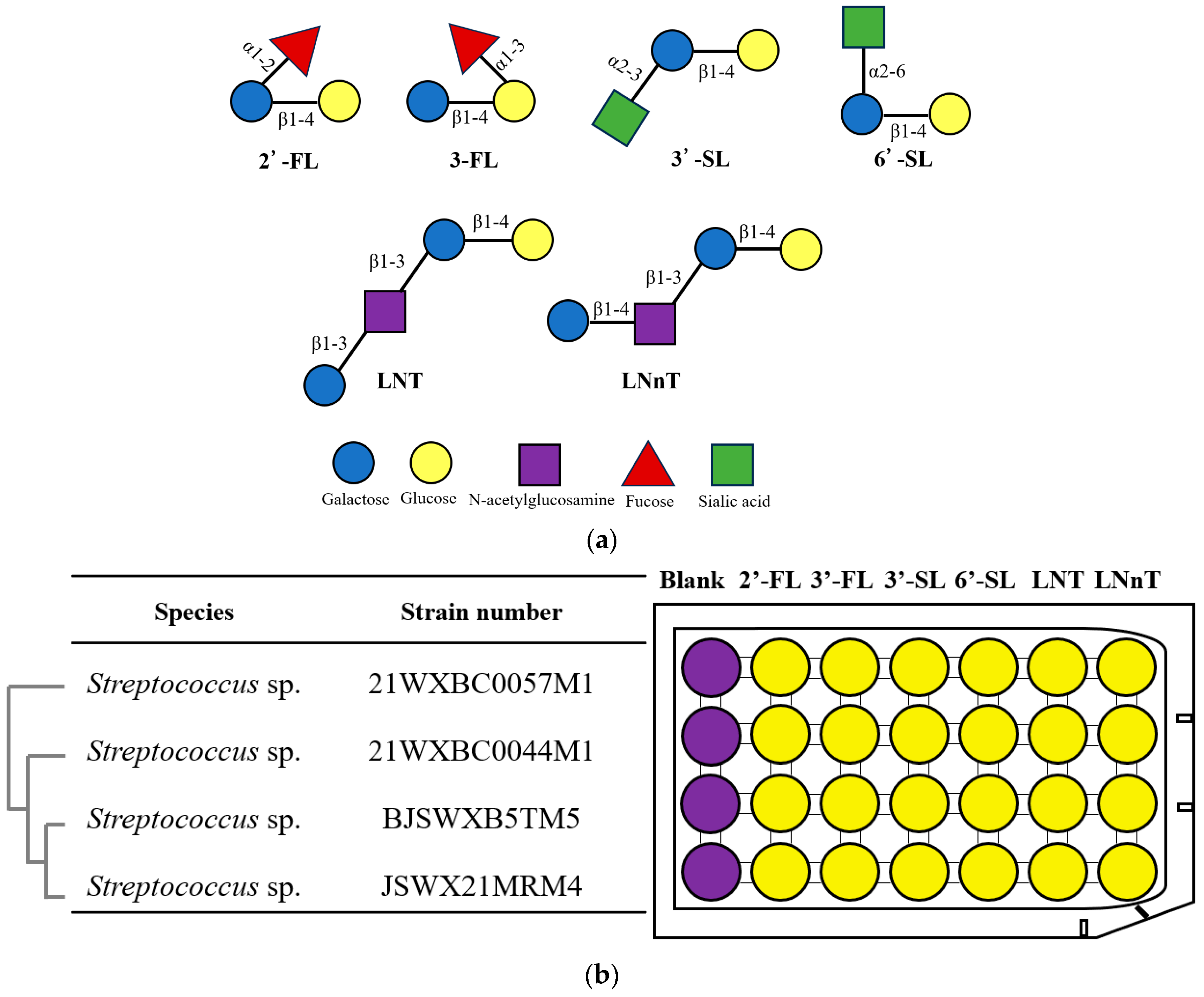
2.1. Isolation and Cultivation of the Strains
2.2. Taxonomic Identification
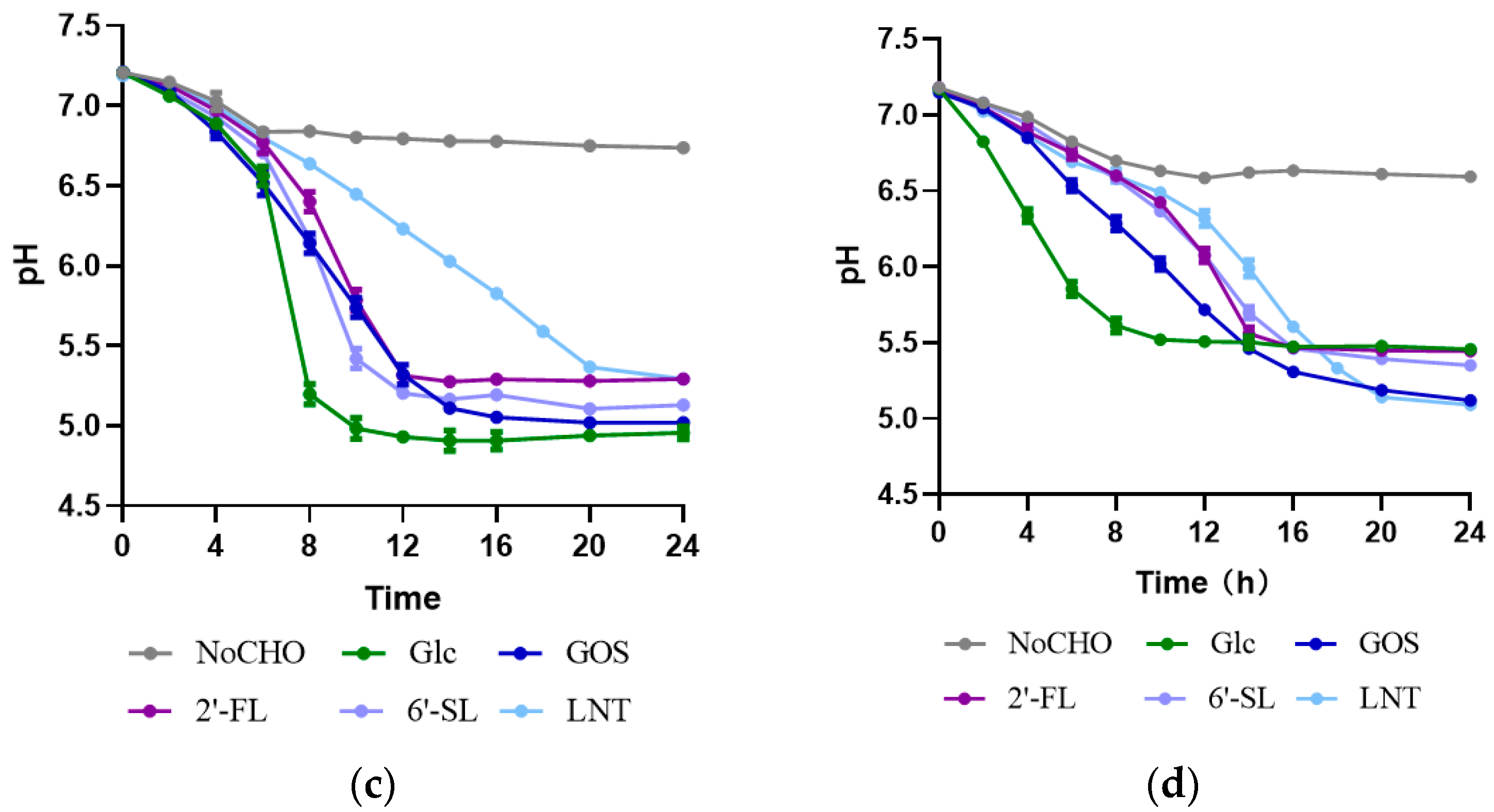
2.3. HMO Fermentation and Growth Curve
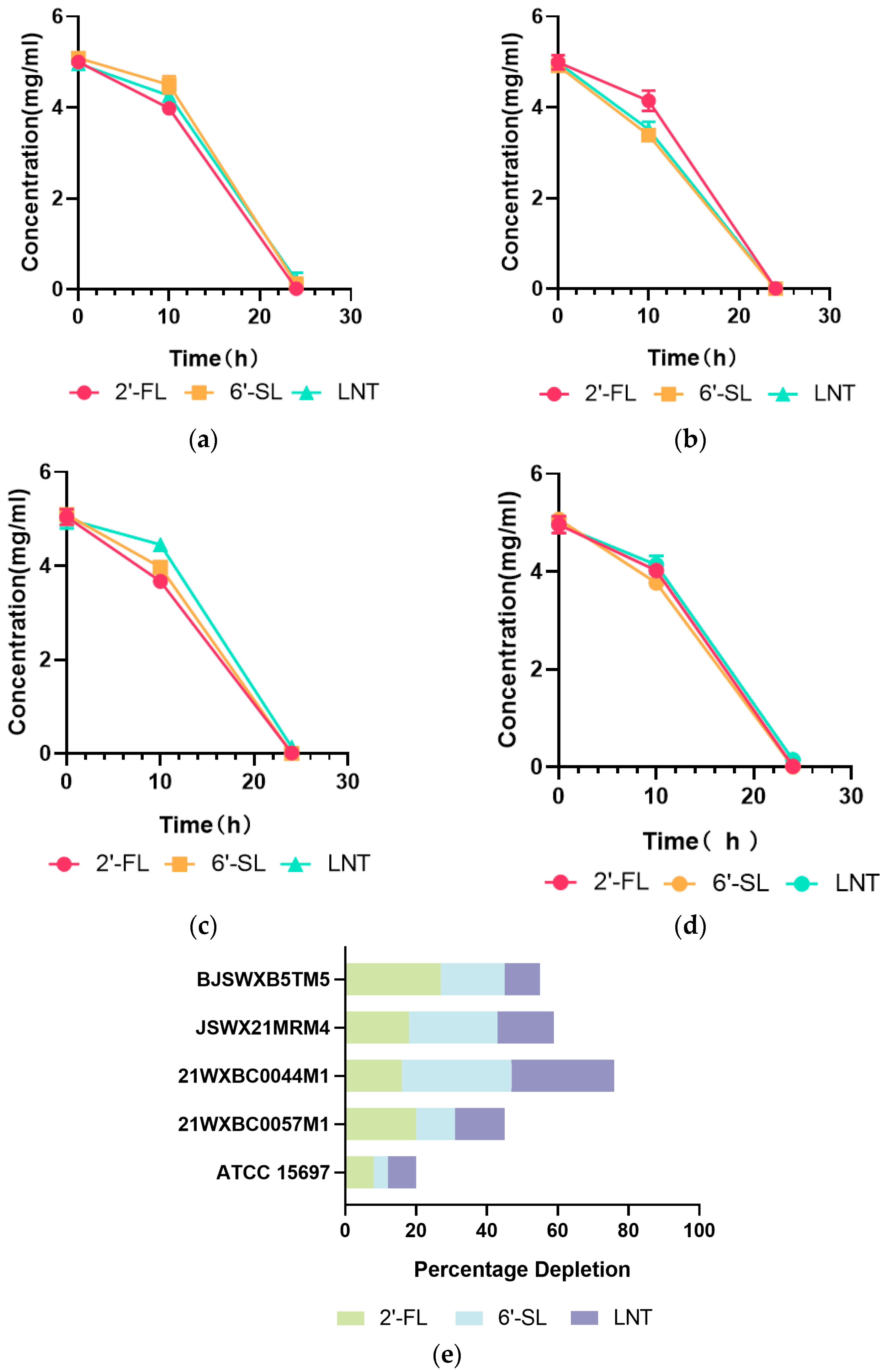
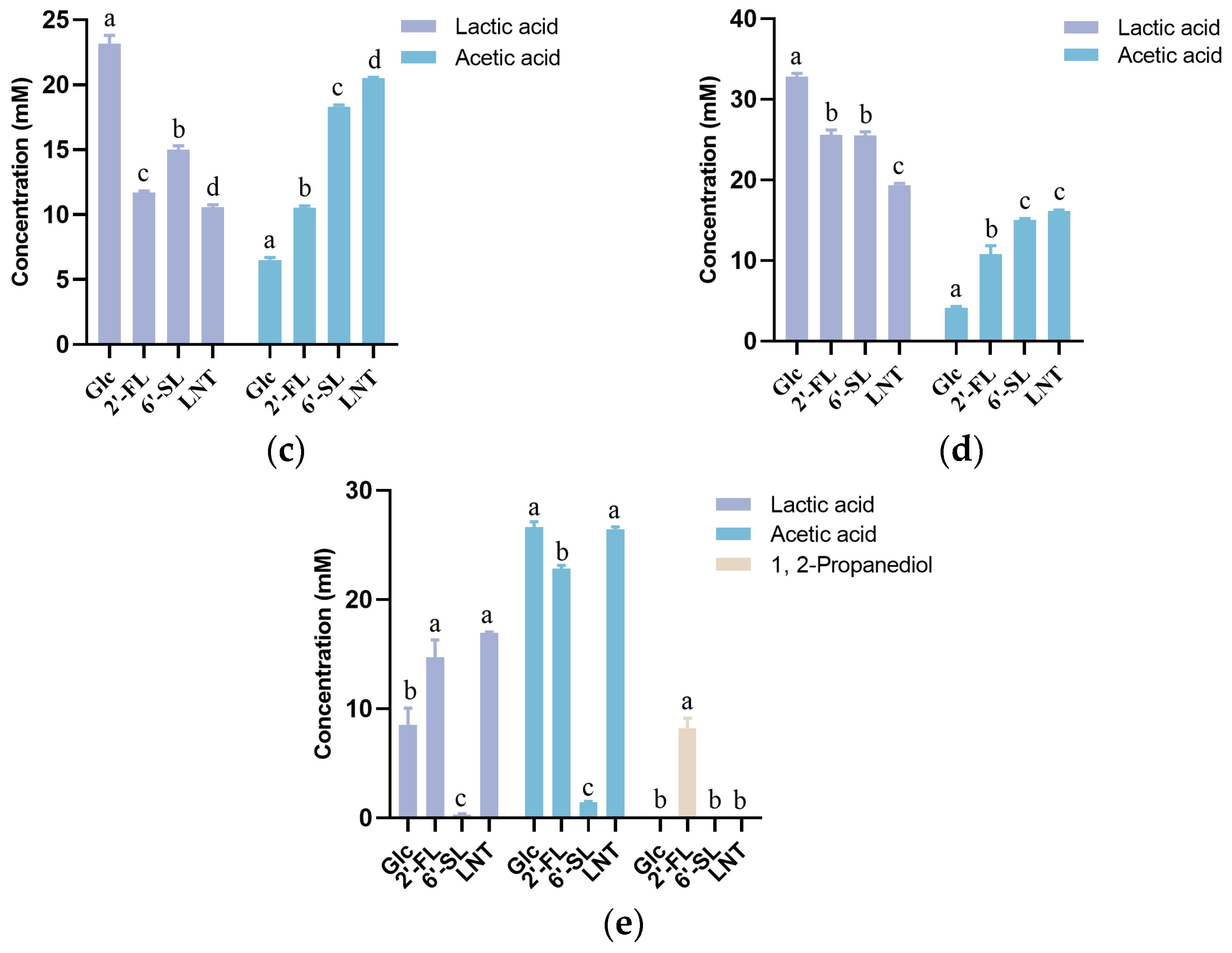
2.4. HMO Consumption and Metabolite Quantitation
2.5. Genome Mining Analysis
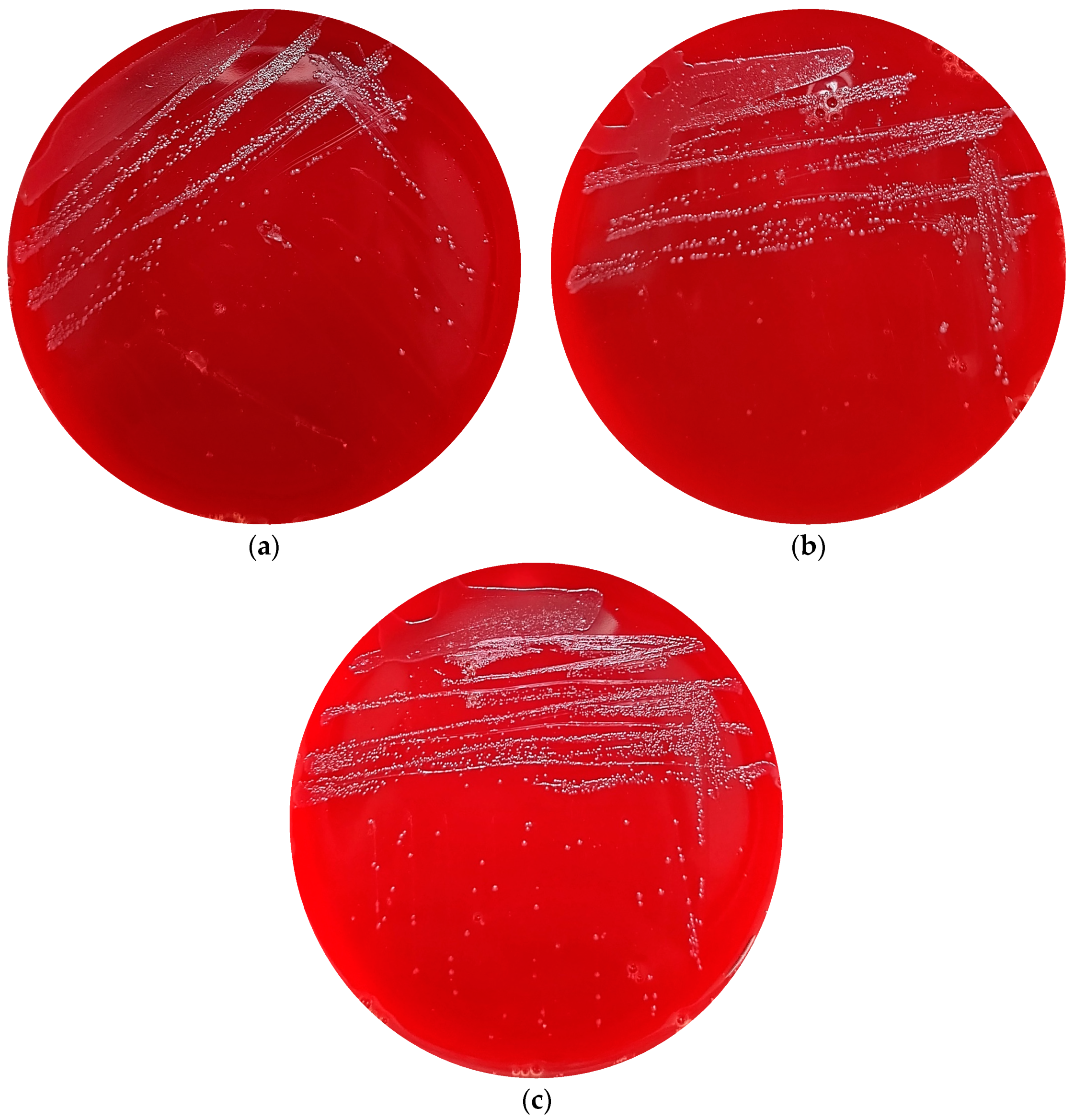
2.6. Haemolysis Activity
3. Results
3.1. Isolation of Streptococcus spp. with HMO Utilization Capability
3.2. Characteristics of HMO Utilization by Streptococcus
3.3. In Silico and In Vitro Safety Assessment of the Three Undefined Streptococci
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koenig, J.E.; Spor, A.; Scalfone, N.; Fricker, A.D.; Stombaugh, J.; Knight, R.; Angenent, L.T.; Ley, R.E. Succession of Microbial Consortia in the Developing Infant Gut Microbiome. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4578–4585. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Ding, J.; Ren, H.; Xin, Q.; Li, Z.; Han, L.; Liu, D.; Zhuo, Z.; Liu, C.; Ren, Z. Distinguishable Influence of the Delivery Mode, Feeding Pattern, and Infant Sex on Dynamic Alterations in the Intestinal Microbiota in the First Year of Life. Microb. Ecol. 2023, 86, 1799–1813. [Google Scholar] [CrossRef] [PubMed]
- Kunz, C.; Rudloff, S.; Baier, W.; Klein, N.; Strobel, S. Oligosaccharides in Human Milk: Structural, Functional, and Metabolic Aspects. Ann. Rev. Nutr. 2000, 20, 699–722. [Google Scholar] [CrossRef] [PubMed]
- Bode, L. Human Milk Oligosaccharides: Structure and Functions. Nestle. Nutr. Inst. Workshop Ser. 2020, 94, 115–123. [Google Scholar] [PubMed]
- Nolan, L.S.; Rimer, J.M.; Good, M. The Role of Human Milk Oligosaccharides and Probiotics on the Neonatal Microbiome and Risk of Necrotizing Enterocolitis: A Narrative Review. Nutrients 2020, 12, 3052. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, M.; Wang, R.; Hu, J.; Liu, S.; Wang, Y.; Xing, X.; Zhang, B.; Liu, J.; Wang, S. Current Advances in Structure-Function Relationships and Dose-Dependent Effects of Human Milk Oligosaccharides. J. Agric. Food. Chem. 2022, 70, 6328–6353. [Google Scholar] [CrossRef]
- Moya-Gonzálvez, E.M.; Rubio-Del-Campo, A.; Rodriguez-Diaz, J.; Yebra, M.J. Infant-gut associated Bifidobacterium dentium Strains Utilize the Galactose Moiety and Release Lacto-N-triose from the Human Milk Oligosaccharides Lacto-N-tetraose and Lacto-N-neotetraose. Sci. Rep. 2021, 11, 23328. [Google Scholar] [CrossRef]
- Yu, Z.; Chen, C.; Kling, D.E.; Liu, B.; Mccoy, J.M.; Merighi, M.; Heidtman, M.; Newburg, D.S. The Principal Fucosylated Oligosaccharides of Human Milk Exhibit Prebiotic Properties on Cultured Infant Microbiota. Glycobiology 2013, 23, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Marcobal, A.; Barboza, M.; Sonnenburg, E.D.; Pudlo, N.; Martens, E.C.; Desai, P.; Lebrilla, C.B.; Weimer, B.C.; Mills, D.A.; German, J.B.; et al. Bacteroides in the Infant Gut Consume Milk Oligosaccharides via Mucus-Utilization Pathways. Cell Host Microbe 2011, 10, 507–514. [Google Scholar] [CrossRef]
- Pichler, M.J.; Yamada, C.; Shuoker, B.; Alvarez-Silva, C.; Gotoh, A.; Leth, M.L.; Schoof, E.; Katoh, T.; Sakanaka, M.; Katayama, T.; et al. Butyrate Producing Colonic Clostridiales Metabolise Human Milk Oligosaccharides and Cross Feed on Mucin via Conserved Pathways. Nat. Commun. 2020, 11, 3285. [Google Scholar] [CrossRef]
- Ward, R.E.; Ninonuevo, M.; Mills, D.A.; Lebrilla, C.B.; German, J.B. In vitro Fermentation of Breast Milk Oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl. Environ. Microbiol. 2006, 72, 4497–4499. [Google Scholar] [CrossRef] [PubMed]
- Reddel, S.; Pascucci, G.R.; Foligno, S.; Del Chierico, F.; Vernocchi, P.; Marzullo, A.; Pattumelli, M.G.; Palma, P.; Salvatori, G.; Putignani, L. A Parallel Tracking of Salivary and Gut Microbiota Profiles can Reveal Maturation and Interplay of Early Life Microbial Communities in Healthy Infants. Microorganisms 2022, 10, 468. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.M.; Engevik, M.; Aagaard, K. Human Milk Oligosaccharides (Hmos) Promote Growth of Commensal Streptococcus spp. Abundant in Human Milk. Am. J. Obstet. Gynecol. 2019, 220, S605–S606. [Google Scholar] [CrossRef]
- Bakir, M.A.; Kitahara, M.; Sakamoto, M.; Matsumoto, M.; Benno, Y. Bacteroides finegoldii sp. nov., Isolated from Human Faeces. Int. J. Syst. Evol. Microbiol. 2006, 56, 931–935. [Google Scholar] [CrossRef] [PubMed]
- Underwood, M.A.; German, B.J.; Lebrilla, C.B.; Mills, D.A. Bifidobacterium longum subspecies infantis: Champion Colonizer of the Infant Gut. Pediatr. Res. 2015, 77, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Perni, S.; Andrew, P.W.; Shama, G. Estimating the Maximum Growth Rate from Microbial Growth Curves: Definition is Everything. Food Microbiol. 2005, 22, 491–495. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, T.; Chen, X.; Pang, X.; Zhang, S.; Ujiroghene, O.J.; Jiang, S.; Lu, J.; Lv, J. Absolute Quantification of Twelve Oligosaccharides in Human Milk using a Targeted Mass Spectrometry-based Approach. Carbohydr. Polym. 2019, 219, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An Empirically Improved Memory-Efficient Short-Read de novo Assembler. GigaScience 2012, 1, 2047–2217. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Ya, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A Reference Database for Bacterial Virulence Factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef]
- Horvath, P.; Barrangou, R. CRISPR/Cas, The Immune System of Bacteria and Archaea. Science. 2010, 327, 167–170. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.Y.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2016, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Sela, D.A.; Chapman, J.; Adeuya, A.; Kim, J.H.; Chen, F.; Whitehead, T.R.; Lapidus, A.; Rokhsar, D.S.; Lebrilla, C.B.; German, J.B.; et al. The Genome Sequence of Bifidobacterium longum subsp. infantis Reveals Adaptations for Milk Utilization within the Infant Microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 18964–18969. [Google Scholar] [PubMed]
- Dedon, L.R.; Özcan, E.; Rani, A.; Sela, D.A. Bifidobacterium infantis Metabolizes 2′Fucosyllactose-Derived and Free Fucose Through a Common Catabolic Pathway Resulting in 1,2-Propanediol Secretion. Front. Nutr. 2020, 7, 583397. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.K.; Dhasmana, N.; Dubey, N.; Kumar, N.; Gangwal, A.; Gupta, M.; Singh, Y. Bacterial Virulence Factors: Secreted for Survival. Indian J. Microbiol. 2017, 57, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.P.; Guo, T.T.; Mu, Y.L.; Kong, J. Identification and Functional Analysis of Potential Prophage-Derived Recombinases for Genome Editing in Lactobacillus casei. FEMS Microbiol. Lett. 2017, 364, fnx243. [Google Scholar] [CrossRef]
- Di Pierro, F.; Campedelli, I.; De Marta, P.; Fracchetti, F.; Del Casale, A.; Cavecchia, I.; Matera, M.; Cazzaniga, M.; Bertuccioli, A.; Guasti, L.; et al. Bifidobacterium breve PRL2020: Antibiotic-Resistant Profile and Genomic Detection of Antibiotic Resistance Determinants. Microorganisms. 2023, 11, 1649. [Google Scholar] [CrossRef] [PubMed]
- Dutta, I.; Reynolds, P.E. The vanC-3 Vancomycin Resistance Gene Cluster of Enterococcus flavescens CCM 439. J. Antimicrob. Chemother. 2003, 51, 703–706. [Google Scholar] [CrossRef] [PubMed]
- WHO; FAO. Report of a Joint FAO/WHO Expert Consultation on Guidelines for the Evaluation of Probiotics in Food; World Health Organization and Food and Agriculture Organization of the United Nations: London, ON, Canada, 2002. [Google Scholar]
- Halder, D.; Mandal, M.; Chatterjee, S.S.; Pal, N.K.; Mandal, S. Indigenous Probiotic Lactobacillus Isolates Presenting Antibiotic like Activity against Human Pathogenic Bacteria. Biomedicines 2017, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Bisson, G.; Marino, M.; Poletti, D.; Innocente, N.; Maifreni, M. Turbidimetric Definition of Growth Limits in Probiotic Lactobacillus Strains from the Perspective of an Adaptation Strategy. J. Dairy Sci. 2021, 104, 12236–12248. [Google Scholar] [CrossRef]
- Supriya, Y.; Snehal, G.; Smita, N.; Vaishali, A. Effect of Supplementation of Micronutrients and Phytochemicals to Fructooligosaccharides on Growth Response of Probiotics and, E. coli. Biofactors 2015, 37, 58–64. [Google Scholar]
- Zabel, B.E.; Gerdes, S.; Evans, K.C.; Nedveck, D.; Singles, S.K.; Volk, B.; Budinoff, C. Strain-Specific Strategies of 2′-fucosyllactose, 3-fucosyllactose, and Difucosyllactose Assimilation by Bifidobacterium longum subsp. infantis Bi-26 and ATCC 15697. Sci. Rep. 2020, 10, 15919. [Google Scholar] [PubMed]
- Marcobal, A.; Barboza, M.; Froehlich, J.W.; Block, D.E.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Consumption of Human Milk Oligosaccharides by Gut-Related Microbes. J. Agric. Food Chem. 2010, 58, 5334–5340. [Google Scholar] [CrossRef] [PubMed]
- LoCascio, R.G.; Niñonuevo, M.R.; Kronewitter, S.R.; Freeman, S.L.; German, J.B.; Lebrilla, C.B.; Mills, D.A. A Versatile and Scalable Strategy for Glycoprofiling Bifidobacterial Consumption of Human Milk Oligosaccharides. Microb. Biotechnol. 2009, 2, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Jae-Han, K.; Hyun, J.A.; Garrido, D.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Proteomic Analysis of Bifidobacterium longum subsp. infantis Reveals the Metabolic Insight on Consumption of Prebiotics and Host Glycans. PLoS ONE 2013, 8, e57535. [Google Scholar]
- Tsukuda, N.; Yahagi, K.; Hara, T.; Watanabe, Y.; Matsumoto, H.; Mori, H.; Higashi, K.; Tsuji, H.; Matsumoto, S.; Kurokawa, K.; et al. Key Bacterial Taxa and Metabolic Pathways Affecting Gut Short-Chain Fatty Acid Profiles in Early Life. ISME J. 2021, 16 (Suppl. 1), 2574–2590. [Google Scholar] [CrossRef]
- Morrison, D.J.; Preston, T. Formation of Short Chain Fatty Acids by the Gut Microbiota and their Impact on Human Metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef]
- Barnett, T.C.; Cole, J.N.; Rivera-Hernandez, T.; Henningham, A.; Paton, J.C.; Nizet, V.; Walker, M.J. Streptococcal Toxins: Role in Pathogenesis and Disease. Cell Microbiol. 2015, 17, 1721–1741. [Google Scholar] [CrossRef]
- Tettelin, H.; Nelson, K.E.; Paulsen, I.T.; Eisen, J.A.; Read, T.D.; Peterson, S.; Heidelberg, J.; DeBoy, R.T.; Haft, D.H.; Dodson, R.J.; et al. Complete Genome Sequence of a Virulent Isolate of Streptococcus pneumoniae. Science 2001, 293, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Ajdic, D.; McShan, W.M.; McLaughlin, R.E.; Savic, G.; Chang, J.; Carson, M.B.; Primeaux, C.; Tian, R.Y.; Kenton, S.; Jia, H.G.; et al. Genome Sequence of Streptococcus mutans UA159, a Cariogenic Dental Pathogen. Proc. Natl. Acad. Sci. USA 2002, 99, 14434–14439. [Google Scholar] [CrossRef]
- Kachroo, P.; Eraso, J.M.; Beres, S.B.; Olsen, R.J.; Zhu, L.C.; Nasser, W.; Bernard, P.E.; Cantu, C.C.; Saavedra, M.O.; Arredondo, M.J.; et al. Integrated Analysis of Population Genomics, Transcriptomics and Virulence Provides Novel Insights into Streptococcus pyogenes Pathogenesis. Nat. Genet 2019, 51, 548–559. [Google Scholar] [CrossRef]
- Bolotin, A.; Quinquis, B.; Renault, P.; Sorokin, A.; Ehrlich, S.D.; Kulakauskas, S.; Lapidus, A.; Goltsman, E.; Mazur, M.; Pusch, G.D.; et al. Complete Sequence and Comparative Genome Analysis of the Dairy Bacterium Streptococcus thermophilus. Nat. Biotechnol. 2004, 22, 1554–1558. [Google Scholar] [CrossRef] [PubMed]
- Senan, S.; Prajapati, J.B.; Joshi, C.G. Feasibility of Genome-Wide Screening for Biosafety Assessment of Probiotics: A Case Study of Lactobacillus helveticus MTCC 5463. Probiotics Antimicrob. Proteins 2015, 7, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Palma, T.H.; Harth-Chú, E.N.; Scott, J.; Stipp, R.N.; Boisvert, H.; Salomao, M.F.; Theobaldo, J.D.; Possobon, R.F.; Nascimento, L.C.; McCafferty, J.W. The Oral Cavities of Healthy Infants Harbor High Proportions of Streptococcus salivarius Strains with Phenotypic and Genotypic Resistance to Multiple Classes of Antibiotics. J. Med. Microbiol. 2016, 65, 1456–1464. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.M.; Li, L.L.; Kromann, S.; Chen, M.R.; Shi, L.; Meng, H.C. Antibiotic Resistance of Lactobacillus spp. and Streptococcus thermophilus Isolated from Chinese Fermented Milk Products. Foodborne Pathog. Dis. 2019, 16, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Teng, D.; Mao, R.Y.; Hao, Y.; Wang, X.M.; Wang, J.H. A Critical Review of Antibiotic Resistance in Probiotic Bacteria. Food Res. Int. 2020, 136, 109571. [Google Scholar] [CrossRef]
- Jose, N.M.; Bunt, C.R.; Hussain, M.A. Implications of Antibiotic Resistance in Probiotics. Food Rev. Int. 2015, 31, 52–62. [Google Scholar] [CrossRef]



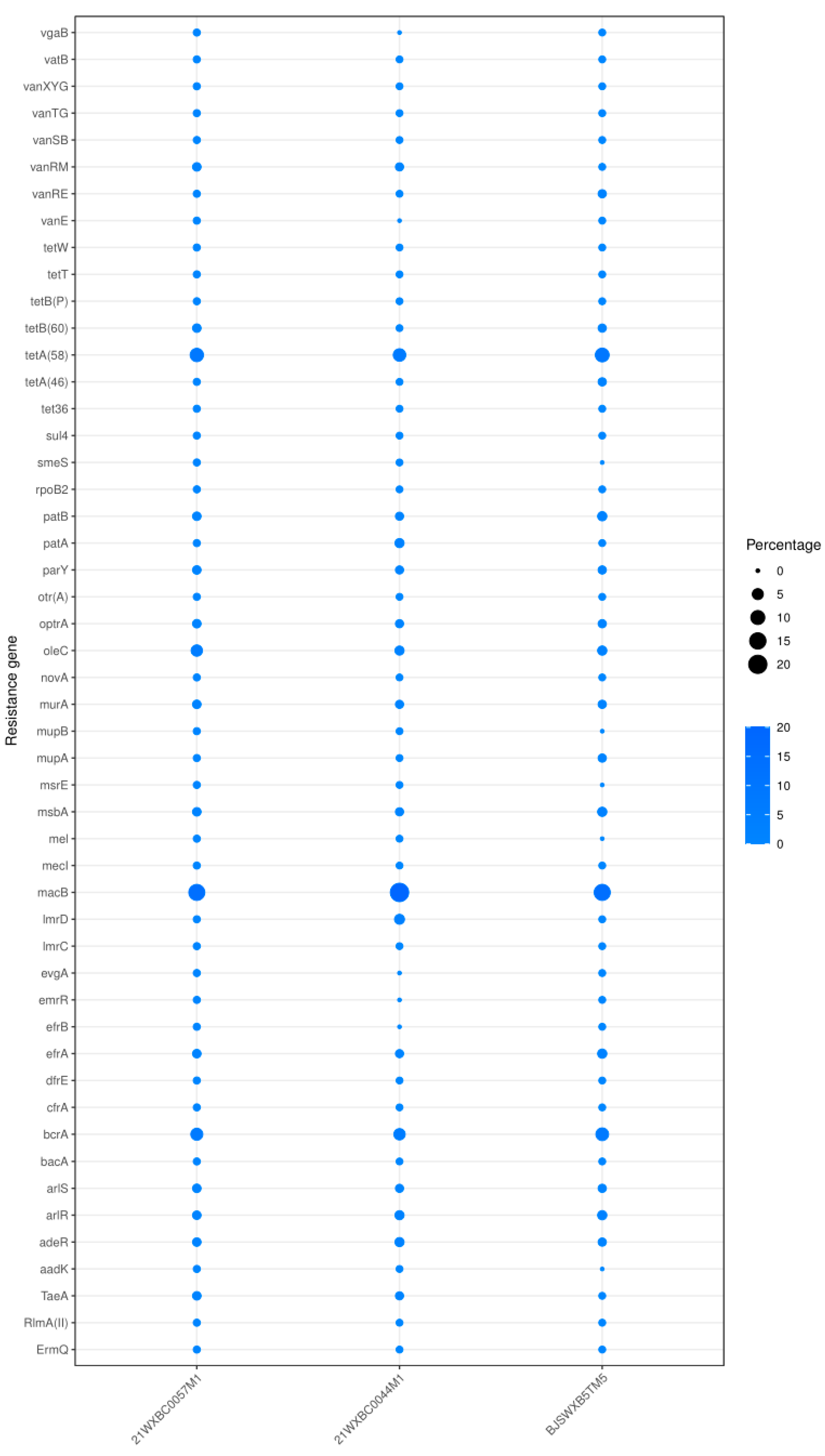
| Strain | VFDB ID | Gene Name | VFs | Identity (%) | Description |
|---|---|---|---|---|---|
| 21WXBC0057M1 | VFG001359 | psaA | PsaA | 91 | Manganese ABC transporter |
| VFG005197 | pavA | PavA | 89.4 | Adherence and virulence protein | |
| VFG000964 | hasC | Hyaluronic acid capsule | 86.8 | Prevents phagocytosis | |
| VFG001365-VFG001368 | cps4A, cps4B, cpsC, cps4D | Capsule | 73.5–74.7 | Resistant to complement deposition | |
| 21WXBC0044M1 | VFG042972 VFG042974 | sipA, srtG1 | PI-2 | >87.7 | Mediates host cell adhesion |
| VFG005197 | pavA | PavA | 94.2 | Adherence and virulence protein | |
| VFG001359 | psaA | PsaA | 93.2 | Manganese ABC transporter | |
| VFG001365- VFG001368 | cps4A, cps4B, cpsC, cps4D | Capsule | 86.4 | Resistant to complement | |
| VFG000964 | hasC | Hyaluronic acid capsule | 79.1 | Prevents phagocytosis | |
| VFG048830 | gndA | Capsule | 70.6 | Protects bacteria from opsonophagocytosis | |
| BJSWXB5TM5 | VFG001359 | psaA | PsaA | 93.5 | Manganese ABC transporter |
| VFG005197 | pavA | PavA | 89.4 | Adherence and virulence protein | |
| VFG000964 | hasC | Hyaluronic acid capsule | 86.5 | Prevents phagocytosis | |
| VFG001378 | / | Neuraminidase | 70 | Contributes to increased adhesion |
| Strain Number | Length | Gene Numbers | The Most Common Prophage | GC Content (%) |
|---|---|---|---|---|
| 21WXBC0057M1 | 32.1Kb | 140 | PHAGE_Strept_PH15_NC_010945(11) | 41.42 |
| BJSWXB5TM5 | 58.7Kb | 140 | PHAGE_Strept_phi3396_NC_009018(10) | 40.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Liu, X.; Chen, H.; Zhao, J.; Zhang, H.; Chen, W.; Yang, B. Isolation and Characterisation of Streptococcus spp. with Human Milk Oligosaccharides Utilization Capacity from Human Milk. Foods 2024, 13, 1291. https://doi.org/10.3390/foods13091291
Zhou Y, Liu X, Chen H, Zhao J, Zhang H, Chen W, Yang B. Isolation and Characterisation of Streptococcus spp. with Human Milk Oligosaccharides Utilization Capacity from Human Milk. Foods. 2024; 13(9):1291. https://doi.org/10.3390/foods13091291
Chicago/Turabian StyleZhou, Ye, Xiaoming Liu, Haiqin Chen, Jianxin Zhao, Hao Zhang, Wei Chen, and Bo Yang. 2024. "Isolation and Characterisation of Streptococcus spp. with Human Milk Oligosaccharides Utilization Capacity from Human Milk" Foods 13, no. 9: 1291. https://doi.org/10.3390/foods13091291





