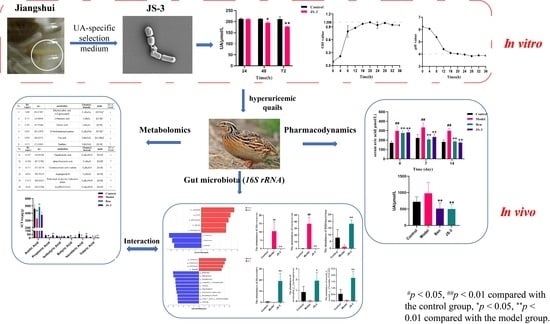
Lacticaseibacillus paracasei JS-3 Isolated from “Jiangshui” Ameliorates Hyperuricemia by Regulating Gut Microbiota and iTS Metabolism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Strains with Uric Acid Degradation Ability
2.2. Determination of Uric Acid Degradation Ability of Cultured Strains In Vitro
2.3. Strain Identification
2.4. Determination of Growth and pH Value of Bacterial Strains
2.5. Animal Treatment
2.6. Determination of UA Degradation by Quail Fecal Microbiota
2.7. 16S-rRNA Gene Sequencing
2.8. Detection of Short-Chain Fatty Acids (SCFAs) in Quail Feces
2.9. Non-Targeted Fecal Metabolomics Analysis
2.10. Statistical Analysis
3. Results
3.1. Screening, Isolation, and Identification of UA-Lowering Strains
3.2. JS-3 Decreases UA Level in Hyperuricemic Quails
3.3. JS-3 Regulated UA-Induced Gut Microbiota Dysbiosis in Hyperuricemic Quails
3.4. JS-3 Increased SCFA Levels in the Gut of Hyperuricemic Quails
3.5. Effect of JS-3 on the Fecal Metabolome
3.6. Correlations between Gut Microbiota, Metabolome, and Hyperuricemia-Related Parameters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Hong, F.; Zheng, A.; Xu, P.; Wang, J.; Xue, T.; Dai, S.; Pan, S.; Guo, Y.; Xie, X.; Li, L.; et al. High-Protein Diet Induces Hyperuricemia in a New Animal Model for Studying Human Gout. Int. J. Mol. Sci. 2020, 21, 2147. [Google Scholar] [CrossRef] [PubMed]
- Hafez, R.M.; Abdel-Rahman, T.M.; Naguib, R.M. Uric acid in plants and microorganisms: Biological applications and genetics—A review. J. Adv. Res. 2017, 8, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Oda, M.; Satta, Y.; Takenaka, O.; Takahata, N. Loss of urate oxidase activity in hominoids and its evolutionary implications. Mol. Biol. Evol. 2002, 19, 640–653. [Google Scholar] [CrossRef] [PubMed]
- Shan, R.; Ning, Y.; Ma, Y.; Gao, X.; Zhou, Z.; Jin, C.; Wu, J.; Lv, J.; Li, L. Incidence and Risk Factors of Hyperuricemia among 2.5 Million Chinese Adults during the Years 2017–2018. Int. J. Environ. Res. Public Health 2021, 18, 2360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhu, X.; Wu, J.; Huang, Z.; Zhao, Z.; Zhang, X.; Xue, Y.; Wan, W.; Li, C.; Zhang, W.; et al. Prevalence of Hyperuricemia Among Chinese Adults: Findings From Two Nationally Representative Cross-Sectional Surveys in 2015–16 and 2018–19. Front. Immunol. 2021, 12, 791983. [Google Scholar] [CrossRef] [PubMed]
- Chen-Xu, M.; Yokose, C.; Rai, S.K.; Pillinger, M.H.; Choi, H.K. Contemporary Prevalence of Gout and Hyperuricemia in the United States and Decadal Trends: The National Health and Nutrition Examination Survey, 2007–2016. Arthritis Rheumatol. 2019, 71, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, Y.; Zeng, C. Update on the epidemiology, genetics, and therapeutic options of hyperuricemia. Am. J. Transl. Res. 2020, 12, 3167–3181. [Google Scholar] [PubMed]
- Lima, W.G.; Martins-Santos, M.E.; Chaves, V.E. Uric acid as a modulator of glucose and lipid metabolism. Biochimie 2015, 116, 17–23. [Google Scholar] [CrossRef]
- Sorensen, L.B. Role of the intestinal tract in the elimination of uric acid. Arthritis Rheum. 1965, 8, 694–706. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, Y.; Liao, W.; Huang, J.; Liu, Y.; Li, Z.; Tang, J. Gut microbiota remodeling: A promising therapeutic strategy to confront hyperuricemia and gout. Front. Cell Infect. Microbiol. 2022, 12, 935723. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, K.; Aoyagi, Y.; Fukuuchi, T.; Inazawa, K.; Yamaoka, N. Total purine and purine base content of common foodstuffs for facilitating nutritional therapy for gout and hyperuricemia. Biol. Pharm. Bull. 2014, 37, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Granados, J.C.; Bhatnagar, V.; Nigam, S.K. Blockade of Organic Anion Transport in Humans After Treatment with the Drug Probenecid Leads to Major Metabolic Alterations in Plasma and Urine. Clin. Pharmacol. Ther. 2022, 112, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Stamp, L.K.; Barclay, M.L. How to prevent allopurinol hypersensitivity reactions? Rheumatology 2018, 57 (Suppl. S1), i35–i41. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Kojima, S.; Uchiyama, K.; Yokota, N.; Tokutake, E.; Wakasa, Y.; Hiramitsu, S.; Waki, M.; Jinnouchi, H.; Kakuda, H.; et al. Effect of febuxostat on clinical outcomes in patients with hyperuricemia and cardiovascular disease. Int. J. Cardiol. 2022, 349, 127–133. [Google Scholar] [CrossRef] [PubMed]
- White, W.B.; Saag, K.G.; Becker, M.A.; Borer, J.S.; Gorelick, P.B.; Whelton, A.; Hunt, B.; Castillo, M.; Gunawardhana, L. Cardiovascular Safety of Febuxostat or Allopurinol in Patients with Gout. N. Engl. J. Med. 2018, 378, 1200–1210. [Google Scholar] [CrossRef] [PubMed]
- Markowiak-Kopeć, P.; Śliżewska, K. The Effect of Probiotics on the Production of Short-Chain Fatty Acids by Human Intestinal Microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef] [PubMed]
- Morales-Ferré, C.; Azagra-Boronat, I.; Massot-Cladera, M.; Tims, S.; Knipping, K.; Garssen, J.; Knol, J.; Franch, À.; Castell, M.; Rodríguez-Lagunas, M.J.; et al. Effects of a Postbiotic and Prebiotic Mixture on Suckling Rats’ Microbiota and Immunity. Nutrients 2021, 13, 2975. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Sestito, S.; D’Auria, E.; Baldassarre, M.E.; Salvatore, S.; Tallarico, V.; Stefanelli, E.; Tarsitano, F.; Concolino, D.; Pensabene, L. The Role of Prebiotics and Probiotics in Prevention of Allergic Diseases in Infants. Front. Pediatr. 2020, 8, 583946. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Mei, L.; Deng, Y.; Liu, Y.; Wei, X.; Liu, M.; Zhou, J.; Ma, H.; Zheng, P.; Yuan, J.; et al. Lactobacillus brevis DM9218 ameliorates fructose-induced hyperuricemia through inosine degradation and manipulation of intestinal dysbiosis. Nutrition 2019, 62, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.W.; Hsieh, S.H.; Chen, J.F.; Liu, C.R.; Chen, C.W.; Huang, Y.F.; Ho, H.H. Lactobacillus reuteri TSR332 and Lactobacillus fermentum TSF331 stabilize serum uric acid levels and prevent hyperuricemia in rats. PeerJ 2021, 9, e11209. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.H.; Liu, T.T.; Liu, X.L.; Wang, C.H. Screening and identification of purine degrading Lactobacillus fermentum 9-4 from Chinese fermented rice-flour noodles. Food Sci. Human. Wellness 2022, 11, 1402–1408. [Google Scholar] [CrossRef]
- Fenster, K.; Freeburg, B.; Hollard, C.; Wong, C.; Rønhave Laursen, R.; Ouwehand, A.C. The Production and Delivery of Probiotics: A Review of a Practical Approach. Microorganisms 2019, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ye, Z.; Feng, P.; Li, R.; Chen, X.; Tian, X.; Han, R.; Kakade, A.; Liu, P.; Li, X. Limosilactobacillus fermentum JL-3 isolated from "Jiangshui" ameliorates hyperuricemia by degrading uric acid. Gut Microbes 2021, 13, 1897211. [Google Scholar] [CrossRef] [PubMed]
- Stojanović-Radić, Z.; Čomić, L.; Radulović, N.; Blagojević, P.; Mihajilov-Krstev, T.; Rajković, J. Commercial Carlinae radix herbal drug: Botanical identity, chemical composition and antimicrobial properties. Pharm. Biol. 2012, 50, 933–940. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-C.; Li, Y.-L.; Chiu, P.-Y.; Chen, C.; Chen, H.-C.; Chen, F.-A. Protective effects of corni fructus extract in mice with potassium oxonate–induced hyperuricemia. J. Vet. Med. Sci. 2022, 84, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Liu, Q.; Hao, H.; Bu, Y.; Tian, X.; Wang, T.; Yi, H. Lactobacillus paracasei X11 Ameliorates Hyperuricemia and Modulates Gut Microbiota in Mice. Front. Immunol. 2022, 13, 940228. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yi, T. Northwest cuisine Jiangshui may lower uric acid levels. Elder. Health Care 2021, 332, 4. [Google Scholar]
- Liu, Y.; Wang, S.S.; Gao, Q.; Zhen, Z.C.; Wang, L.; Lu, H.Z.; Zhang, T. Microbial community succession and function prediction of Jiangshui in Hanzhong during the fermentation process. China Brew. 2023, 42, 142–149. [Google Scholar]
- Xiang, S.Y.; Zhai, R.; Zhang, H.Y.; Yu, S.L.; Pan, L. Comparison of microbial community structure and identification of dominant microflora in fermented broth “Jiangshui” from different regions. Mod. Food Sci. Technol. 2023, 39, 121–128. [Google Scholar]
- Hu, Y.Y.; Zhao, L.; Shi, L.X.; Li, Q.H. Isolation and identification of dominant lactic acid bacteria in Fermented pickle juice. Gansu Agric. Sci. Technol. 2021, 52, 43–48. [Google Scholar]
- Zhang, L.K.; Zhou, G.G.; Meng, X.G. Isolation and initiative identification of anaerobic microorganisms from traditional fermentative food pickle juice. Food Sci. Technol. 2010, 35, 39–41. [Google Scholar]
- Jingjing, F.; Weilin, J.; Shaochen, S.; Aman, K.; Ying, W.; Yanyi, C.; Pengya, F.; Byong-Hun, J.; El-Sayed, S.; Zhenmin, L.; et al. A Probiotic Targets Bile Acids Metabolism to Alleviate Ulcerative Colitis by Reducing Conjugated Bile Acids. Mol. Nutr. Food Res. 2024, 68, e2300731. [Google Scholar] [CrossRef]
- Hu, Y.; Zhao, Y.; Jia, X.; Liu, D.; Huang, X.; Wang, C.; Zhu, Y.; Yue, C.; Deng, S.; Lyu, Y. Lactic acid bacteria with a strong antioxidant function isolated from “Jiangshui”, pickles, and feces. Front. Microbiol. 2023, 14, 1163662. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Khan, I.; Li, Y.; Yang, Y.; Lu, X.; Wang, Y.; Li, J.; Zhang, C. Overcoming Anxiety Disorder by Probiotic Lactiplantibacillus plantarum LZU-J-TSL6 through Regulating Intestinal Homeostasis. Foods 2022, 11, 3596. [Google Scholar] [CrossRef] [PubMed]
- El Ridi, R.; Tallima, H. Physiological functions and pathogenic potential of uric acid: A review. J. Adv. Res. 2017, 8, 487–493. [Google Scholar] [CrossRef]
- Song, N.; Wang, M.; Zhong, G.; Zhu, K.; Chen, P.; Zhang, N.; Liu, X.; Zhang, W. Bacteroides xylanisolvens possesses a potent anti-hyperuricemia effect in goslings fed on a high-protein diet. Front. Microbiol. 2023, 14, 1173856. [Google Scholar] [CrossRef] [PubMed]
- Budden, K.F.; Gellatly, S.L.; Vaughan, A.; Amorim, N.; Horvat, J.C.; Hansbro, N.G.; Wood, D.L.A.; Hugenholtz, P.; Dennis, P.G.; Wark, P.A.B.; et al. Probiotic Bifidobacterium longum subsp. longum Protects against Cigarette Smoke-Induced Inflammation in Mice. Int. J. Mol. Sci. 2022, 24, 252. [Google Scholar] [CrossRef] [PubMed]
- Ćesić, D.; Lugović Mihić, L.; Ozretić, P.; Lojkić, I.; Buljan, M.; Šitum, M.; Zovak, M.; Vidović, D.; Mijić, A.; Galić, N.; et al. Association of Gut Lachnospiraceae and Chronic Spontaneous Urticaria. Life 2023, 13, 1280. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Ma, Y.; Yang, S.; Zhang, S.; Liu, S.; Xiao, J.; Wang, Y.; Wang, W.; Yang, H.; Li, S.; et al. Gut microbiota-derived ursodeoxycholic acid from neonatal dairy calves improves intestinal homeostasis and colitis to attenuate extended-spectrum β-lactamase-producing enteroaggregative Escherichia coli infection. Microbiome 2022, 10, 79. [Google Scholar] [CrossRef] [PubMed]
- Phillippi, D.T.; Daniel, S.; Nguyen, K.N.; Penaredondo, B.A.; Lund, A.K. Probiotics Function as Immunomodulators in the Intestine in C57Bl/6 Male Mice Exposed to Inhaled Diesel Exhaust Particles on a High-Fat Diet. Cells 2022, 11, 1445. [Google Scholar] [CrossRef] [PubMed]
- Hosomi, A.; Nakanishi, T.; Fujita, T.; Tamai, I. Extra-renal elimination of uric acid via intestinal efflux transporter BCRP/ABCG2. PLoS ONE 2012, 7, e30456. [Google Scholar] [CrossRef] [PubMed]
- Nieuwdorp, M.; Gilijamse, P.W.; Pai, N.; Kaplan, L.M. Role of the microbiome in energy regulation and metabolism. Gastroenterology 2014, 146, 1525–1533. [Google Scholar] [CrossRef] [PubMed]
- Hamer, H.M.; Jonkers, D.M.; Bast, A.; Vanhoutvin, S.A.; Fischer, M.A.; Kodde, A.; Troost, F.J.; Venema, K.; Brummer, R.J. Butyrate modulates oxidative stress in the colonic mucosa of healthy humans. Clin. Nutr. 2009, 28, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Bo, L.; Zhang, M.; Li, S.; Zhao, X.; Sun, C. Metabonomics analysis of serum from rats given long-term and low-level cadmium by ultra-performance liquid chromatography-mass spectrometry. Xenobiotica 2018, 48, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Yu, H.; Fu, J.; Hu, J.; Xu, H.; Zhang, Z.; Bu, M.; Yang, X.; Zhang, H.; Lu, J.; et al. Berberine ameliorates chronic kidney disease through inhibiting the production of gut-derived uremic toxins in the gut microbiota. Acta Pharm. Sin. B 2023, 13, 1537–1553. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Gao, X.; Hu, H.; Le, J.; Chen, Y.; Shu, X.; Liang, Z.; Xu, Y.; Wang, Y.; Zhang, T. Exclusive Enteral Nutrition Exerts Anti-Inflammatory Effects through Modulating Microbiota, Bile Acid Metabolism, and Immune Activities. Nutrients 2022, 14, 4463. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xu, K.; Cai, X.; Wang, C.; Cao, Y.; Xiao, J. Rosmarinic Acid Restores Colonic Mucus Secretion in Colitis Mice by Regulating Gut Microbiota-Derived Metabolites and the Activation of Inflammasomes. J. Agric. Food Chem. 2023, 71, 4571–4585. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, H.; Yan, F.; He, Y.; Ji, A.; Liu, Z.; Li, M.; Ji, X.; Li, C. Kidney and plasma metabolomics provide insights into the molecular mechanisms of urate nephropathy in a mouse model of hyperuricemia. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166374. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Cho, K.; Kim, J.S.; Jung, H.C.; Kim, B.; Park, M.S.; Ji, G.E.; Cho, J.Y.; Hong, K.S. Probiotic treatment induced change of inflammation related metabolites in IBS-D patients/double-blind, randomized, placebo-controlled trial. Food Sci. Biotechnol. 2020, 29, 837–844. [Google Scholar] [CrossRef] [PubMed]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Aga, L.; Tang, L.; Li, H.; Wang, N.; Yang, L.; Zhang, N.; Wang, X.; Wang, X. Lacticaseibacillus paracasei JS-3 Isolated from “Jiangshui” Ameliorates Hyperuricemia by Regulating Gut Microbiota and iTS Metabolism. Foods 2024, 13, 1371. https://doi.org/10.3390/foods13091371
Wu J, Aga L, Tang L, Li H, Wang N, Yang L, Zhang N, Wang X, Wang X. Lacticaseibacillus paracasei JS-3 Isolated from “Jiangshui” Ameliorates Hyperuricemia by Regulating Gut Microbiota and iTS Metabolism. Foods. 2024; 13(9):1371. https://doi.org/10.3390/foods13091371
Chicago/Turabian StyleWu, Jiahui, Lvbu Aga, Leimengyuan Tang, Houxier Li, Nan Wang, Li Yang, Nan Zhang, Xiang Wang, and Xueyong Wang. 2024. "Lacticaseibacillus paracasei JS-3 Isolated from “Jiangshui” Ameliorates Hyperuricemia by Regulating Gut Microbiota and iTS Metabolism" Foods 13, no. 9: 1371. https://doi.org/10.3390/foods13091371