Gene Expression, Protein Function and Pathways of Arabidopsis thaliana Responding to Silver Nanoparticles in Comparison to Silver Ions, Cold, Salt, Drought, and Heat
Abstract
:1. Introduction
2. Results
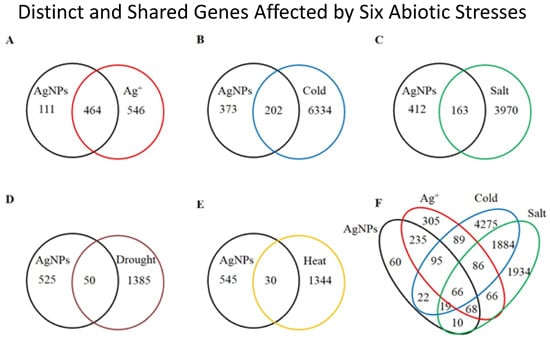
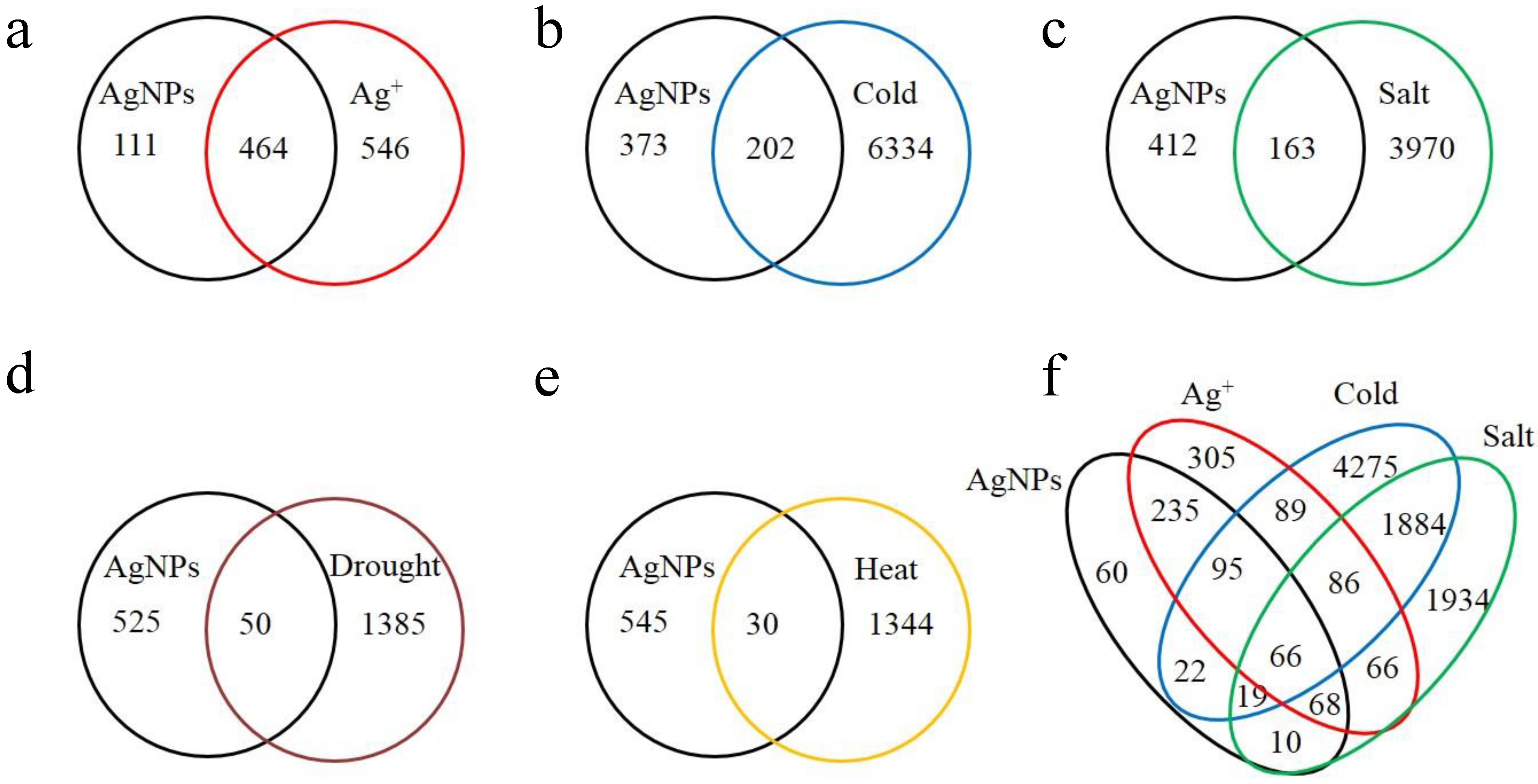
2.1. Overview of the Affected Genes by the Six Abiotic Stresses
| Stress | Number of up regulated genes (% a) | Number of down regulated genes (% a) | Number of total affected genes | Percentage of total affected genes in genome |
|---|---|---|---|---|
| AgNPs | 439 (76.34) | 136 (23.65) | 575 | 2.10 |
| Ag+ | 780 (77.22) | 230 (22.77) | 1010 | 3.68 |
| Cold | 2514 (38.46) | 4022 (61.54) | 6536 | 23.84 |
| Salt | 2057 (49.77) | 2076 (50.23) | 4133 | 15.08 |
| Drought | 814 (56.72) | 621 (43.28) | 1435 | 5.23 |
| Heat | 694 (50.50) | 680 (49.50) | 1374 | 5.01 |
2.2. Gene Ontology Term Enrichment
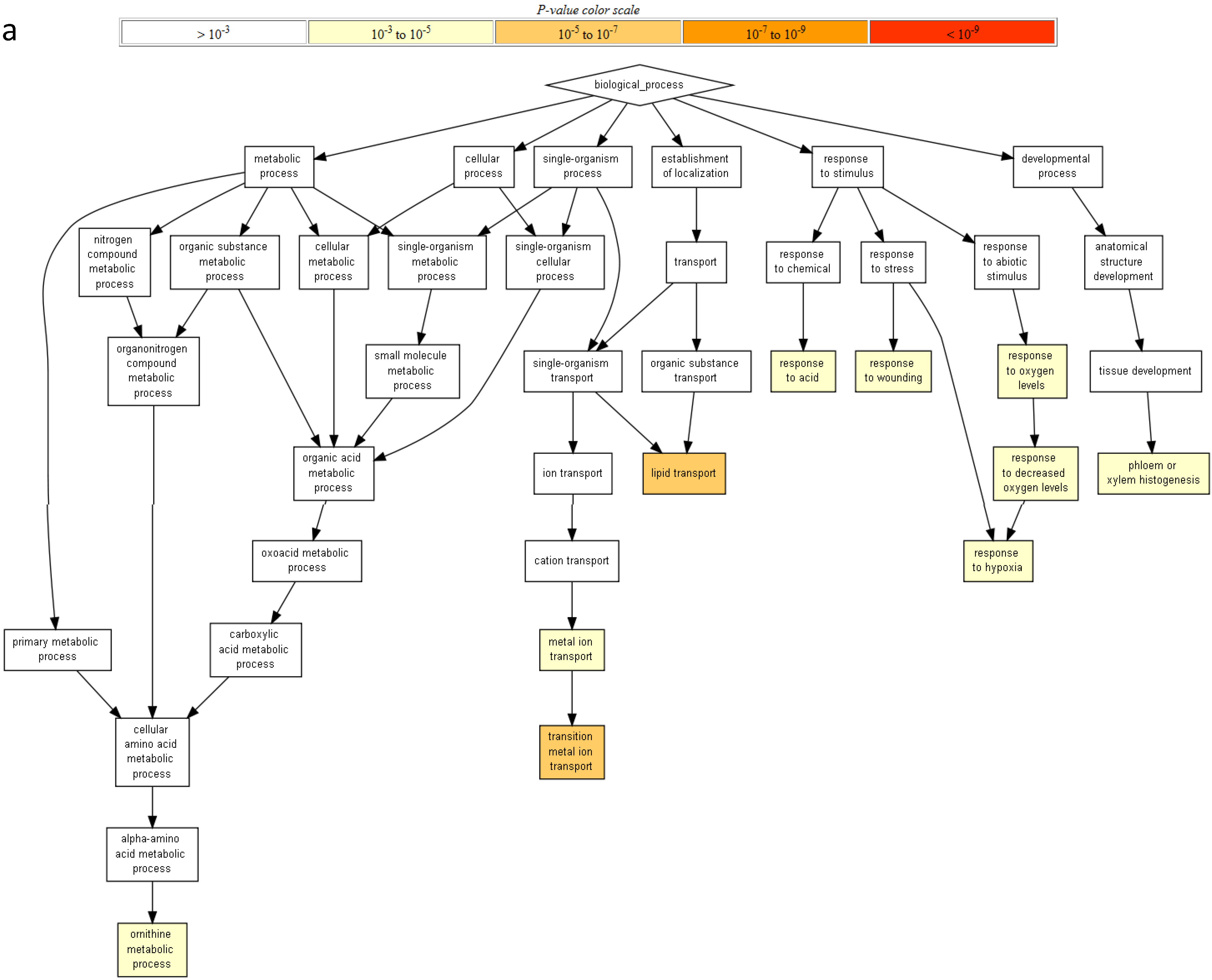
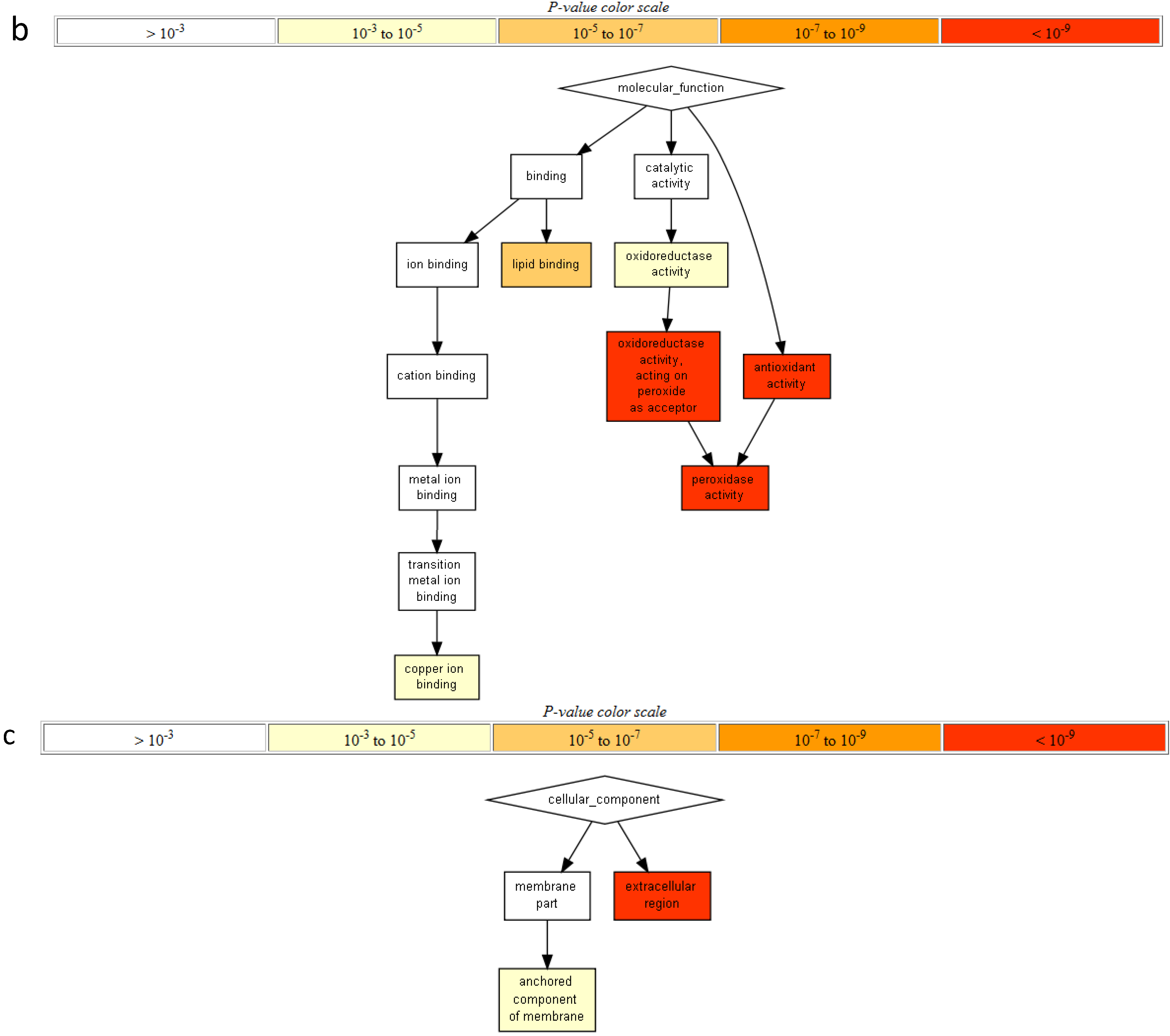
2.3. Protein Domain Enrichment
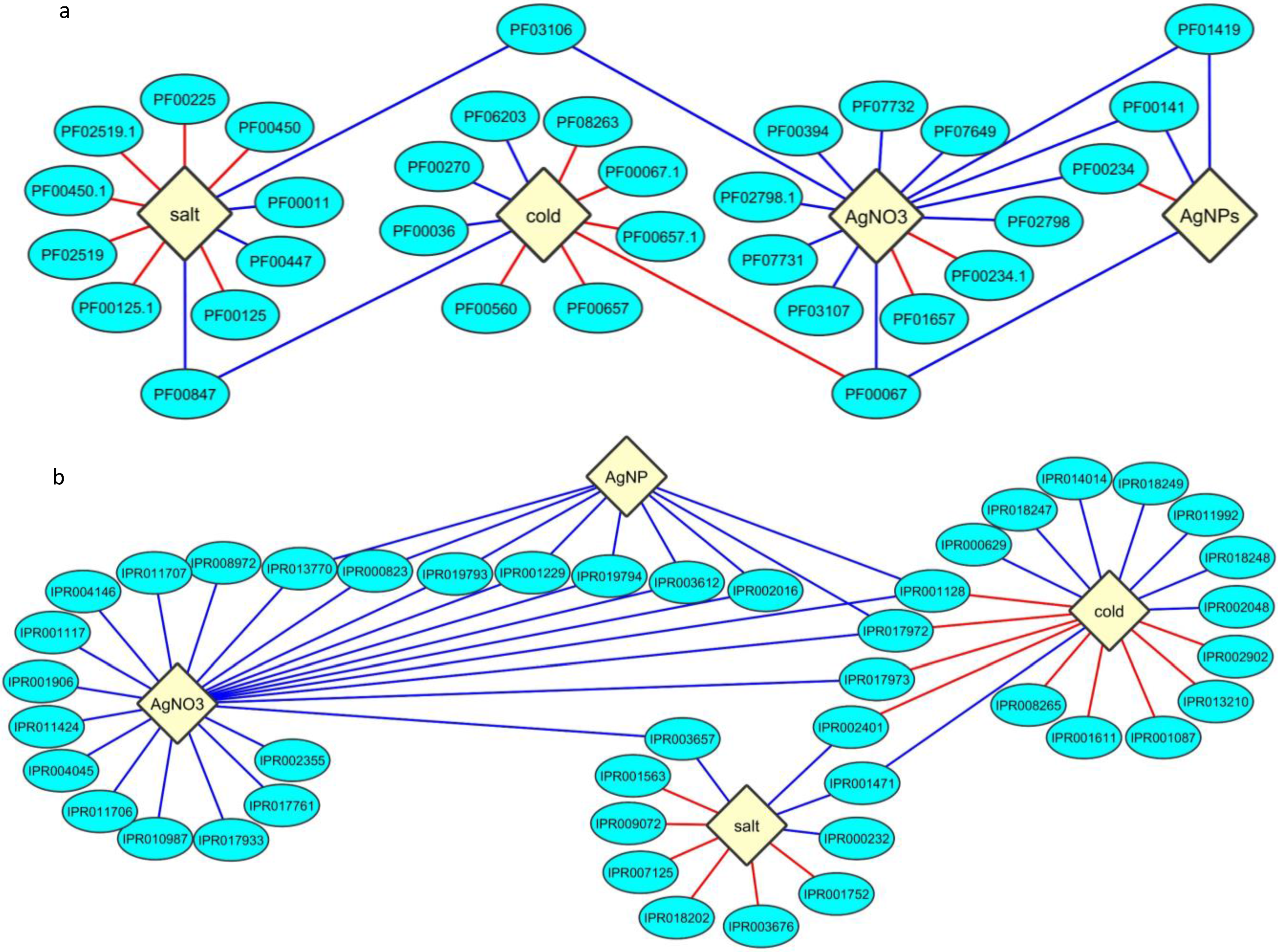
2.4. Enrichment of InterPro Protein Classes
2.5. Enrichment within KEGG Pathways
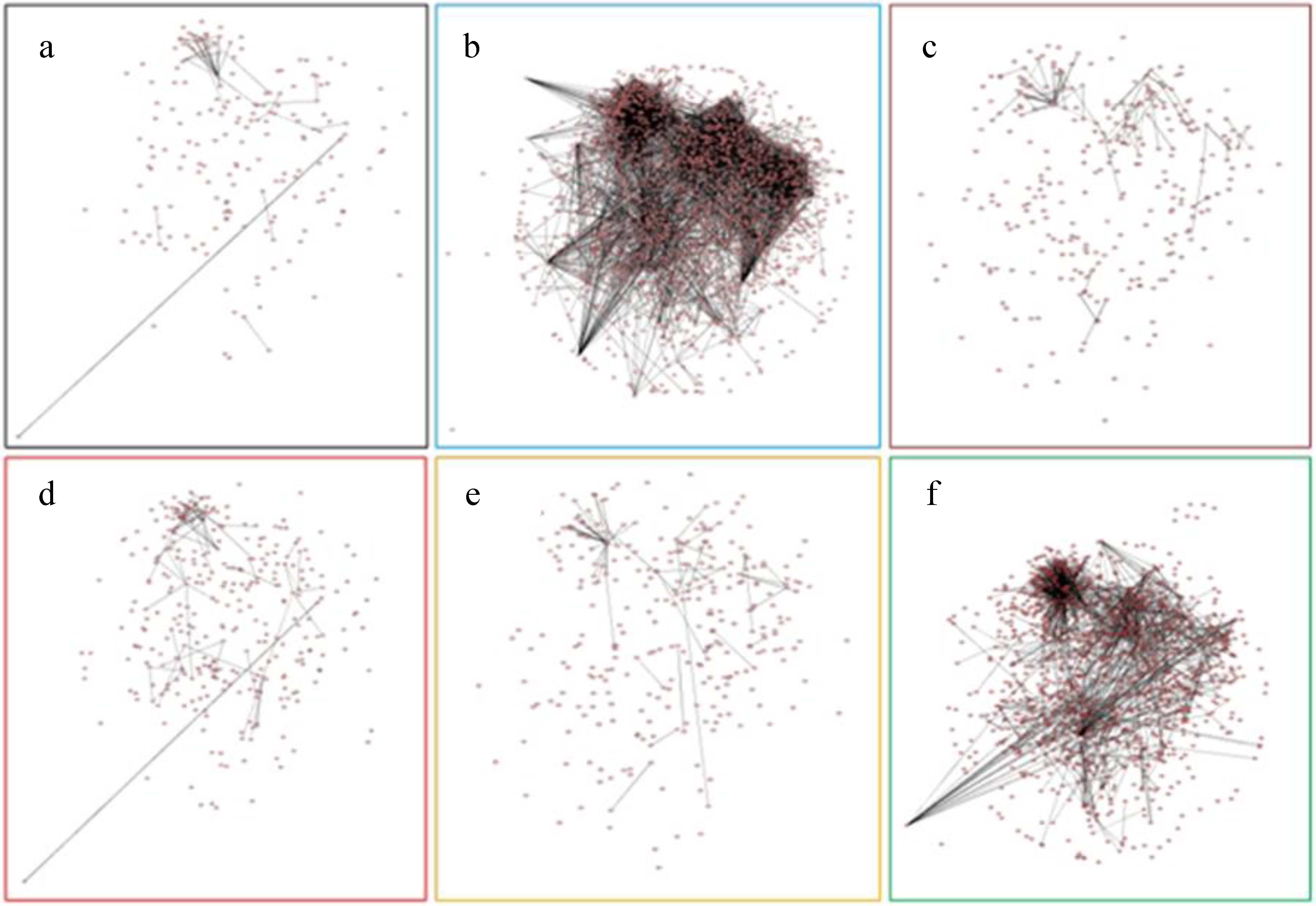
2.7. Protein-Protein Interaction Networks of Affected Genes by Six Abiotic Stresses
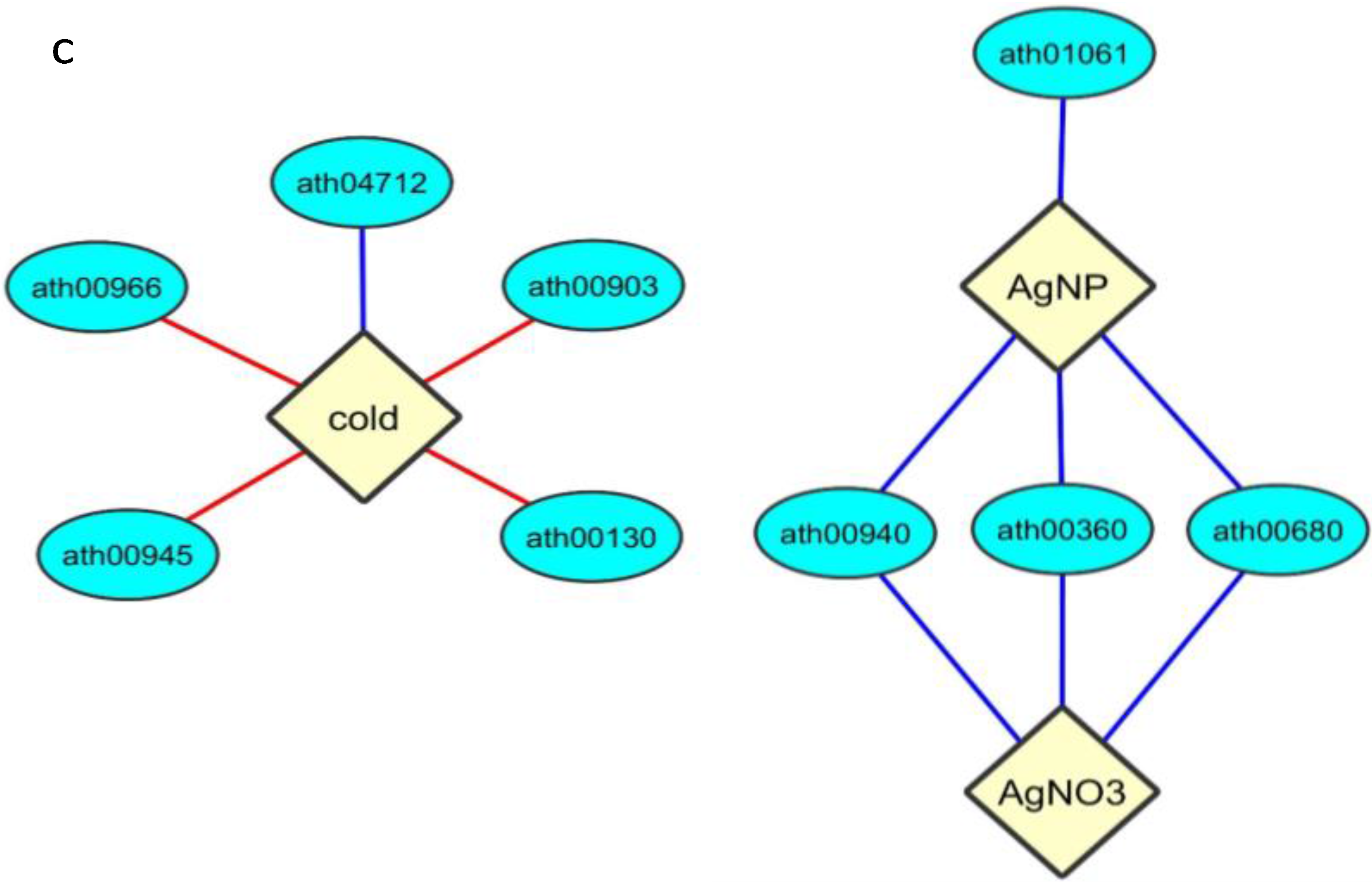
3. Discussion
3.1. Similarities and Differences of AgNP Stress and Five Other Abiotic Stresses
3.2. Similarity and Difference of AgNP and Ag+ Stresses
3.3. Comparison of AgNP and Cold Stresses
3.4. AgNP-Specific Responses in Genes and Functions
4. Experimental Section
4.1. Microarray Data and Data Processing
- E-MEXP-3950. AgNP and Ag+ stresses after 10-day treatment [29].
- GSE5620. Control after 24 h treatment [137].
- GSE5621. Cold stress after 24 h treatment [137].
- GSE5623. Salt (NaCl) stress after 24 h treatment [137].
- GSE5624. Drought stress after 24 h treatment [137].
- GSE5628. Heat stress after 24 h treatment [137].
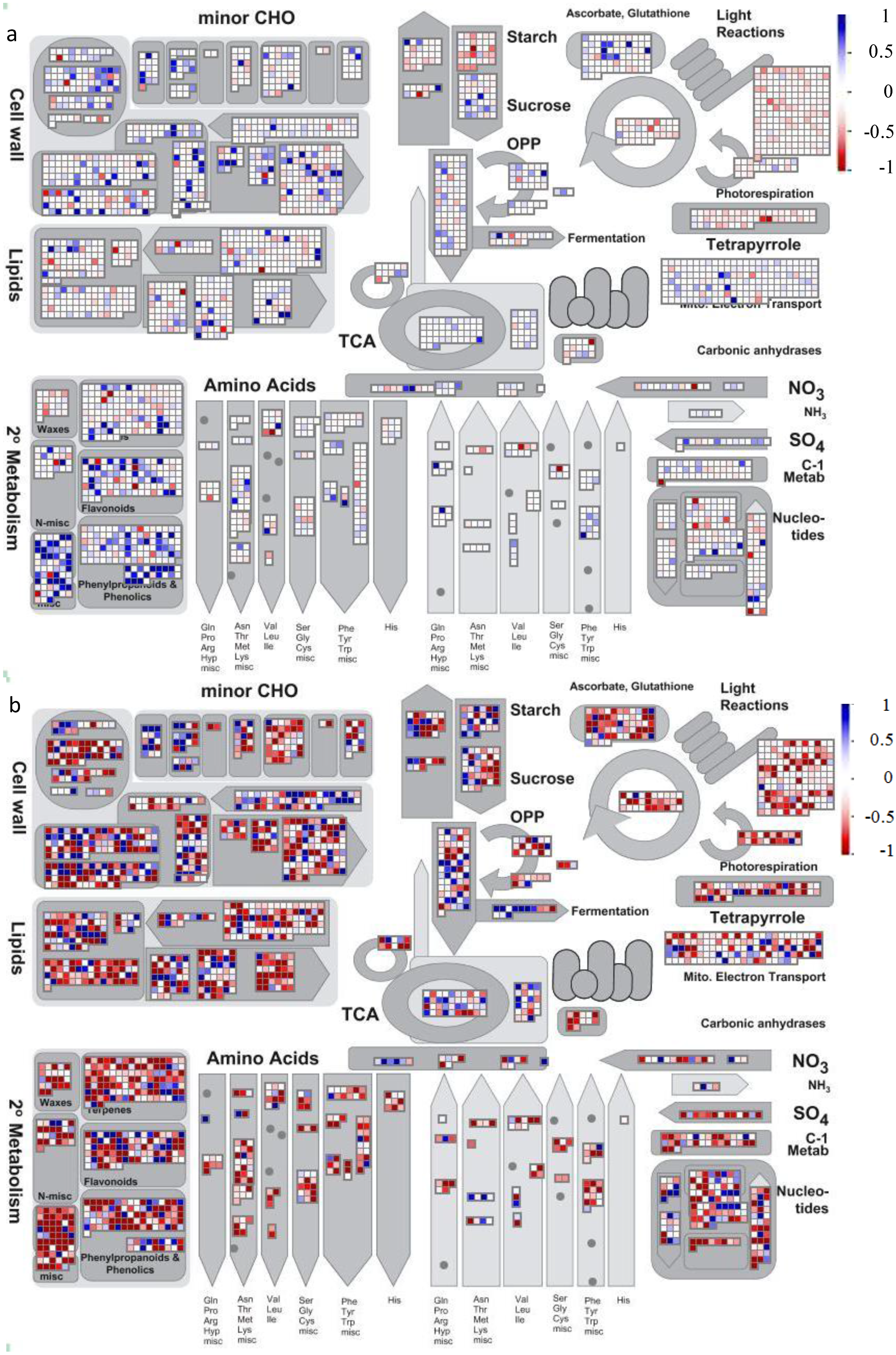
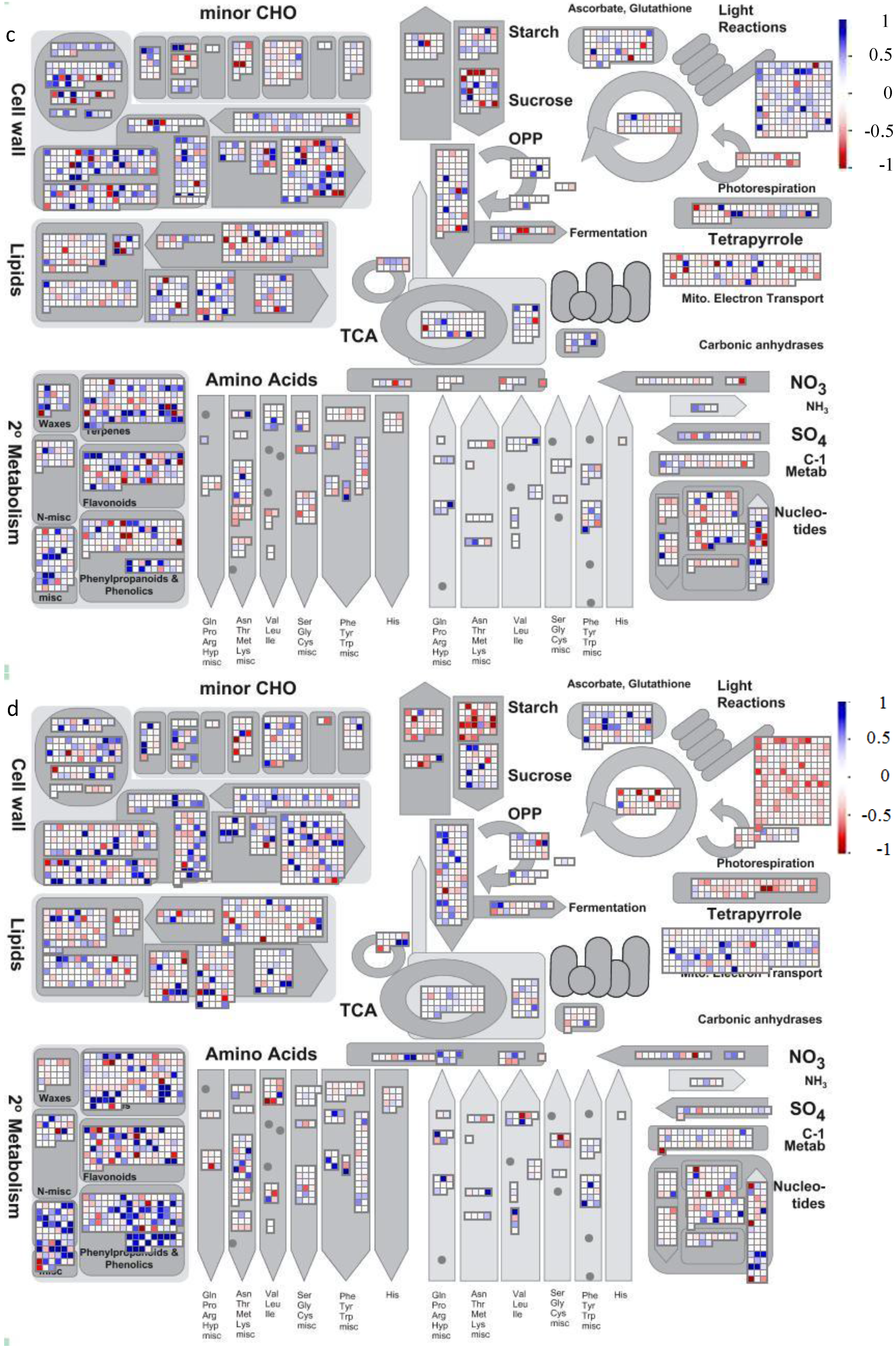
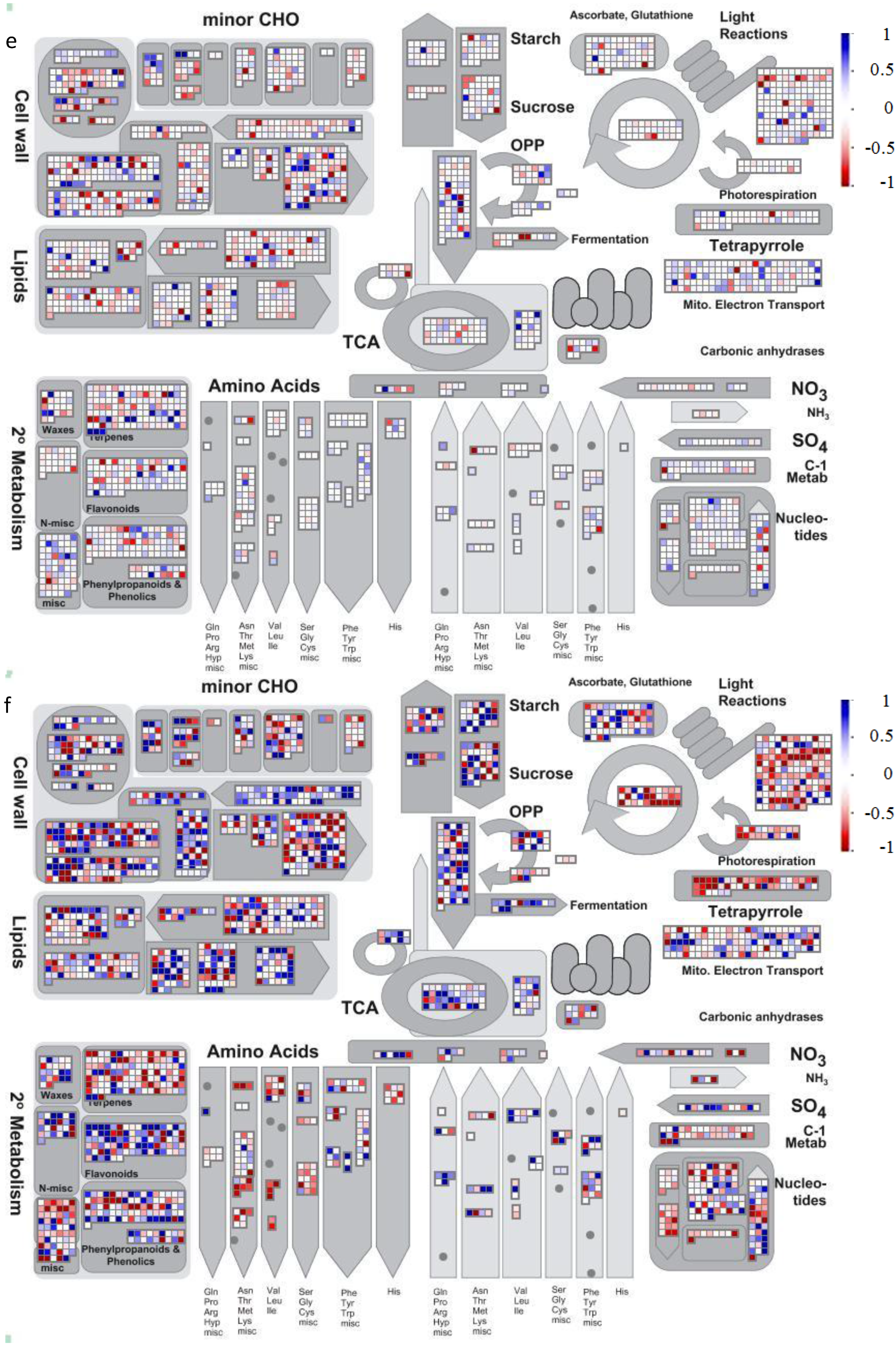
4.2. Visualization of Affected Genes in Metabolic Pathways
4.3. Coded Proteins of Affected Genes by the Stresses in Protein-Protein Interaction Networks
4.4. Enrichment Analyses of Differentially Expressed Genes in Six Abiotic Stresses
4.6. Plasmodesmata Related Genes Expressed in AgNP and Ag+ Stresses
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Oberdörster, G.; Oberdörster, E.; Oberdörster, J. Nanotoxicology: An emerging discipline evolving from studies of ultrafine particles. Environ. Health Perspect. 2005, 113, 823–839. [Google Scholar] [CrossRef] [PubMed]
- EPA. Module 3: Characteristics of particles-particle size categories. Available online: http://web.archive.org/web/20101203205130/http://www.epa.gov/apti/bces/module3/category/category.htm (accessed on 6 December 2014).
- Taylor, R.; Coulombe, S.; Otanicar, T.; Phelan, P.; Gunawan, A.; Lv, W.; Rosengarten, G.; Prasher, R.; Tyagi, H. Small particles, big impacts: A review of the diverse applications of nanofluids. J. Appl. Phys. 2013, 113, 011301. [Google Scholar] [CrossRef]
- Keller, A.; McFerran, S.; Lazareva, A.; Suh, S. Global life cycle releases of engineered nanomaterials. J. Nanopart. Res. 2013, 15, 1–17. [Google Scholar] [CrossRef]
- Ahamed, M.; AlSalhi, M.S.; Siddiqui, M. Silver nanoparticle applications and human health. Clin. Chim. Acta 2010, 411, 1841–1848. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.-K.; Kim, B.H.; Jung, G. Antifungal activity of silver ions and nanoparticles on phytopathogenic fungi. Plant Dis. 2009, 93, 1037–1043. [Google Scholar] [CrossRef]
- Kim, S.W.; Jung, J.H.; Lamsal, K.; Kim, Y.S.; Min, J.S.; Lee, Y.S. Antifungal effects of silver nanoparticles (AgNPs) against various plant pathogenic fungi. Mycobiology 2012, 40, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Bechert, T.; Böswald, M.; Lugauer, S.; Regenfus, A.; Greil, J.; Guggenbichler, J.P. The erlanger silver catheter: In vitro results for antimicrobial activity. Infection 1999, 27, S24–S29. [Google Scholar] [CrossRef] [PubMed]
- Liong, M.; Lu, J.; Kovochich, M.; Xia, T.; Ruehm, S.G.; Nel, A.E.; Tamanoi, F.; Zink, J.I. Multifunctional inorganic nanoparticles for imaging, targeting, and drug delivery. ACS Nano 2008, 2, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Korkin, A.; Rosei, F. Nanoelectronics and Photonics: From Atoms to Materials, Devices, and Architecture; Springer Science & Business Media: Berlin, Germany, 2008. [Google Scholar]
- Geranio, L.; Heuberger, M.; Nowack, B. The behavior of silver nanotextiles during washing. Environ. Sci. Technol. 2009, 43, 8113–8118. [Google Scholar] [CrossRef] [PubMed]
- Gardea-Torresdey, J.L.; Rico, C.M.; White, J.C. Trophic transfer, transformation, and impact of engineered nanomaterials in terrestrial environments. Environ. Sci. Technol. 2014, 48, 2526–2540. [Google Scholar] [CrossRef] [PubMed]
- Tomczyk, M. Nanoinnovation: What Every Manager Needs to Know; John Wiley & Sons: Hoboken, NJ, USA, 2014. [Google Scholar]
- Watson, C.; Ge, J.; Cohen, J.; Pyrgiotakis, G.; Engelward, B.P.; Demokritou, P. High-throughput screening platform for engineered nanoparticle-mediated genotoxicity using cometchip technology. ACS Nano 2014, 8, 2118–2133. [Google Scholar] [CrossRef] [PubMed]
- Colman, B.P.; Arnaout, C.L.; Anciaux, S.; Gunsch, C.K.; Hochella, M.F., Jr.; Kim, B.; Lowry, G.V.; McGill, B.M.; Reinsch, B.C.; Richardson, C.J.; et al. Low concentrations of silver nanoparticles in biosolids cause adverse ecosystem responses under realistic field scenario. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Sur, I.; Cam, D.; Kahraman, M.; Baysal, A.; Culha, M. Interaction of multi-functional silver nanoparticles with living cells. Nanotechnology 2010, 21, 175104. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Choi, J.E.; Choi, J.; Chung, K.-H.; Park, K.; Yi, J.; Ryu, D.-Y. Oxidative stress-dependent toxicity of silver nanoparticles in human hepatoma cells. Toxicol. In Vitro 2009, 23, 1076–1084. [Google Scholar] [CrossRef] [PubMed]
- Park, E.-J.; Yi, J.; Kim, Y.; Choi, K.; Park, K. Silver nanoparticles induce cytotoxicity by a trojan-horse type mechanism. Toxicol. In Vitro 2010, 24, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Geisler-Lee, J.; Wang, Q.; Yao, Y.; Zhang, W.; Geisler, M.; Li, K.; Huang, Y.; Chen, Y.; Kolmakov, A.; Ma, X. Phytotoxicity, accumulation and transport of silver nanoparticles by Arabidopsis thaliana. Nanotoxicology 2013, 7, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-M.; Kwak, J.I.; An, Y.-J. Effect of silver nanoparticles in crop plants phaseolus radiatus and sorghum bicolor: Media effect on phytotoxicity. Chemosphere 2012, 86, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Peng, X.; Han, X.; Ren, J.; Sun, L.; Fu, Z. Comparison of the toxicity of silver nanoparticles and silver ions on the growth of terrestrial plant model Arabidopsis thaliana. J. Environ. Sci. 2013, 25, 1947–1956. [Google Scholar] [CrossRef]
- Geisler-Lee, J.; Brooks, M.; Gerfen, J.; Wang, Q.; Fotis, C.; Sparer, A.; Ma, X.; Berg, R.; Geisler, M. Reproductive toxicity and life history study of silver nanoparticle effect, uptake and transport in Arabidopsis thaliana. Nanomaterials 2014, 4, 301–318. [Google Scholar] [CrossRef]
- Ma, X.; Geisler-Lee, J.; Deng, Y.; Kolmakov, A. Interactions between engineered nanoparticles (ENPs) and plants: Phytotoxicity, uptake and accumulation. Sci. Total Environ. 2010, 408, 3053–3061. [Google Scholar] [CrossRef] [PubMed]
- Miralles, P.; Church, T.L.; Harris, A.T. Toxicity, uptake, and translocation of engineered nanomaterials in vascular plants. Environ. Sci. Technol. 2012, 46, 9224–9239. [Google Scholar] [CrossRef] [PubMed]
- Mirzajani, F.; Askari, H.; Hamzelou, S.; Farzaneh, M.; Ghassempour, A. Effect of silver nanoparticles on Oryza sativa l. And its rhizosphere bacteria. Ecotoxicol. Environ. Saf. 2013, 88, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Colman, B.P.; McGill, B.M.; Wright, J.P.; Bernhardt, E.S. Effects of silver nanoparticle exposure on germination and early growth of eleven wetland plants. PLoS ONE 2012, 7, e47674. [Google Scholar] [CrossRef] [PubMed]
- Dimkpa, C.O.; McLean, J.E.; Martineau, N.; Britt, D.W.; Haverkamp, R.; Anderson, A.J. Silver nanoparticles disrupt wheat (Triticum aestivum l.) growth in a sand matrix. Environ. Sci. Technol. 2012, 47, 1082–1090. [Google Scholar] [CrossRef]
- Vannini, C.; Domingo, G.; Onelli, E.; de Mattia, F.; Bruni, I.; Marsoni, M.; Bracale, M. Phytotoxic and genotoxic effects of silver nanoparticles exposure on germinating wheat seedlings. J. Plant Physiol. 2014, 171, 1142–1148. [Google Scholar] [CrossRef] [PubMed]
- Kaveh, R.; Li, Y.-S.; Ranjbar, S.; Tehrani, R.; Brueck, C.L.; van Aken, B. Changes in Arabidopsis thaliana gene expression in response to silver nanoparticles and silver ions. Environ. Sci. Technol. 2013, 47, 10637–10644. [Google Scholar] [PubMed]
- Larue, C.; Castillo-Michel, H.; Sobanska, S.; Cécillon, L.; Bureau, S.; Barthès, V.; Ouerdane, L.; Carrière, M.; Sarret, G. Foliar exposure of the crop Lactuca sativa to silver nanoparticles: Evidence for internalization and changes in Ag speciation. J. Hazard. Mater. 2014, 264, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Cheng, Y.; Espinasse, B.; Colman, B.P.; Auffan, M.; Wiesner, M.; Rose, J.; Liu, J.; Bernhardt, E.S. More than the ions: The effects of silver nanoparticles on Lolium multiflorum. Environ. Sci. Technol. 2011, 45, 2360–2367. [Google Scholar] [CrossRef] [PubMed]
- Romera, F.J.; Alcantara, E. Iron-deficiency stress responses in cucumber (Cucumis sativus l.) roots (a possible role for ethylene?). Plant Physiol. 1994, 105, 1133–1138. [Google Scholar] [PubMed]
- Yoshioka, K.; Shinozaki, K. Signal Crosstalk in Plant Stress Responses; Wiley: Hoboken, Germany, 2009. [Google Scholar]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Duque, A.S.; de Almeida, A.M.; da Silva, A.B.; da Silva, J.M.; Farinha, A.P.; Santos, D.; Fevereiro, P.; de Sousa Araújo, S. Abiotic Stress Responses in Plants: Unraveling the Complexity of Genes and Networks to Survive. In Abiotic Stress-Plant Responses and Applications in Agriculture; Vahdati, K., Leslie, C., Eds.; INTECH: Rijeka, Croatia, 2013. [Google Scholar]
- Bailey-Serres, J.; Mittler, R. The roles of reactive oxygen species in plant cells. Plant Physiol. 2006, 141, 311. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.; Mittler, R.; Suzuki, N. Ros as key players in plant stress signalling. J. Exp. Botany 2013, 65, 1229–1240. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, H.; Pitzschke, A.; Hirt, H. Emerging map kinase pathways in plant stress signalling. Trends Plant Sci. 2005, 10, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Hirt, H. Multiple roles of map kinases in plant signal transduction. Trends Plant Sci. 1997, 2, 11–15. [Google Scholar] [CrossRef]
- Alcázar-Román, A.; Wente, S. Inositol polyphosphates: A new frontier for regulating gene expression. Chromosoma 2008, 117, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Schumaker, K.S.; Zhu, J.-K. Cell signaling during cold, drought, and salt stress. Plant Cell Online 2002, 14, S165–S183. [Google Scholar] [CrossRef]
- Mahalingam, R.; Fedoroff, N. Stress response, cell death and signalling: The many faces of reactive oxygen species. Physiol. Plant. 2003, 119, 56–68. [Google Scholar] [CrossRef]
- Dixon, R.A.; Achnine, L.; Kota, P.; Liu, C.J.; Reddy, M.; Wang, L. The phenylpropanoid pathway and plant defence—A genomics perspective. Mol. Plant Pathol. 2002, 3, 371–390. [Google Scholar] [CrossRef] [PubMed]
- Edreva, A.; Velikova, V.; Tsonev, T.; Dagnon, S.; Gürel, A.; Aktaş, L.; Gesheva, E. Stress-protective role of secondary metabolites: Diversity of functions and mechanisms. Gen. Appl. Plant Physiol. 2008, 34, 67–78. [Google Scholar]
- Oh, M.-M.; Trick, H.N.; Rajashekar, C. Secondary metabolism and antioxidants are involved in environmental adaptation and stress tolerance in lettuce. J. Plant Physiol. 2009, 166, 180–191. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Degenhardt, B.; Gimmler, H. Cell wall adaptations to multiple environmental stresses in maize roots. J. Exp. Bot. 2000, 51, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Rouet, M.; Mathieu, Y.; Barbier-Brygoo, H.; Laurière, C. Characterization of active oxygen-producing proteins in response to hypo-osmolarity in tobacco and Arabidopsis cell suspensions: Identification of a cell wall peroxidase. J. Exp. Bot. 2006, 57, 1323–1332. [Google Scholar] [CrossRef] [PubMed]
- Daudi, A.; Cheng, Z.; O’Brien, J.A.; Mammarella, N.; Khan, S.; Ausubel, F.M.; Bolwell, G.P. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell Online 2012, 24, 275–287. [Google Scholar] [CrossRef]
- Passardi, F.; Penel, C.; Dunand, C. Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 2004, 9, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Minibayeva, F.; Kolesnikov, O.; Chasov, A.; Beckett, R.; Lüthje, S.; Vylegzhanina, N.; Buck, F.; Böttger, M. Wound-induced apoplastic peroxidase activities: Their roles in the production and detoxification of reactive oxygen species. Plant Cell Environ. 2009, 32, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Lucas, W.J.; Ham, B.-K.; Kim, J.-Y. Plasmodesmata–bridging the gap between neighboring plant cells. Trends Cell Biol. 2009, 19, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Furuta, K.; Lichtenberger, R.; Helariutta, Y. The role of mobile small RNA species during root growth and development. Curr. Opin. Cell Biol. 2012, 24, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ferrer, V.; Voinnet, O. Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 2009, 60, 485–510. [Google Scholar] [CrossRef] [PubMed]
- Zhai, G.; Walters, K.S.; Peate, D.W.; Alvarez, P.J.J.; Schnoor, J.L. Transport of gold nanoparticles through plasmodesmata and precipitation of gold ions in woody poplar. Environ. Sci. Technol. Lett. 2014, 1, 146–151. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Jones, A.M.; Garg, S.; Pham, A.N.; Waite, T.D. Silver nanoparticle−reactive oxygen species interactions: Application of a charging−discharging model. J. Phys. Chem. C 2011, 115, 5461–5468. [Google Scholar] [CrossRef]
- Xu, H.; Qu, F.; Xu, H.; Lai, W.; Andrew Wang, Y.; Aguilar, Z.; Wei, H. Role of reactive oxygen species in the antibacterial mechanism of silver nanoparticles on Escherichia coli O157:H7. Biometals 2012, 25, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.-S.; Qiu, X.-N.; Li, G.-B.; Li, W.; Yin, L.-Y. Silver nanoparticles induced accumulation of reactive oxygen species and alteration of antioxidant systems in the aquatic plant Spirodela polyrhiza. Environ. Toxicol. Chem. 2014, 33, 1398–1405. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Jung, H.; Lim, J.-S. Cell death by polyvinylpyrrolidine-coated silver nanoparticles is mediated by ROS-dependent signaling. Biomol. Ther. 2012, 20, 399–405. [Google Scholar] [CrossRef]
- Lim, D.; Roh, J.Y.; Eom, H.J.; Choi, J.Y.; Hyun, J.; Choi, J. Oxidative stress-related PMK-1 P38 MAPK activation as a mechanism for toxicity of silver nanoparticles to reproduction in the nematode Caenorhabditis elegans. Environ. Toxicol. Chem. 2012, 31, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.K.; Asharani, P.V.; Hande, M.P. Enhanced genotoxicity of silver nanoparticles in DNA repair deficient mammalian cells. Front. Genet. 2012, 3, 104. [Google Scholar] [CrossRef] [PubMed]
- Gliga, A.R.; Skoglund, S.; Wallinder, I.O.; Fadeel, B.; Karlsson, H.L. Size-dependent cytotoxicity of silver nanoparticles in human lung cells: The role of cellular uptake, agglomeration and Ag release. Part. Fibre Toxicol. 2014, 11. [Google Scholar] [CrossRef]
- AshaRani, P.; Sethu, S.; Lim, H.; Balaji, G.; Valiyaveettil, S.; Hande, M.P. Differential regulation of intracellular factors mediating cell cycle, DNA repair and inflammation following exposure to silver nanoparticles in human cells. Genome Integr. 2012, 3. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L. The pfam protein families database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef] [PubMed]
- Jeyaprakash, A.A.; Rani, P.G.; Reddy, G.B.; Banumathi, S.; Betzel, C.; Sekar, K.; Surolia, A.; Vijayan, M. Crystal structure of the jacalin-t-antigen complex and a comparative study of lectin-T-antigen complexes. J. Mol. Biol. 2002, 321, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Raval, S.; Gowda, S.B.; Singh, D.D.; Chandra, N.R. A database analysis of jacalin-like lectins: Sequence-structure-function relationships. Glycobiology 2004, 14, 1247–1263. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, R.; Sekar, K.; Banerjee, R.; Sharma, V.; Surolia, A.; Vijayan, M. A novel mode of carbohydrate recognition in jacalin, a Moraceae plant lectin with a β-prism fold. Nat. Struct. Mol. Biol. 1996, 3, 596–603. [Google Scholar] [CrossRef]
- Welinder, K.G. Superfamily of plant, fungal and bacterial peroxidases. Curr. Opin. Struct. Biol. 1992, 2, 388–393. [Google Scholar] [CrossRef]
- Campa, A. Biological Roles of Plant Peroxidases: Known and Potential Function. In Peroxidases in Chemistry and Biology; Everse, J., Everse, K.E., Grisham, M.B., Eds.; CRC Press, Taylor & Francis Group, Inc.: London, UK, 1990; Volume 2, pp. 25–50. [Google Scholar]
- Kader, J.-C. Lipid-transfer proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 627–654. [Google Scholar] [CrossRef] [PubMed]
- Kader, J.-C. Lipid-transfer proteins: A puzzling family of plant proteins. Trends Plant Sci. 1997, 2, 66–70. [Google Scholar] [CrossRef]
- Lamb, D.C.; Lei, L.; Warrilow, A.G.S.; Lepesheva, G.I.; Mullins, J.G.L.; Waterman, M.R.; Kelly, S.L. The first virally encoded cytochrome p450. J. Virol. 2009, 83, 8266–8269. [Google Scholar] [CrossRef] [PubMed]
- Werck-Reichhart, D.; Feyereisen, R. Cytochromes p450: A success story. Genome Biol. 2000, 1, REVIEWS3003. [Google Scholar] [CrossRef] [PubMed]
- Pinot, F.; Beisson, F. Cytochrome p450 metabolizing fatty acids in plants: Characterization and physiological roles. FEBS J. 2011, 278, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Schuler, M.A. The role of cytochrome p450 monooxygenases in plant-insect interactions. Plant Physiol. 1996, 112, 1411–1419. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Narusaka, M.; Ishida, J.; Nanjo, T.; Fujita, M.; Oono, Y.; Kamiya, A.; Nakajima, M.; Enju, A.; Sakurai, T.; et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cdna microarray. Plant J. 2002, 31, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Narusaka, Y.; Narusaka, M.; Seki, M.; Umezawa, T.; Ishida, J.; Nakajima, M.; Enju, A.; Shinozaki, K. Crosstalk in the responses to abiotic and biotic stresses in Arabidopsis: Analysis of gene expression in cytochrome p450 gene superfamily by cdna microarray. Plant Mol. Biol. 2004, 55, 327–342. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. Wrky transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Torres, J.T.; Parniske, M.; Wernert, P.; Hahlbrock, K.; Somssich, I.E. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996, 15, 5690–5700. [Google Scholar] [PubMed]
- Riechmann, J.L.; Meyerowitz, E.M. The AP2/EREBP family of plant transcription factors. Biol. Chem. 1998, 379, 633–646. [Google Scholar] [PubMed]
- Mitchell, A.; Chang, H.Y.; Daugherty, L.; Fraser, M.; Hunter, S.; Lopez, R.; McAnulla, C.; McMenamin, C.; Nuka, G.; Pesseat, S.; et al. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2015, 43, D213–D221. [Google Scholar] [CrossRef] [PubMed]
- The Homepage of InterPro. Available online: http://www.ebi.ac.uk/interpro (accessed on 30 March 2014).
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009, 37, D211–D215. [Google Scholar] [CrossRef] [PubMed]
- Hunter, S.; Jones, P.; Mitchell, A.; Apweiler, R.; Attwood, T.K.; Bateman, A.; Bernard, T.; Binns, D.; Bork, P.; Burge, S.; et al. InterPro in 2011: New developments in the family and domain prediction database. Nucleic Acids Res. 2011, 40, D306–D312. [Google Scholar] [CrossRef] [PubMed]
- Rico, M.; Bruix, M.; González, C.; Monsalve, R.I.; Rodríguez, R. 1H NMR assignment and global fold of napin Bnib, a representative 2S albumin seed protein. Biochemistry 1996, 35, 15672–15682. [Google Scholar] [CrossRef] [PubMed]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.; Hans, J.; Matern, U. Biosynthesis of Phenylpropanoids and Related Compounds. In Annual Plant Reviews Volume 40: Biochemistry of Plant Secondary Metabolism; Wiley-Blackwell: Hoboken, JN, USA, 2010; pp. 182–257. [Google Scholar]
- Ferrer, J.L.; Austin, M.B.; Stewart, C.; Noel, J.P. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 2008, 46, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Fraser, C.M.; Chapple, C. The phenylpropanoid pathway in Arabidopsis. Arabidopsis Book 2011, 9, e0152. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Kim, J.; Oh, J.; Bae, S.; Lee, S.; Hong, I.S.; Kim, S.H. Ion-release kinetics and ecotoxicity effects of silver nanoparticles. Environ. Toxicol. Chem. 2012, 31, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Gubbins, E.J.; Batty, L.C.; Lead, J.R. Phytotoxicity of silver nanoparticles to Lemna minor l. Environ. Pollut. 2011, 159, 1551–1559. [Google Scholar] [CrossRef] [PubMed]
- Jaspers, P.; Kangasjärvi, J. Reactive oxygen species in abiotic stress signaling. Physiol. Plant. 2010, 138, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.-J.; Herth, S. Plant nanotoxicology. Trends Plant Sci. 2011, 16, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Wuest, S.E.; Vijverberg, K.; Schmidt, A.; Weiss, M.; Gheyselinck, J.; Lohr, M.; Wellmer, F.; Rahnenführer, J.; von Mering, C.; Grossniklaus, U. Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr. Biol. 2010, 20, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Zhang, Z.; Cao, J. Pollen wall development: The associated enzymes and metabolic pathways. Plant Biol. 2013, 15, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Bock, K.W.; Honys, D.; Ward, J.M.; Padmanaban, S.; Nawrocki, E.P.; Hirschi, K.D.; Twell, D.; Sze, H. Integrating membrane transport with male gametophyte development and function through transcriptomics. Plant Physiol. 2006, 140, 1151–1168. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.H.; Goodman, H.M. A novel gene family in Arabidopsis encoding putative heptahelical transmembrane proteins homologous to human adiponectin receptors and progestin receptors. J. Exp. Bot. 2005, 56, 3137–3147. [Google Scholar] [CrossRef] [PubMed]
- Sanghera, G.S.; Wani, S.H.; Hussain, W.; Singh, N.B. Engineering cold stress tolerance in crop plants. Curr. Genomics 2011, 12, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Lata, C.; Prasad, M. Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 2011, 62, 4731–4748. [Google Scholar] [CrossRef] [PubMed]
- Musante, C. Nanoparticle Contamination of Agricultural Crops; Chemistry, D.O.A., Ed.; The Connecticut Agricultural Experiment Station: New Haven, CT, USA, 2011; p. 32. [Google Scholar]
- Atkinson, N.J.; Urwin, P.E. The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef] [PubMed]
- Oukarroum, A.; Barhoumi, L.; Pirastru, L.; Dewez, D. Silver nanoparticle toxicity effect on growth and cellular viability of the aquatic plant Lemna gibba. Environ. Toxicol. Chem. 2013, 32, 902–907. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Nemhauser, J.L.; Hong, F.; Chory, J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 2006, 126, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Eom, H.-J.; Choi, J. p38 MAPK activation, DNA damage, cell cycle arrest and apoptosis as mechanisms of toxicity of silver nanoparticles in Jurkat T cells. Environ. Sci. Technol. 2010, 44, 8337–8342. [Google Scholar] [CrossRef] [PubMed]
- Hare, P.; Cress, W.; van Staden, J. Dissecting the roles of osmolyte accumulation during stress. Plant Cell Environ. 1998, 21, 535–553. [Google Scholar] [CrossRef]
- Kalamaki, M.S.; Merkouropoulos, G.; Kanellis, A.K. Can ornithine accumulation modulate abiotic stress tolerance in Arabidopsis. Plant Signal. Behav. 2009, 4, 1099–1101. [Google Scholar] [CrossRef] [PubMed]
- Nayak, L.; Raval, M.; Biswal, B.; Biswal, U. Silver nitrate influences in vitro shoot multiplication and root formation in Vanilla planifolia Andr. Curr. Sci. India 2001, 14, 1166. [Google Scholar]
- Verma, A.; Uzun, O.; Hu, Y.; Hu, Y.; Han, H.-S.; Watson, N.; Chen, S.; Irvine, D.J.; Stellacci, F. Surface-structure-regulated cell-membrane penetration by monolayer-protected nanoparticles. Nat. Mater. 2008, 7, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Koo, Y.; Alexander, A.; Yang, Y.; Westerhof, S.; Zhang, Q.; Schnoor, J.L.; Colvin, V.L.; Braam, J.; Alvarez, P.J. Phytostimulation of poplars and Arabidopsis exposed to silver nanoparticles and Ag+ at sublethal concentrations. Environ. Sci. Technol. 2013, 47, 5442–5449. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Prasad, S.; Rai, L. Inhibition of photosynthetic electron transport in Nostoc muscorum by Ni2+ and Ag+. J. Gen. Appl. Microbiol. 1991, 37, 167–174. [Google Scholar] [CrossRef]
- Kumar, V.; Parvatam, G.; Ravishankar, G.A. AgNO3: A potential regulator of ethylene activity and plant growth modulator. Electron. J. Biotechnol. 2009, 12, 8–9. [Google Scholar] [CrossRef]
- Shinwari, Z.K.; Nakashima, K.; Miura, S.; Kasuga, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem. Biophys. Res. Commun. 1998, 250, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Kohan, E.; Bagherieh-Najjar, M.B. DRE-binding transcription factor (DREB1A) as a master regulator induced a broad range of abiotic stress tolerance in plant. Afr. J. Biotechnol. 2013, 10, 15100–15108. [Google Scholar]
- Maruyama, K.; Sakuma, Y.; Kasuga, M.; Ito, Y.; Seki, M.; Goda, H.; Shimada, Y.; Yoshida, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 2004, 38, 982–993. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Narusaka, M.; Abe, H.; Kasuga, M.; Yamaguchi-Shinozaki, K.; Carninci, P.; Hayashizaki, Y.; Shinozaki, K. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell Online 2001, 13, 61–72. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.-K.; Sunkar, R. Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 2010, 639, 39–55. [Google Scholar] [PubMed]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.-H.; Hong, X.; Agarwal, M.; Zhu, J.-K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Zarka, D.G.; Stockinger, E.J.; Salazar, M.P.; Houghton, J.M.; Thomashow, M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-inducedcorgene expression. Plant J. 1998, 16, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Zhang, L.; Zhang, T.; Zhang, H.; Xu, S.; An, L. Transcriptional regulation network of cold-responsive genes in higher plants. Plant Sci. 2005, 169, 987–995. [Google Scholar] [CrossRef]
- Uemura, M.; Tominaga, Y.; Nakagawara, C.; Shigematsu, S.; Minami, A.; Kawamura, Y. Responses of the plasma membrane to low temperatures. Physiol. Plant. 2006, 126, 81–89. [Google Scholar] [CrossRef]
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Piett, C.; Farkas, S.; Qazzaz, M.; Syed, N. Silver nanoparticles (AgNPs) cause degeneration of cytoskeleton and disrupt synaptic machinery of cultured cortical neurons. Mol. Brain 2013, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Amanda, S.M.; Laura, B.-S.K.; John, S.J.; Liming, D.; Saber, H.M. Can silver nanoparticles be useful as potential biological labels? Nanotechnology 2008, 19, 235104. [Google Scholar] [CrossRef] [PubMed]
- Corredor, E.; Testillano, P.; Coronado, M.-J.; Gonzalez-Melendi, P.; Fernandez-Pacheco, R.; Marquina, C.; Ibarra, M.R.; de la Fuente, J.; Rubiales, D.; Perez-de-Luque, A.; et al. Nanoparticle penetration and transport in living pumpkin plants: In situ subcellular identification. BMC Plant Biol. 2009, 9. [Google Scholar] [CrossRef] [PubMed]
- Kendall, E.J.; McKersie, B.D. Free radical and freezing injury to cell membranes of winter wheat. Physiol. Plant. 1989, 76, 86–94. [Google Scholar] [CrossRef]
- Ceron-Garcia, A.; Vargas-Arispuro, I.; Aispuro-Hernandez, E.; Martinez-Tellez, M.A. Oligoglucan Elicitor Effects during Plant Oxidative Stress, Cell Metabolism. In Cell Homeostasis and Stress Response; Bubulya, P., Ed.; InTech: Rijeka, Croatia, 2012. [Google Scholar]
- Bak, S.; Beisson, F.; Bishop, G.; Hamberger, B.; Höfer, R.; Paquette, S.; Werck-Reichhart, D. Cytochrome p450; The American Society of Plant Biologists: Rockville, MD, USA, 2011; Volume 9, p. e0144. [Google Scholar]
- Vranova, E.; Inzé, D.; van Breusegem, F. Signal transduction during oxidative stress. J. Exp. Bot. 2002, 53, 1227–1236. [Google Scholar] [CrossRef] [PubMed]
- Gene Expression Omnibus (GEO). Available online: http://www.ncbi.nlm.nih.gov/geo/ (accessed on 28 February 2014).
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- ArrayExpress. Available online: https://www.ebi.ac.uk/arrayexpress/ (accessed on 28 February 2014).
- Kolesnikov, N.; Hastings, E.; Keays, M.; Melnichuk, O.; Tang, Y.A.; Williams, E.; Dylag, M.; Kurbatova, N.; Brandizi, M.; Burdett, T.; et al. ArrayExpress update—Simplifying data submissions. Nucleic Acids Res. 2015, 43, D1113–D1116. [Google Scholar] [CrossRef] [PubMed]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The atgenexpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007, 50, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Hetu, M.; Tremblay, L.J.; Lefebvre, D.D. High root biomass production in anchored Arabidopsis plants grown in axenic sucrose supplemented liquid culture. Biotechniques 2005, 39, 345. [Google Scholar] [CrossRef] [PubMed]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. Mapman: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis information resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef] [PubMed]
- Geisler-Lee, J.; O’Toole, N.; Ammar, R.; Provart, N.J.; Millar, A.H.; Geisler, M. A predicted interactome for Arabidopsis. Plant Physiol. 2007, 145, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Stark, C.; Breitkreutz, B.-J.; Chatr-Aryamontri, A.; Boucher, L.; Oughtred, R.; Livstone, M.S.; Nixon, J.; van Auken, K.; Wang, X.; Shi, X. The biogrid interaction database: 2011 update. Nucleic Acids Res. 2011, 39, D698–D704. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. Gorilla: A tool for discovery and visualization of enriched go terms in ranked gene lists. BMC Bioinform. 2009, 10, 48. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.O. The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [CrossRef] [PubMed]
- Mulder, N.J.; Apweiler, R. The InterPro Database and Tools for Protein Domain Analysis. In Current Protocols in Bioinformatics; John Wiley & Sons, Inc.: Berlin, Germany, 2002. [Google Scholar]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2011, 40, D109–D114. [Google Scholar] [CrossRef] [PubMed]
- Van de Peer, Y. Calculate and draw custom Venn diagrams. Available online: http://bioinformatics.psb.ugent.be/webtools/Venn/ (accessed on 30 April 2014).
- Kasprzyk, A. BioMart: Driving a paradigm change in biological data management. Database 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.G.O. Gene Ontology. 2014. Available online: http://geneontology.org/ (accessed on 30 April 2014).
- Consortium, T.G.O. AmiGO 2. Available online: http://amigo2.berkeleybop.org/amigo (accessed on 30 April 2014).
- Consortium, T.G.O. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Carbon, S.; Ireland, A.; Mungall, C.J.; Shu, S.; Marshall, B.; Lewis, S. AmiGO: Online access to ontology and annotation data. Bioinformatics 2009, 25, 288–289. [Google Scholar] [CrossRef] [PubMed]
- Hu, J. The gene ontology normal usage tracking system. Available online: http://gowiki.tamu.edu/wiki/ (accessed on 30 March 2014).
- Yadav, S.R.; Yan, D.; Sevilem, I.; Helariutta, Y. Plasmodesmata-mediated intercellular signaling during plant growth and development. Front. Plant Sci. 2014, 5, 44. [Google Scholar] [CrossRef] [PubMed]
- Maule, A.J.; Gaudioso-Pedraza, R.; Benitez-Alfonso, Y. Callose deposition and symplastic connectivity are regulated prior to lateral root emergence. Commun. Integr. Biol. 2013, 6, e26531. [Google Scholar] [CrossRef] [PubMed]
- Vilaine, F.; Kerchev, P.; Clément, G.; Batailler, B.; Cayla, T.; Bill, L.; Gissot, L.; Dinant, S. Increased expression of a phloem membrane protein encoded by NHL26 alters phloem export and sugar partitioning in Arabidopsis. Plant Cell 2013, 25, 1689–1708. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.K.; Sieburth, L.E. Plasmodesmata formation: Poking holes in walls with ise. Curr. Biol. 2010, 20, R488–R490. [Google Scholar] [CrossRef] [PubMed]
- Burch-Smith, T.M.; Cui, Y.; Zambryski, P.C. Reduced levels of class 1 reversibly glycosylated polypeptide increase intercellular transport via plasmodesmata. Plant Signal. Behav. 2012, 7, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Zavaliev, R.; Levy, A.; Gera, A.; Epel, B.L. Subcellular dynamics and role of arabidopsis β-1,3-glucanases in cell-to-cell movement of tobamoviruses. Mol. Plant Microbe Interact. 2013, 26, 1016–1030. [Google Scholar] [CrossRef] [PubMed]
- Zalepa-King, L.; Citovsky, V. A plasmodesmal glycosyltransferase-like protein. PLoS ONE 2013, 8, e58025. [Google Scholar] [CrossRef] [PubMed]
- De Storme, N.; de Schrijver, J.; van Criekinge, W.; Wewer, V.; Dörmann, P.; Geelen, D. Glucan synthase-like8 and sterol methyltransferase2 are required for ploidy consistency of the sexual reproduction system in arabidopsis. Plant Cell 2013, 25, 387–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, M.; Cho, E.; Burch-Smith, T.M.; Zambryski, P.C. Plasmodesmata formation and cell-to-cell transport are reduced in decreased size exclusion limit 1 during embryogenesis in arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 5098–5103. [Google Scholar] [CrossRef] [PubMed]
- Burch-Smith, T.M.; Brunkard, J.O.; Choi, Y.G.; Zambryski, P.C. Organelle-nucleus cross-talk regulates plant intercellular communication via plasmodesmata. Proc. Natl. Acad. Sci. USA 2011, 108, E1451–E1460. [Google Scholar] [CrossRef] [PubMed]
- Burch-Smith, T.M.; Zambryski, P.C. Loss of increased size exclusion limit (ise)1 or ise2 increases the formation of secondary plasmodesmata. Curr. Biol. 2010, 20, 989–993. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Wang, X.; Zhu, M.; Zhang, Z.; Hong, Z. CalS7 encodes a callose synthase responsible for callose deposition in the phloem. Plant J. Cell Mol. Biol. 2011, 65, 1–14. [Google Scholar] [CrossRef]
- Benitez-Alfonso, Y.; Cilia, M.; San Roman, A.; Thomas, C.; Maule, A.; Hearn, S.; Jackson, D. Control of arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc. Natl. Acad. Sci. USA 2009, 106, 3615–3620. [Google Scholar] [CrossRef] [PubMed]
- Golomb, L.; Abu-Abied, M.; Belausov, E.; Sadot, E. Different subcellular localizations and functions of Arabidopsis myosin VIII. BMC Plant Biol. 2008, 8. [Google Scholar] [CrossRef] [PubMed]
- Maule, A.J. Plasmodesmata: Structure, function and biogenesis. Curr. Opin. Plant Biol. 2008, 11, 680–686. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kohan-Baghkheirati, E.; Geisler-Lee, J. Gene Expression, Protein Function and Pathways of Arabidopsis thaliana Responding to Silver Nanoparticles in Comparison to Silver Ions, Cold, Salt, Drought, and Heat. Nanomaterials 2015, 5, 436-467. https://doi.org/10.3390/nano5020436
Kohan-Baghkheirati E, Geisler-Lee J. Gene Expression, Protein Function and Pathways of Arabidopsis thaliana Responding to Silver Nanoparticles in Comparison to Silver Ions, Cold, Salt, Drought, and Heat. Nanomaterials. 2015; 5(2):436-467. https://doi.org/10.3390/nano5020436
Chicago/Turabian StyleKohan-Baghkheirati, Eisa, and Jane Geisler-Lee. 2015. "Gene Expression, Protein Function and Pathways of Arabidopsis thaliana Responding to Silver Nanoparticles in Comparison to Silver Ions, Cold, Salt, Drought, and Heat" Nanomaterials 5, no. 2: 436-467. https://doi.org/10.3390/nano5020436