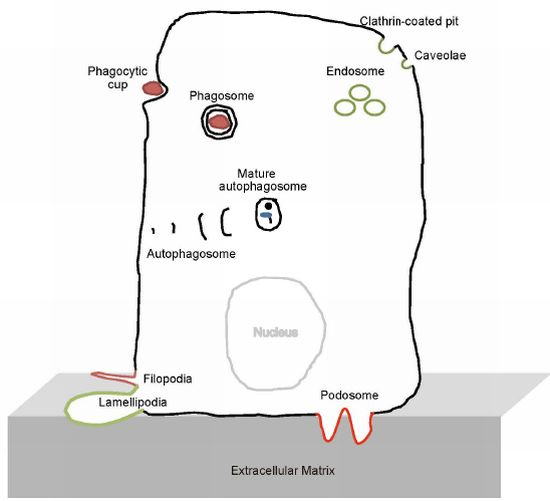
The BAR Domain Superfamily Proteins from Subcellular Structures to Human Diseases
Abstract
:1. Introduction

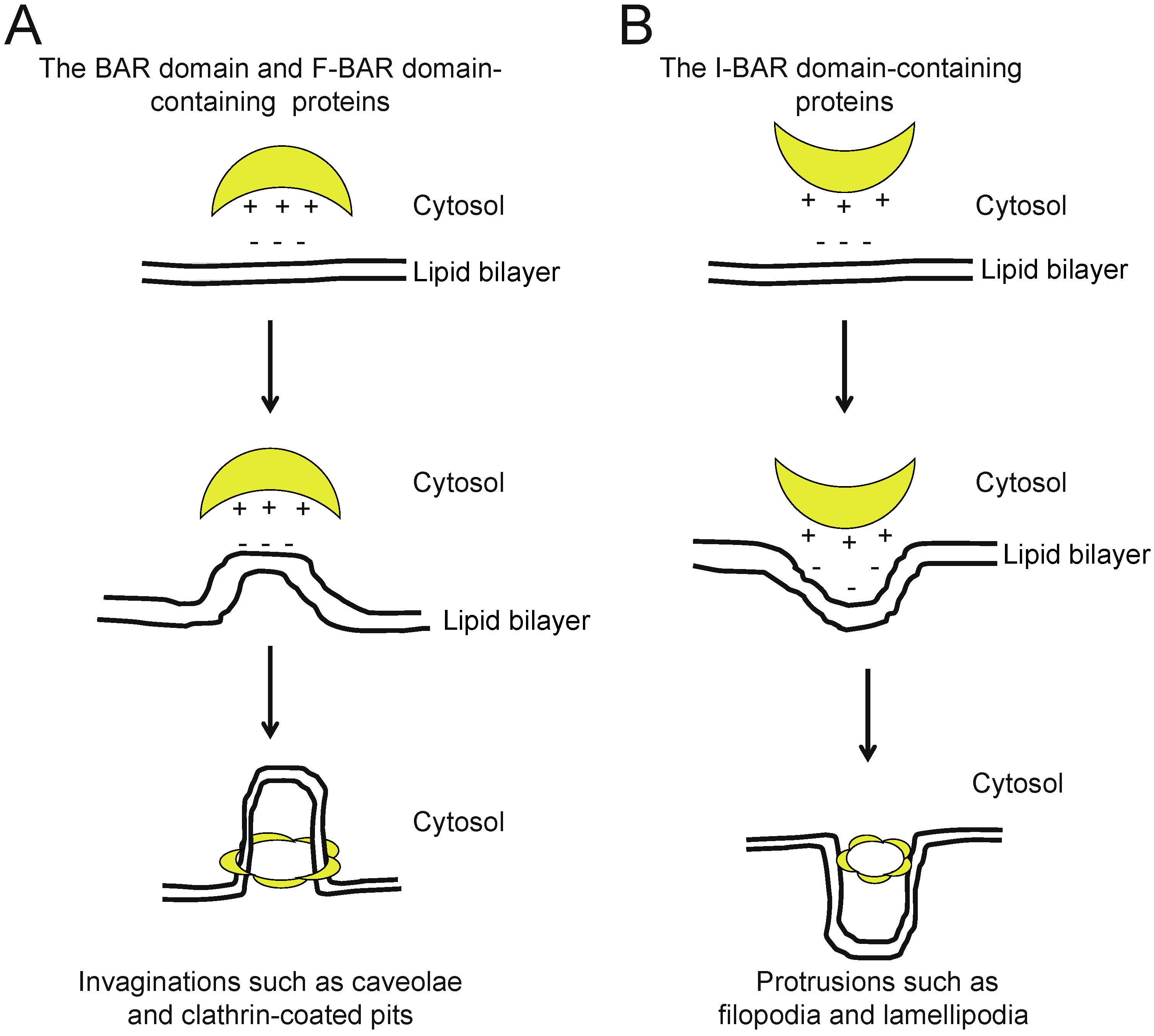
2. The BAR and N-BAR Domain Subfamily

2.1. Amphiphysin
2.2. Endophilin
2.3. Sorting Nexins
2.4. Tuba
2.5. APPL1
2.6. ASAP1
2.7. PICK1
3. F-BAR Domain Subfamily
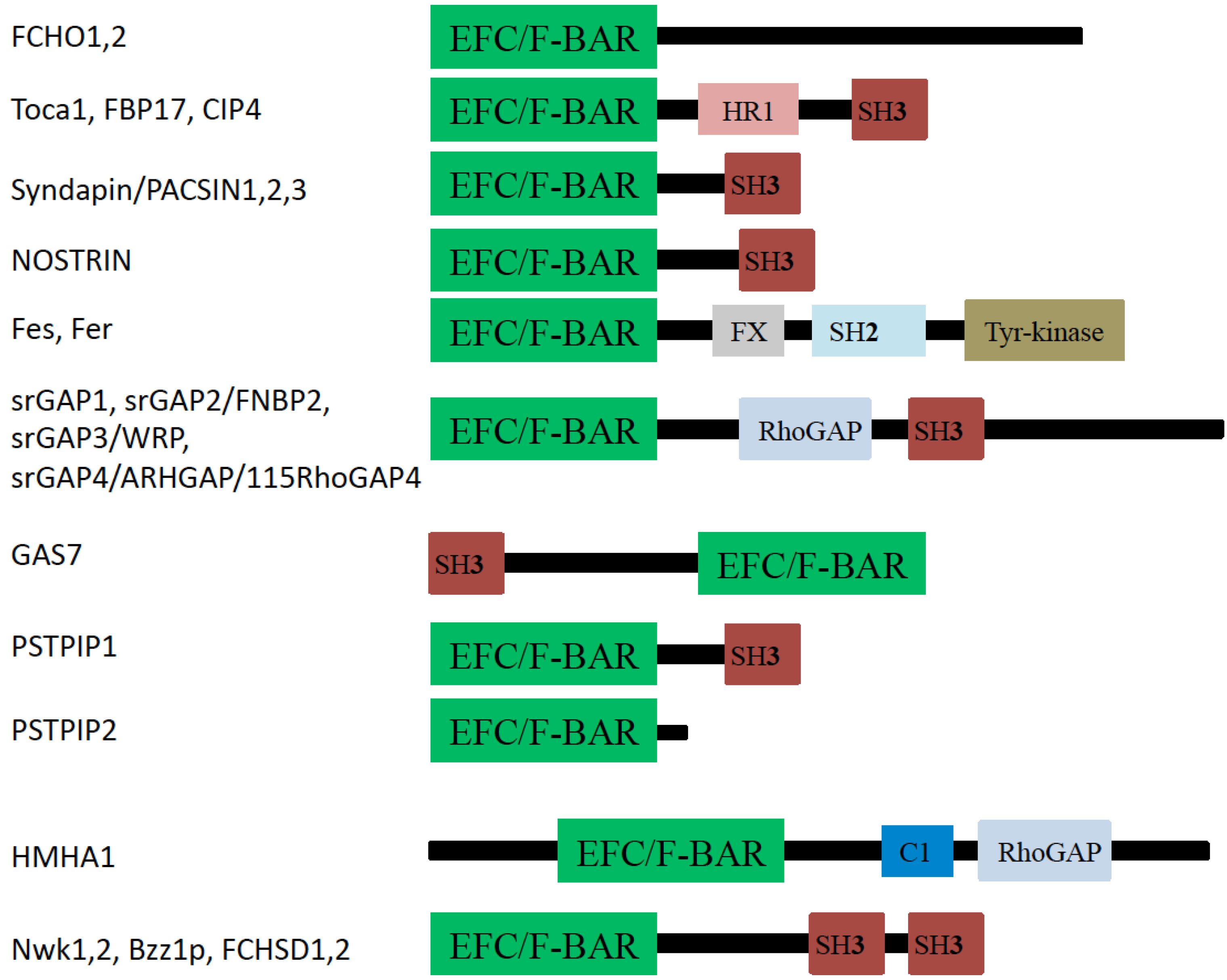
3.1. FCHo1 and FCHo2
3.2. FBP17, CIP4, and Toca-1
3.3. PACSIN (Syndapin)
3.4. NOSTRIN
3.5. Fes and Fer
3.6. srGAPs
3.7. GAS7
3.8. PSTPIP1/2 and Cdc15
4. I-BAR Subfamily

4.1. IRSp53
4.2. MIM
4.3. Pinkbar
5. Conclusions
Acknowledgements
References
- Qualmann, B.; Koch, D.; Kessels, M.M. Let’s go bananas: Revisiting the endocytic BAR code. EMBO J. 2011, 30, 3501–3515. [Google Scholar] [CrossRef]
- Suetsugu, S.; Toyooka, K.; Senju, Y. Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin. Cell Dev. Biol. 2010, 21, 340–349. [Google Scholar]
- Frost, A.; Unger, V.M.; De Camilli, P. The BAR domain superfamily: Membrane-molding macromolecules. Cell 2009, 137, 191–196. [Google Scholar] [CrossRef]
- Doherty, G.J.; McMahon, H.T. Mediation, modulation, and consequences of membrane-cytoskeleton interactions. Annu. Rev. Biophys. 2008, 37, 65–95. [Google Scholar] [CrossRef]
- Takenawa, T.; Suetsugu, S. The WASP-WAVE protein network: Connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2007, 8, 37–48. [Google Scholar] [CrossRef]
- Praefcke, G.J.; McMahon, H.T. The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 2004, 5, 133–147. [Google Scholar] [CrossRef]
- Schmid, S.L.; Frolov, V.A. Dynamin: Functional design of a membrane fission catalyst. Annu. Rev. Cell Dev. Biol. 2011, 27, 79–105. [Google Scholar] [CrossRef]
- Milosevic, I.; Giovedi, S.; Lou, X.; Raimondi, A.; Collesi, C.; Shen, H.; Paradise, S.; O’Toole, E.; Ferguson, S.; Cremona, O.; De Camilli, P. Recruitment of endophilin to clathrin-coated pit necks is required for efficient vesicle uncoating after fission. Neuron. 2011, 72, 587–601. [Google Scholar] [CrossRef]
- Peter, B.J.; Kent, H.M.; Mills, I.G.; Vallis, Y.; Butler, P.J.; Evans, P.R.; McMahon, H.T. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science 2004, 303, 495–499. [Google Scholar] [CrossRef]
- Gallop, J.L.; Jao, C.C.; Kent, H.M.; Butler, P.J.; Evans, P.R.; Langen, R.; McMahon, H.T. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006, 25, 2898–2910. [Google Scholar] [CrossRef]
- Tarricone, C.; Xiao, B.; Justin, N.; Walker, P.A.; Rittinger, K.; Gamblin, S.J.; Smerdon, S.J. The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature 2001, 411, 215–219. [Google Scholar] [CrossRef]
- Yamada, H.; Padilla-Parra, S.; Park, S.J.; Itoh, T.; Chaineau, M.; Monaldi, I.; Cremona, O.; Benfenati, F.; De Camilli, P.; Coppey-Moisan, M.; Tramier, M.; Galli, T.; Takei, K. Dynamic interaction of amphiphysin with N-WASP regulates actin assembly. J. Biol. Chem. 2009, 284, 34244–34256. [Google Scholar]
- Takei, K.; Slepnev, V.I.; Haucke, V.; De Camilli, P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat. Cell Biol. 1999, 1, 33–39. [Google Scholar] [CrossRef]
- McMahon, H.T.; Wigge, P.; Smith, C. Clathrin interacts specifically with amphiphysin and is displaced by dynamin. FEBS Lett. 1997, 413, 319–322. [Google Scholar] [CrossRef]
- Taylor, M.J.; Perrais, D.; Merrifield, C.J. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011, 9, e1000604. [Google Scholar] [CrossRef]
- Yamada, H.; Ohashi, E.; Abe, T.; Kusumi, N.; Li, S.A.; Yoshida, Y.; Watanabe, M.; Tomizawa, K.; Kashiwakura, Y.; Kumon, H.; Matsui, H.; Takei, K. Amphiphysin 1 is important for actin polymerization during phagocytosis. Mol. Biol. Cell 2007, 18, 4669–4680. [Google Scholar] [CrossRef]
- Bokoch, G.M. Regulation of innate immunity by Rho GTPases. Trends Cell Biol. 2005, 15, 163–171. [Google Scholar] [CrossRef]
- Chimini, G.; Chavrier, P. Function of Rho family proteins in actin dynamics during phagocytosis and engulfment. Nat. Cell Biol. 2000, 2, E191–196. [Google Scholar] [CrossRef]
- Soderling, S.H. Grab your partner with both hands: Cytoskeletal remodeling by Arp2/3 signaling. Sci. Signal 2009, 2, pe5. [Google Scholar] [CrossRef]
- Padrick, S.B.; Cheng, H.C.; Ismail, A.M.; Panchal, S.C.; Doolittle, L.K.; Kim, S.; Skehan, B.M.; Umetani, J.; Brautigam, C.A.; Leong, J.M.; Rosen, M.K. Hierarchical regulation of WASP/WAVE proteins. Mol. Cell 2008, 32, 426–438. [Google Scholar] [CrossRef]
- Razzaq, A.; Robinson, I.M.; McMahon, H.T.; Skepper, J.N.; Su, Y.; Zelhof, A.C.; Jackson, A.P.; Gay, N.J.; O’Kane, C.J. Amphiphysin is necessary for organization of the excitation-contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila. Gene. Develop. 2001, 15, 2967–2979. [Google Scholar] [CrossRef]
- Lee, E.; Marcucci, M.; Daniell, L.; Pypaert, M.; Weisz, O.A.; Ochoa, G.C.; Farsad, K.; Wenk, M.R.; De Camilli, P. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science 2002, 297, 1193–1196. [Google Scholar] [CrossRef]
- Nicot, A.S.; Toussaint, A.; Tosch, V.; Kretz, C.; Wallgren-Pettersson, C.; Iwarsson, E.; Kingston, H.; Garnier, J.M.; Biancalana, V.; Oldfors, A.; Mandel, J.L.; Laporte, J. Mutations in amphiphysin 2 (BIN1) disrupt interaction with dynamin 2 and cause autosomal recessive centronuclear myopathy. Nat. Genet. 2007, 39, 1134–1139. [Google Scholar] [CrossRef]
- Bitoun, M.; Maugenre, S.; Jeannet, P.Y.; Lacène, E.; Ferrer, X.; Laforêt, P.; Martin, J.J.; Laporte, J.; Lochmüller, H.; Beggs, A.H.; Fardeau, M.; Eymard, B.; Romero, N.B.; Guicheney, P. Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat. Genet. 2005, 37, 1207–1209. [Google Scholar] [CrossRef]
- Kojima, C.; Hashimoto, A.; Yabuta, I.; Hirose, M.; Hashimoto, S.; Kanaho, Y.; Sumimoto, H.; Ikegami, T.; Sabe, H. Regulation of Bin1 SH3 domain binding by phosphoinositides. EMBO J. 2004, 23, 4413–4422. [Google Scholar] [CrossRef]
- Jao, C.C.; Hegde, B.G.; Gallop, J.L.; Hegde, P.B.; McMahon, H.T.; Haworth, I.S.; Langen, R. Roles of amphipathic helices and the bin/amphiphysin/rvs (BAR) domain of endophilin in membrane curvature generation. J. Biol. Chem. 2010, 285, 20164–20170. [Google Scholar]
- Masuda, M.; Takeda, S.; Sone, M.; Ohki, T.; Mori, H.; Kamioka, Y.; Mochizuki, N. Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms. EMBO J. 2006, 25, 2889–2897. [Google Scholar] [CrossRef]
- Llobet, A.; Gallop, J.L.; Burden, J.J.; Camdere, G.; Chandra, P.; Vallis, Y.; Hopkins, C.R.; Lagnado, L.; McMahon, H.T. Endophilin drives the fast mode of vesicle retrieval in a ribbon synapses. J. Neurosci. 2011, 31, 8512–8519. [Google Scholar]
- Otsuki, M.; Itoh, T.; Takenawa, T. Neural Wiskott-Aldrich syndrome protein is recruited to rafts and associates with endophilin A in response to epidermal growth factor. J. Biol. Chem. 2003, 278, 6461–6469. [Google Scholar] [CrossRef]
- Gortat, A.; Jouve San-Roman, M.; Vannier, C.; Schmidt, A.A. Single point mutation in the bin/amphiphysin/RVS (BAR) sequence of endophilin impairs dimerization, membrane shaping, and SRC homology 3 domain-mediated partnership. J. Biol. Chem. 2012, 287, 4232–4247. [Google Scholar] [CrossRef]
- Trempe, J.F.; Chen, C.X.; Grenier, K.; Camacho, E.M.; Kozlov, G.; McPherson, P.S.; Gehring, K.; Fon, E.A. SH3 domains from a subset of BAR proteins define a Ubl-binding domain and implicate parkin in synaptic ubiquitination. Mol. Cell 2009, 36, 1034–1047. [Google Scholar] [CrossRef]
- Ralser, M.; Nonhoff, U.; Albrecht, M.; Lengauer, T.; Wanker, E.E.; Lehrach, H.; Krobitsch, S. Ataxin-2 and huntingtin interact with endophilin-A complexes to function in plastin-associated pathways. Hum. Mol. Genet. 2005, 14, 2893–2909. [Google Scholar] [CrossRef]
- Takahashi, Y.; Coppola, D.; Matsushita, N.; Cualing, H.D.; Sun, M.; Sato, Y.; Liang, C.; Jung, J.U.; Cheng, J.Q.; Mulé, J.J.; Pledger, W.J.; Wang, H.G. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 2007, 9, 1142–1151. [Google Scholar] [CrossRef]
- Karbowski, M.; Jeong, S.Y.; Youle, R.J. Endophilin B1 is required for the maintenance of mitochondrial morphology. J. Cell Biol. 2004, 166, 1027–1039. [Google Scholar] [CrossRef]
- Takahashi, Y.; Karbowski, M.; Yamaguchi, H.; Kazi, A.; Wu, J.; Sebti, S.M.; Youle, R.J.; Wang, H.G. Loss of Bif-1 suppresses Bax/Bak conformational change and mitochondrial apoptosis. Mol. Cell Biol. 2005, 25, 9369–9382. [Google Scholar]
- Pylypenko, O.; Lundmark, R.; Rasmuson, E.; Carlsson, S.R.; Rak, A. The PX-BAR membrane-remodeling unit of sorting nexin 9. EMBO J. 2007, 26, 4788–4800. [Google Scholar] [CrossRef]
- Lundmark, R.; Carlsson, S.R. SNX9-a prelude to vesicle release. J. Cell Sci. 2009, 122, 5–11. [Google Scholar] [CrossRef]
- Seet, L.F.; Hong, W. The Phox (PX) domain proteins and membrane traffic. Biochim. Biophys. Acta 2006, 1761, 878–896. [Google Scholar] [CrossRef]
- Yarar, D.; Surka, M.C.; Leonard, M.C.; Schmid, S.L. SNX9 activities are regulated by multiple phosphoinositides through both PX and BAR domains. Traffic 2008, 9, 133–146. [Google Scholar] [CrossRef]
- Håberg, K.; Lundmark, R.; Carlsson, S.R. SNX18 is an SNX9 paralog that acts as a membrane tubulator in AP-1-positive endosomal trafficking. J. Cell Sci. 2008, 121, 1495–1505. [Google Scholar] [CrossRef]
- Yarar, D.; Waterman-Storer, C.M.; Schmid, S.L. SNX9 couples actin assembly to phosphoinositide signals and is required for membrane remodeling during endocytosis. Dev. Cell 2007, 13, 43–56. [Google Scholar] [CrossRef]
- Kovacs, E.M.; Makar, R.S.; Gertler, F.B. Tuba stimulates intracellular N-WASP-dependent actin assembly. J. Cell Sci. 2006, 119, 2715–2726. [Google Scholar] [CrossRef]
- Salazar, M.A.; Kwiatkowski, A.V.; Pellegrini, L.; Cestra, G.; Butler, M.H.; Rossman, K.L.; Serna, D.M.; Sondek, J.; Gertler, F.B.; De Camilli, P. Tuba, a novel protein containing bin/amphiphysin/Rvs and Dbl homology domains, links dynamin to regulation of the actin cytoskeleton. J. Biol. Chem. 2003, 278, 49031–49043. [Google Scholar]
- Otani, T.; Ichii, T.; Aono, S.; Takeichi, M. Cdc42 GEF Tuba regulates the junctional configuration of simple epithelial cells. J. Cell Biol. 2006, 175, 135–146. [Google Scholar] [CrossRef]
- Kovacs, E.M.; Verma, S.; Thomas, S.G.; Yap, A.S. Tuba and N-WASP function cooperatively to position the central lumen during epithelial cyst morphogenesis. Cell Adh. Migr. 2011, 5, 344–350. [Google Scholar] [CrossRef]
- Inoue, H.; Ha, V.L.; Prekeris, R.; Randazzo, P.A. Arf GTPase-activating protein ASAP1 interacts with Rab11 effector FIP3 and regulates pericentrosomal localization of transferrin receptor-positive recycling endosome. Mol. Biol. Cell 2008, 19, 4224–4237. [Google Scholar] [CrossRef]
- Zoncu, R.; Perera, R.M.; Balkin, D.M.; Pirruccello, M.; Toomre, D.; De Camilli, P.A. Phosphoinositide switch controls the maturation and signaling properties of APPL endosomes. Cell 2009, 136, 1110–1121. [Google Scholar] [CrossRef]
- Mao, X.; Kikani, C.K.; Riojas, R.A.; Langlais, P.; Wang, L.; Ramos, F.J.; Fang, Q.; Christ-Roberts, C.Y.; Hong, J.Y.; Kim, R.Y.; Liu, F.; Dong, L.Q. APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nat. Cell Biol. 2006, 8, 516–523. [Google Scholar] [CrossRef]
- Miaczynska, M.; Christoforidis, S.; Giner, A.; Shevchenko, A.; Uttenweiler-Joseph, S.; Habermann, B.; Wilm, M.; Parton, R.G.; Zerial, M. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell 2004, 116, 445–456. [Google Scholar] [CrossRef]
- Kam, J.L.; Miura, K.; Jackson, T.R.; Gruschus, J.; Roller, P.; Stauffer, S.; Clark, J.; Aneja, R.; Randazzo, P.A. Phosphoinositide-dependent activation of the ADP-ribosylation factor GTPase-activating protein ASAP1. Evidence for the pleckstrin homology domain functioning as an allosteric site. J. Biol. Chem. 2000, 275, 9653–9663. [Google Scholar]
- Bharti, S.; Inoue, H.; Bharti, K.; Hirsch, D.S.; Nie, Z.; Yoon, H.Y.; Artym, V.; Yamada, K.M.; Mueller, S.C.; Barr, V.A.; Randazzo, P.A. Src-dependent phosphorylation of ASAP1 regulates podosomes. Mol. Cell Biol. 2007, 27, 8271–8283. [Google Scholar] [CrossRef]
- Brown, M.T.; Andrade, J.; Radhakrishna, H.; Donaldson, J.G.; Cooper, J.A.; Randazzo, P.A. ASAP1, a phospholipid-dependent arf GTPase-activating protein that associates with and is phosphorylated by Src. Mol. Cell Biol. 1998, 18, 7038–7051. [Google Scholar]
- Oda, A.; Wada, I.; Miura, K.; Okawa, K.; Kadoya, T.; Kato, T.; Nishihara, H.; Maeda, M.; Tanaka, S.; Nagashima, K.; Nishitani, C.; Matsuno, K.; Ishino, M.; Machesky, L.M.; Fujita, H.; Randazzo, P. CrkL directs ASAP1 to peripheral focal adhesions. J. Biol. Chem. 2003, 278, 6456–6460. [Google Scholar]
- Shiba, Y.; Randazzo, P.A. GEFH1 binds ASAP1 and regulates podosome formation. Biochem. Biophys. Res. Commun. 2011, 406, 574–579. [Google Scholar] [CrossRef]
- Ha, V.L.; Bharti, S.; Inoue, H.; Vass, W.C.; Campa, F.; Nie, Z.; de Gramont, A.; Ward, Y.; Randazzo, P.A. ASAP3 is a focal adhesion-associated Arf GAP that functions in cell migration and invasion. J. Biol. Chem. 2008, 283, 14915–14926. [Google Scholar]
- Kaverina, I.; Stradal, T.E.; Gimona, M. Podosome formation in cultured A7r5 vascular smooth muscle cells requires Arp2/3-dependent de-novo actin polymerization at discrete microdomains. J. Cell Sci. 2003, 116, 4915–4924. [Google Scholar] [CrossRef]
- Ochoa, G.C.; Slepnev, V.I.; Neff, L.; Ringstad, N.; Takei, K.; Daniell, L.; Kim, W.; Cao, H.; McNiven, M.; Baron, R.; De Camilli, P.A. Functional link between dynamin and the actin cytoskeleton at podosomes. J. Cell Biol. 2000, 150, 377–389. [Google Scholar] [CrossRef]
- Rocca, D.L.; Martin, S.; Jenkins, E.L.; Hanley, J.G. Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat. Cell Biol. 2008, 10, 259–271. [Google Scholar] [CrossRef]
- Suh, Y.H.; Pelkey, K.A.; Lavezzari, G.; Roche, P.A.; Huganir, R.L.; McBain, C.J.; Roche, K.W. Corequirement of PICK1 binding and PKC phosphorylation for stable surface expression of the metabotropic glutamate receptor mGluR7. Neuron 2008, 58, 736–748. [Google Scholar] [CrossRef]
- Pan, L.; Wu, H.; Shen, C.; Shi, Y.; Jin, W.; Xia, J.; Zhang, M. Clustering and synaptic targeting of PICK1 requires direct interaction between the PDZ domain and lipid membranes. EMBO J. 2007, 26, 4576–4587. [Google Scholar] [CrossRef]
- He, Y.; Liwo, A.; Weinstein, H.; Scheraga, H.A. PDZ binding to the BAR domain of PICK1 is elucidated by coarse-grained molecular dynamics. J. Mol. Biol. 2011, 405, 298–314. [Google Scholar] [CrossRef]
- Lu, W.; Ziff, E.B. PICK1 interacts with ABP/GRIP to regulate AMPA receptor trafficking. Neuron 2005, 47, 407–421. [Google Scholar] [CrossRef]
- Perez, J.L.; Khatri, L.; Chang, C.; Srivastava, S.; Osten, P.; Ziff, E.B. PICK1 targets activated protein kinase Calpha to AMPA receptor clusters in spines of hippocampal neurons and reduces surface levels of the AMPA-type glutamate receptor subunit 2. J. Neurosci. 2001, 21, 5417–5428. [Google Scholar]
- Henne, W.M.; Kent, H.M.; Ford, M.G.; Hegde, B.G.; Daumke, O.; Butler, P.J.; Mittal, R.; Langen, R.; Evans, P.R.; McMahon, H.T. Structure and analysis of FCHo2 F-BAR domain: A dimerizing and membrane recruitment module that effects membrane curvature. Structure 2007, 15, 839–852. [Google Scholar] [CrossRef]
- Henne, W.M.; Boucrot, E.; Meinecke, M.; Evergren, E.; Vallis, Y.; Mittal, R.; McMahon, H.T. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science 2010, 328, 1281–1284. [Google Scholar] [CrossRef]
- Shimada, A.; Niwa, H.; Tsujita, K.; Suetsugu, S.; Nitta, K.; Hanawa-Suetsugu, K.; Akasaka, R.; Nishino, Y.; Toyama, M.; Chen, L.; Liu, Z.J.; Wang, B.C.; Yamamoto, M.; Terada, T.; Miyazawa, A.; Tanaka, A.; Sugano, S.; Shirouzu, M.; Nagayama, K.; Takenawa, T.; Yokoyama, S. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell 2007, 129, 761–772. [Google Scholar] [CrossRef]
- Tsujita, K.; Suetsugu, S.; Sasaki, N.; Furutani, M.; Oikawa, T.; Takenawa, T. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J. Cell Biol. 2006, 172, 269–279. [Google Scholar] [CrossRef]
- Itoh, T.; Erdmann, K.S.; Roux, A.; Habermann, B.; Werner, H.; De Camilli, P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev. Cell 2005, 9, 791–804. [Google Scholar] [CrossRef]
- Ho, H.Y.; Rohatgi, R.; Lebensohn, A.M.; Le, Ma; Li, J.; Gygi, S.P.; Kirschner, M.W. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell 2004, 118, 203–216. [Google Scholar] [CrossRef]
- Chang, L.; Adams, R.D.; Saltiel, A.R. The TC10-interacting protein CIP4/2 is required for insulin-stimulated Glut4 translocation in 3T3L1 adipocytes. Proc. Natl. Acad. Sci. USA 2002, 99, 12835–12840. [Google Scholar] [CrossRef]
- Hartig, S.M.; Ishikura, S.; Hicklen, R.S.; Feng, Y.; Blanchard, E.G.; Voelker, K.A.; Pichot, C.S.; Grange, R.W.; Raphael, R.M.; Klip, A.; Corey, S.J. The F-BAR protein CIP4 promotes GLUT4 endocytosis through bidirectional interactions with N-WASp and Dynamin-2. J. Cell Sci. 2009, 122, 2283–2291. [Google Scholar] [CrossRef]
- Lodhi, I.J.; Chiang, S.H.; Chang, L.; Vollenweider, D.; Watson, R.T.; Inoue, M.; Pessin, J.E.; Saltiel, A.R. Gapex-5, a Rab31 guanine nucleotide exchange factor that regulates Glut4 trafficking in adipocytes. Cell Metab. 2007, 5, 59–72. [Google Scholar] [CrossRef]
- Feng, Y.; Hartig, S.M.; Bechill, J.E.; Blanchard, E.G.; Caudell, E.; Corey, S.J. The Cdc42-interacting protein-4 (CIP4) gene knock-out mouse reveals delayed and decreased endocytosis. J. Biol. Chem. 2010, 285, 4348–4354. [Google Scholar]
- Kakimoto, T.; Katoh, H.; Negishi, M. Identification of splicing variants of Rapostlin, a novel RND2 effector that interacts with neural Wiskott-Aldrich syndrome protein and induces neurite branching. J. Biol. Chem. 2004, 279, 14104–14110. [Google Scholar] [CrossRef]
- Aspenström, P.A. Cdc42 target protein with homology to the non-kinase domain of FER has a potential role in regulating the actin cytoskeleton. Curr. Biol. 1997, 7, 479–487. [Google Scholar] [CrossRef]
- Takano, K.; Toyooka, K.; Suetsugu, S. EFC/F-BAR proteins and the N-WASP-WIP complex induce membrane curvature-dependent actin polymerization. EMBO J. 2008, 27, 2817–2828. [Google Scholar] [CrossRef]
- Tian, L.; Nelson, D.L.; Stewart, D.M. Cdc42-interacting protein 4 mediates binding of the Wiskott-Aldrich syndrome protein to microtubules. J. Biol. Chem. 2000, 275, 7854–7861. [Google Scholar] [CrossRef]
- Linder, S.; Higgs, H.; Hüfner, K.; Schwarz, K.; Pannicke, U.; Aepfelbacher, M. The polarization defect of Wiskott-Aldrich syndrome macrophages is linked to dislocalization of the Arp2/3 complex. J. Immunol. 2000, 165, 221–225. [Google Scholar]
- Tsuboi, S.; Takada, H.; Hara, T.; Mochizuki, N.; Funyu, T.; Saitoh, H.; Terayama, Y.; Yamaya, K.; Ohyama, C.; Nonoyama, S.; Ochs, H.D. FBP17 mediates a common molecular step in the formation of podosomes and phagocytic cups in macrophages. J. Biol. Chem. 2009, 284, 8548–8556. [Google Scholar]
- Linder, S.; Hufner, K.; Wintergerst, U.; Aepfelbacher, M. Microtubule-dependent formation of podosomal adhesion structures in primary human macrophages. J. Cell Sci. 2000, 113, 4165–4176. [Google Scholar]
- Kamioka, Y.; Fukuhara, S.; Sawa, H.; Nagashima, K.; Masuda, M.; Matsuda, M.; Mochizuki, N. A novel dynamin-associating molecule, formin-binding protein 17, induces tubular membrane invaginations and participates in endocytosis. J. Biol. Chem. 2004, 279, 40091–40099. [Google Scholar]
- Kakimoto, T.; Katoh, H.; Negishi, M. Regulation of neuronal morphology by Toca-1, an F-BAR/EFC protein that induces plasma membrane invagination. J. Biol. Chem. 2006, 281, 29042–29053. [Google Scholar] [CrossRef]
- Pichot, C.S.; Arvanitis, C.; Hartig, S.M.; Jensen, S.A.; Bechill, J.; Marzouk, S.; Yu, J.; Frost, J.A.; Corey, S.J. Cdc42-interacting protein 4 promotes breast cancer cell invasion and formation of invadopodia through activation of N-WASp. Cancer Res. 2010, 70, 8347–8356. [Google Scholar]
- Hu, J.; Mukhopadhyay, A.; Truesdell, P.; Chander, H.; Mukhopadhyay, U.K.; Mak, A.S.; Craig, A.W. Cdc42-interacting protein 4 is a Src substrate that regulates invadopodia and invasiveness of breast tumors by promoting MT1-MMP endocytosis. J. Cell Sci. 2011, 124, 1739–1751. [Google Scholar] [CrossRef]
- Wu, X.; Gan, B.; Yoo, Y.; Guan, J.L. FAK-mediated Src phosphorylation of endophilin A2 inhibits endocytosis of MT1-MMP and promotes ECM degradation. Dev. Cell 2005, 9, 185–196. [Google Scholar] [CrossRef]
- Lee, K.; Gallop, J.L.; Rambani, K.; Kirschner, M.W. Self-assembly of filopodia-like structures on supported lipid bilayers. Science 2010, 329, 1341–1345. [Google Scholar] [CrossRef]
- Suetsugu, S. The proposed functions of membrane curvatures mediated by the BAR domain superfamily proteins. J. Biochem. 2010, 148, 1–12. [Google Scholar] [CrossRef]
- Toguchi, M.; Richnau, N.; Ruusala, A.; Aspenström, P. Members of the CIP4 family of proteins participate in the regulation of platelet-derived growth factor receptor-beta-dependent actin reorganization and migration. Biol. Cell 2010, 102, 215–230. [Google Scholar] [CrossRef]
- Koduru, S.; Kumar, L.; Massaad, M.J.; Ramesh, N.; Le Bras, S.; Ozcan, E.; Oyoshi, M.K.; Kaku, M.; Fujiwara, Y.; Kremer, L.; King, S.; Fuhlbrigge, R.; Rodig, S.; Sage, P.; Carman, C.; Alcaide, P.; Luscinskas, F.W.; Geha, R.S. Cdc42 interacting protein 4 (CIP4) is essential for integrin-dependent T-cell trafficking. Proc. Natl. Acad. Sci. USA 2010, 107, 16252–16256. [Google Scholar]
- Bu, W.; Chou, A.M.; Lim, K.B.; Sudhaharan, T.; Ahmed, S. The Toca-1-N-WASP complex links filopodial formation to endocytosis. J. Biol. Chem. 2009, 284, 11622–11636. [Google Scholar]
- Bu, W.; Lim, K.B.; Yu, Y.H.; Chou, A.M.; Sudhaharan, T.; Ahmed, S. Cdc42 interaction with N-WASP and Toca-1 regulates membrane tubulation, vesicle formation and vesicle motility: Implications for endocytosis. PLoS One 2010, 5, e12153. [Google Scholar]
- Hu, J.; Mukhopadhyay, A.; Craig, A.W. Transducer of Cdc42-dependent actin assembly promotes epidermal growth factor-induced cell motility and invasiveness. J. Biol. Chem. 2011, 286, 2261–2272. [Google Scholar]
- Huett, A.; Ng, A.; Cao, Z.; Kuballa, P.; Komatsu, M.; Daly, M.J.; Podolsky, D.K.; Xavier, R.J. A novel hybrid yeast-human network analysis reveals an essential role for FNBP1L in antibacterial autophagy. J. Immunol. 2009, 182, 4917–4930. [Google Scholar] [CrossRef]
- Hu, J.; Troglio, F.; Mukhopadhyay, A.; Everingham, S.; Kwok, E.; Scita, G.; Craig, A.W. F-BAR-containing adaptor CIP4 localizes to early endosomes and regulates Epidermal Growth Factor Receptor trafficking and downregulation. Cell. Signal. 2009, 21, 1686–1697. [Google Scholar] [CrossRef]
- Holbert, S.; Dedeoglu, A.; Humbert, S.; Saudou, F.; Ferrante, R.J.; Néri, C. Cdc42-interacting protein 4 binds to huntingtin: neuropathologic and biological evidence for a role in Huntington’s disease. Proc. Natl. Acad. Sci. USA 2003, 100, 2712–2717. [Google Scholar] [CrossRef]
- Tsuji, E.; Tsuji, Y.; Fujiwara, T.; Ogata, S.; Tsukamoto, K.; Saku, K. Splicing variant of Cdc42 interacting protein-4 disrupts beta-catenin-mediated cell-cell adhesion: Expression and function in renal cell carcinoma. Biochem. Biophys. Res. Commun. 2006, 339, 1083–1088. [Google Scholar] [CrossRef]
- Modregger, J.; Ritter, B.; Witter, B.; Paulsson, M.; Plomann, M. All three PACSIN isoforms bind to endocytic proteins and inhibit endocytosis. J. Cell Sci. 2000, 113, 4511–4521. [Google Scholar]
- Kessels, M.M.; Qualmann, B. Syndapins integrate N-WASP in receptor-mediated endocytosis. EMBO J. 2002, 21, 6083–6094. [Google Scholar] [CrossRef]
- Qualmann, B.; Kessels, M.M.; Kelly, R.B. Molecular links between endocytosis and the actin cytoskeleton. J. Cell Biol. 2000, 150, F111–116. [Google Scholar] [CrossRef]
- Senju, Y.; Itoh, Y.; Takano, K.; Hamada, S.; Suetsugu, S. Essential role of PACSIN2/syndapin-II in caveolae membrane sculpting. J. Cell Sci. 2011, 124, 2032–2040. [Google Scholar] [CrossRef]
- Hansen, C.G.; Howard, G.; Nichols, B.J. Pacsin 2 is recruited to caveolae and functions in caveolar biogenesis. J. Cell Sci. 2011, 124, 2777–2785. [Google Scholar] [CrossRef]
- Shimada, A.; Takano, K.; Shirouzu, M.; Hanawa-Suetsugu, K.; Terada, T.; Toyooka, K.; Umehara, T.; Yamamoto, M.; Yokoyama, S.; Suetsugu, S. Mapping of the basic amino-acid residues responsible for tubulation and cellular protrusion by the EFC/F-BAR domain of pacsin2/Syndapin II. FEBS Lett. 2010, 584, 1111–1118. [Google Scholar] [CrossRef]
- Wang, Q.; Navarro, M.V.; Peng, G.; Molinelli, E.; Goh, S.L.; Judson, B.L.; Rajashankar, K.R.; Sondermann, H. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc. Natl. Acad. Sci. USA 2009, 106, 12700–12705. [Google Scholar]
- Simpson, F.; Hussain, N.K.; Qualmann, B.; Kelly, R.B.; Kay, B.K.; McPherson, P.S.; Schmid, S.L. SH3-domain-containing proteins function at distinct steps in clathrin-coated vesicle formation. Nat. Cell Biol. 1999, 1, 119–124. [Google Scholar] [CrossRef]
- Qualmann, B.; Kelly, R.B. Syndapin isoforms participate in receptor-mediated endocytosis and actin organization. J. Cell Biol. 2000, 148, 1047–1062. [Google Scholar] [CrossRef]
- Rao, Y.; Ma, Q.; Vahedi-Faridi, A.; Sundborger, A.; Pechstein, A.; Puchkov, D.; Luo, L.; Shupliakov, O.; Saenger, W.; Haucke, V. Molecular basis for SH3 domain regulation of F-BAR-mediated membrane deformation. Proc. Natl. Acad. Sci. USA 2010, 107, 8213–8218. [Google Scholar]
- Zarrinpar, A.; Bhattacharyya, R.P.; Lim, W.A. The structure and function of proline recognition domains. Sci. STKE 2003, 2003, RE8. [Google Scholar]
- Modregger, J.; DiProspero, N.A.; Charles, V.; Tagle, D.A.; Plomann, M. PACSIN 1 interacts with huntingtin and is absent from synaptic varicosities in presymptomatic Huntington’s disease brains. Hum. Mol. Genet. 2002, 11, 2547–2558. [Google Scholar] [CrossRef]
- Schilling, K.; Opitz, N.; Wiesenthal, A.; Oess, S.; Tikkanen, R.; Müller-Esterl, W.; Icking, A. Translocation of endothelial nitric-oxide synthase involves a ternary complex with caveolin-1 and NOSTRIN. Mol. Biol. Cell 2006, 17, 3870–3880. [Google Scholar] [CrossRef]
- Icking, A.; Matt, S.; Opitz, N.; Wiesenthal, A.; Müller-Esterl, W.; Schilling, K. NOSTRIN functions as a homotrimeric adaptor protein facilitating internalization of eNOS. J. Cell Sci. 2005, 118, 5059–5069. [Google Scholar] [CrossRef]
- Mookerjee, R.P.; Wiesenthal, A.; Icking, A.; Hodges, S.J.; Davies, N.A.; Schilling, K.; Sen, S.; Williams, R.; Novelli, M.; Müller-Esterl, W.; Jalan, R. Increased gene and protein expression of the novel eNOS regulatory protein NOSTRIN and a variant in alcoholic hepatitis. Gastroenterology 2007, 132, 2533–2541. [Google Scholar] [CrossRef]
- Greer, P. Closing in on the biological functions of Fps/Fes and Fer. Nat. Rev. Mol. Cell Biol. 2002, 3, 278–289. [Google Scholar] [CrossRef]
- Ahmed, S.; Bu, W.; Lee, R.T.; Maurer-Stroh, S.; Goh, W.I. F-BAR domain proteins: Families and function. Commun. Integr. Biol. 2010, 3, 116–121. [Google Scholar] [CrossRef]
- Craig, A.W. FES/FER kinase signaling in hematopoietic cells and leukemias. Front. Biosci. 2012, 17, 861–875. [Google Scholar] [CrossRef]
- Itoh, T.; Hasegawa, J.; Tsujita, K.; Kanaho, Y.; Takenawa, T. The tyrosine kinase Fer is a downstream target of the PLD-PA pathway that regulates cell migration. Sci. Signal. 2009, 2, ra52. [Google Scholar] [CrossRef]
- Di Fulvio, M.; Frondorf, K.; Henkels, K.M.; Grunwald, W.C.; Cool, D.; Gomez-Cambronero, J. Phospholipase D2 (PLD2) Shortens the Time Required for Myeloid Leukemic Cell Differentiation: Mechanism of Action. J. Biol. Chem. 2012, 287, 393–407. [Google Scholar]
- Kogata, N.; Masuda, M.; Kamioka, Y.; Yamagishi, A.; Endo, A.; Okada, M.; Mochizuki, N. Identification of Fer tyrosine kinase localized on microtubules as a platelet endothelial cell adhesion molecule-1 phosphorylating kinase in vascular endothelial cells. Mol. Biol. Cell 2003, 14, 3553–3564. [Google Scholar] [CrossRef]
- Zhang, S.; Chitu, V.; Stanley, E.R.; Elliott, B.E.; Greer, P.A. Fes tyrosine kinase expression in the tumor niche correlates with enhanced tumor growth, angiogenesis, circulating tumor cells, metastasis, and infiltrating macrophages. Cancer Res. 2011, 71, 1465–1473. [Google Scholar] [CrossRef]
- Miyata, Y.; Watanabe, S.; Matsuo, T.; Hayashi, T.; Sakai, H.; Xuan, J.W.; Greer, P.A.; Kanda, S. Pathological significance and predictive value for biochemical recurrence of c-Fes expression in prostate cancer. Prostate 2012, 72, 201–208. [Google Scholar] [CrossRef]
- Guo, C.; Stark, G.R. FER tyrosine kinase (FER) overexpression mediates resistance to quinacrine through EGF-dependent activation of NF-kappaB. Proc. Natl. Acad. Sci. USA 2011, 108, 7968–7973. [Google Scholar] [CrossRef]
- Makovski, A.; Yaffe, E.; Shpungin, S.; Nir, U. Intronic promoter drives the BORIS-regulated expression of FerT in colon carcinoma cells. J. Biol. Chem. 2013, in press. [Google Scholar]
- Soderling, S.H.; Binns, K.L.; Wayman, G.A.; Davee, S.M.; Ong, S.H.; Pawson, T.; Scott, J.D. The WRP component of the WAVE-1 complex attenuates Rac-mediated signalling. Nat. Cell Biol. 2002, 4, 970–975. [Google Scholar] [CrossRef]
- Guerrier, S.; Coutinho-Budd, J.; Sassa, T.; Gresset, A.; Jordan, N.V.; Chen, K.; Jin, W.L.; Frost, A.; Polleux, F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell 2009, 138, 990–1004. [Google Scholar] [CrossRef]
- Neukomm, L.J.; Kinchen, J.M. SRGP-1 regulation, targets, and contribution to cell killing in C. elegans. Small Gtpases 2011, 2, 177–181. [Google Scholar] [CrossRef]
- Linkermann, A.; Gelhaus, C.; Lettau, M.; Qian, J.; Kabelitz, D.; Janssen, O. Identification of interaction partners for individual SH3 domains of Fas ligand associated members of the PCH protein family in T lymphocytes. Biochim. Biophys. Acta 2009, 1794, 168–176. [Google Scholar] [CrossRef]
- Carlson, B.R.; Lloyd, K.E.; Kruszewski, A.; Kim, I.H.; Rodriguiz, R.M.; Heindel, C.; Faytell, M.; Dudek, S.M.; Wetsel, W.C.; Soderling, S.H. WRP/srGAP3 facilitates the initiation of spine development by an inverse F-BAR domain, and its loss impairs long-term memory. J. Neurosci. 2011, 31, 2447–2460. [Google Scholar]
- Endris, V.; Wogatzky, B.; Leimer, U.; Bartsch, D.; Zatyka, M.; Latif, F.; Maher, E.R.; Tariverdian, G.; Kirsch, S.; Karch, D.; Rappold, G.A. The novel Rho-GTPase activating gene MEGAP/ srGAP3 has a putative role in severe mental retardation. Proc. Natl. Acad. Sci. USA 2002, 99, 11754–11759. [Google Scholar]
- You, J.J.; Lin-Chao, S. Gas7 functions with N-WASP to regulate the neurite outgrowth of hippocampal neurons. J. Biol. Chem. 2010, 285, 11652–11666. [Google Scholar] [CrossRef]
- She, B.R.; Liou, G.G.; Lin-Chao, S. Association of the growth-arrest-specific protein Gas7 with F-actin induces reorganization of microfilaments and promotes membrane outgrowth. Exp. Cell Res. 2002, 273, 34–44. [Google Scholar] [CrossRef]
- Yeung, Y.G.; Soldera, S.; Stanley, E.R. A novel macrophage actin-associated protein (MAYP) is tyrosine-phosphorylated following colony stimulating factor-1 stimulation. J. Biol. Chem. 1998, 273, 30638–30642. [Google Scholar] [CrossRef]
- Wu, Y.; Spencer, S.D.; Lasky, L.A. Tyrosine phosphorylation regulates the SH3-mediated binding of the Wiskott-Aldrich syndrome protein to PSTPIP, a cytoskeletal-associated protein. J. Biol. Chem. 1998, 273, 5765–5770. [Google Scholar] [CrossRef]
- Wise, C.A.; Gillum, J.D.; Seidman, C.E.; Lindor, N.M.; Veile, R.; Bashiardes, S.; Lovett, M. Mutations in CD2BP1 disrupt binding to PTP PEST and are responsible for PAPA syndrome, an autoinflammatory disorder. Hum. Mol. Genet. 2002, 11, 961–969. [Google Scholar] [CrossRef]
- Chitu, V.; Pixley, F.J.; Macaluso, F.; Larson, D.R.; Condeelis, J.; Yeung, Y.G.; Stanley, E.R. The PCH family member MAYP/PSTPIP2 directly regulates F-actin bundling and enhances filopodia formation and motility in macrophages. Mol. Biol. Cell 2005, 16, 2947–2959. [Google Scholar] [CrossRef]
- Ferguson, P.J.; Bing, X.; Vasef, M.A.; Ochoa, L.A.; Mahgoub, A.; Waldschmidt, T.J.; Tygrett, L.T.; Schlueter, A.J.; El-Shanti, H. A missense mutation in pstpip2 is associated with the murine autoinflammatory disorder chronic multifocal osteomyelitis. Bone 2006, 38, 41–47. [Google Scholar] [CrossRef]
- Grosse, J.; Chitu, V.; Marquardt, A.; Hanke, P.; Schmittwolf, C.; Zeitlmann, L.; Schropp, P.; Barth, B.; Yu, P.; Paffenholz, R.; Stumm, G.; Nehls, M.; Stanley, E.R. Mutation of mouse Mayp/Pstpip2 causes a macrophage autoinflammatory disease. Blood 2006, 107, 3350–3358. [Google Scholar] [CrossRef]
- Chitu, V.; Ferguson, P.J.; de Bruijn, R.; Schlueter, A.J.; Ochoa, L.A.; Waldschmidt, T.J.; Yeung, Y.G.; Stanley, E.R. Primed innate immunity leads to autoinflammatory disease in PSTPIP2-deficient cmo mice. Blood 2009, 114, 2497–2505. [Google Scholar]
- Spencer, S.; Dowbenko, D.; Cheng, J.; Li, W.; Brush, J.; Utzig, S.; Simanis, V.; Lasky, L.A. PSTPIP: A tyrosine phosphorylated cleavage furrow-associated protein that is a substrate for a PEST tyrosine phosphatase. J. Cell Biol. 1997, 138, 845–860. [Google Scholar] [CrossRef]
- Fankhauser, C.; Reymond, A.; Cerutti, L.; Utzig, S.; Hofmann, K.; Simanis, V. The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell 1995, 82, 435–444. [Google Scholar]
- Wu, J.Q.; Kuhn, J.R.; Kovar, D.R.; Pollard, T.D. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell 2003, 5, 723–734. [Google Scholar] [CrossRef]
- Wu, J.Q.; Pollard, T.D. Counting cytokinesis proteins globally and locally in fission yeast. Science 2005, 310, 310–314. [Google Scholar] [CrossRef]
- Roberts-Galbraith, R.H.; Ohi, M.D.; Ballif, B.A.; Chen, J.S.; McLeod, I.; McDonald, W.H.; Gygi, S.P.; Yates, J.R.; Gould, K.L. Dephosphorylation of F-BAR protein Cdc15 modulates its conformation and stimulates its scaffolding activity at the cell division site. Mol. Cell 2010, 39, 86–99. [Google Scholar] [CrossRef]
- Carnahan, R.H.; Gould, K.L. The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. J. Cell Biol. 2003, 162, 851–862. [Google Scholar] [CrossRef]
- Roberts-Galbraith, R.H.; Chen, J.S.; Wang, J.; Gould, K.L. The SH3 domains of two PCH family members cooperate in assembly of the Schizosaccharomyces pombe contractile ring. J. Cell Biol. 2009, 184, 113–127. [Google Scholar] [CrossRef]
- Itoh, T.; De Camilli, P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim. Biophys. Acta 2006, 1761, 897–912. [Google Scholar] [CrossRef]
- Aspenström, P. Roles of F-BAR/PCH proteins in the regulation of membrane dynamics and actin reorganization. Int. Rev. Cell Mol. Biol. 2009, 272, 1–31. [Google Scholar] [CrossRef]
- Meitinger, F.; Boehm, M.E.; Hofmann, A.; Hub, B.; Zentgraf, H.; Lehmann, W.D.; Pereira, G. Phosphorylation-dependent regulation of the F-BAR protein Hof1 during cytokinesis. Genes Dev. 2011, 25, 875–888. [Google Scholar] [CrossRef]
- Millard, T.H.; Bompard, G.; Heung, M.Y.; Dafforn, T.R.; Scott, D.J.; Machesky, L.M.; Fütterer, K. Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J. 2005, 24, 240–250. [Google Scholar] [CrossRef]
- Suetsugu, S.; Murayama, K.; Sakamoto, A.; Hanawa-Suetsugu, K.; Seto, A.; Oikawa, T.; Mishima, C.; Shirouzu, M.; Takenawa, T.; Yokoyama, S. The RAC binding domain/IRSp53-MIM homology domain of IRSp53 induces RAC-dependent membrane deformation. J. Biol. Chem. 2006, 281, 35347–35358. [Google Scholar]
- Mattila, P.K.; Pykäläinen, A.; Saarikangas, J.; Paavilainen, V.O.; Vihinen, H.; Jokitalo, E.; Lappalainen, P. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J. Cell Biol. 2007, 176, 953–964. [Google Scholar] [CrossRef]
- Pykäläinen, A.; Boczkowska, M.; Zhao, H.; Saarikangas, J.; Rebowski, G.; Jansen, M.; Hakanen, J.; Koskela, E.V.; Peränen, J.; Vihinen, H.; Jokitalo, E.; Salminen, M.; Ikonen, E.; Dominguez, R.; Lappalainen, P. Pinkbar is an epithelial-specific BAR domain protein that generates planar membrane structures. Nat. Struct. Mol. Biol. 2011, 18, 902–907. [Google Scholar]
- Saarikangas, J.; Zhao, H.; Pykäläinen, A.; Laurinmäki, P.; Mattila, P.K.; Kinnunen, P.K.; Butcher, S.J.; Lappalainen, P. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr. Biol. 2009, 19, 95–107. [Google Scholar]
- Yamagishi, A.; Masuda, M.; Ohki, T.; Onishi, H.; Mochizuki, N.A. Novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J. Biol. Chem. 2004, 279, 14929–14936. [Google Scholar]
- Yang, C.; Hoelzle, M.; Disanza, A.; Scita, G.; Svitkina, T. Coordination of membrane and actin cytoskeleton dynamics during filopodia protrusion. PLoS One 2009, 4, e5678. [Google Scholar]
- Nakagawa, H.; Miki, H.; Nozumi, M.; Takenawa, T.; Miyamoto, S.; Wehland, J.; Small, J.V. IRSp53 is colocalised with WAVE2 at the tips of protruding lamellipodia and filopodia independently of Mena. J. Cell Sci. 2003, 116, 2577–2583. [Google Scholar] [CrossRef]
- Miki, H.; Yamaguchi, H.; Suetsugu, S.; Takenawa, T. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature 2000, 408, 732–735. [Google Scholar]
- Suetsugu, S.; Kurisu, S.; Oikawa, T.; Yamazaki, D.; Oda, A.; Takenawa, T. Optimization of WAVE2 complex-induced actin polymerization by membrane-bound IRSp53, PIP(3), and Rac. J. Cell Biol. 2006, 173, 571–585. [Google Scholar] [CrossRef]
- Lim, K.B.; Bu, W.; Goh, W.I.; Koh, E.; Ong, S.H.; Pawson, T.; Sudhaharan, T.; Ahmed, S. The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J. Biol. Chem. 2008, 283, 20454–20472. [Google Scholar]
- Krugmann, S.; Jordens, I.; Gevaert, K.; Driessens, M.; Vandekerckhove, J.; Hall, A. Cdc42 induces filopodia by promoting the formation of an IRSp53: Mena complex. Curr. Biol. 2001, 11, 1645–1655. [Google Scholar] [CrossRef]
- Govind, S.; Kozma, R.; Monfries, C.; Lim, L.; Ahmed, S. Cdc42Hs facilitates cytoskeletal reorganization and neurite outgrowth by localizing the 58-kD insulin receptor substrate to filamentous actin. J. Cell Biol. 2001, 152, 579–594. [Google Scholar] [CrossRef]
- Hori, K.; Konno, D.; Maruoka, H.; Sobue, K. MALS is a binding partner of IRSp53 at cell-cell contacts. FEBS Lett. 2003, 554, 30–34. [Google Scholar] [CrossRef]
- Hori, K.; Yasuda, H.; Konno, D.; Maruoka, H.; Tsumoto, T.; Sobue, K. NMDA receptor-dependent synaptic translocation of insulin receptor substrate p53 via protein kinase C signaling. J. Neurosci. 2005, 25, 2670–2681. [Google Scholar] [CrossRef]
- Massari, S.; Perego, C.; Padovano, V.; D’Amico, A.; Raimondi, A.; Francolini, M.; Pietrini, G. LIN7 mediates the recruitment of IRSp53 to tight junctions. Traffic 2009, 10, 246–257. [Google Scholar] [CrossRef]
- Soltau, M.; Berhörster, K.; Kindler, S.; Buck, F.; Richter, D.; Kreienkamp, H.J. Insulin receptor substrate of 53 kDa links postsynaptic shank to PSD-95. J. Neurochem. 2004, 90, 659–665. [Google Scholar] [CrossRef]
- Disanza, A.; Mantoani, S.; Hertzog, M.; Gerboth, S.; Frittoli, E.; Steffen, A.; Berhoerster, K.; Kreienkamp, H.J.; Milanesi, F.; Di Fiore, P.P.; Ciliberto, A.; Stradal, T.E.; Scita, G. Regulation of cell shape by Cdc42 is mediated by the synergic actin-bundling activity of the Eps8-IRSp53 complex. Nat. Cell Biol. 2006, 8, 1337–1347. [Google Scholar] [CrossRef]
- Abou-Kheir, W.; Isaac, B.; Yamaguchi, H.; Cox, D. Membrane targeting of WAVE2 is not sufficient for WAVE2-dependent actin polymerization: A role for IRSp53 in mediating the interaction between Rac and WAVE2. J. Cell Sci. 2008, 121, 379–390. [Google Scholar] [CrossRef]
- Shi, J.; Scita, G.; Casanova, J.E. WAVE2 signaling mediates invasion of polarized epithelial cells by Salmonella typhimurium. J. Biol. Chem. 2005, 280, 29849–29855. [Google Scholar]
- Clarke, M.; Engel, U.; Giorgione, J.; Müller-Taubenberger, A.; Prassler, J.; Veltman, D.; Gerisch, G. Curvature recognition and force generation in phagocytosis. BMC Biol. 2010, 8, 154. [Google Scholar] [CrossRef]
- Mattila, P.K.; Salminen, M.; Yamashiro, T.; Lappalainen, P. Mouse MIM, a tissue-specific regulator of cytoskeletal dynamics, interacts with ATP-actin monomers through its C-terminal WH2 domain. J. Biol. Chem. 2003, 278, 8452–8459. [Google Scholar]
- Machesky, L.M.; Johnston, S.A. MIM: A multifunctional scaffold protein. J. Mol. Med. (Berl.) 2007, 85, 569–576. [Google Scholar]
- Callahan, C.A.; Ofstad, T.; Horng, L.; Wang, J.K.; Zhen, H.H.; Coulombe, P.A.; Oro, A.E. MIM/BEG4, a Sonic hedgehog-responsive gene that potentiates Gli-dependent transcription. Genes Dev. 2004, 18, 2724–2729. [Google Scholar] [CrossRef]
- Saarikangas, J.; Mattila, P.K.; Varjosalo, M.; Bovellan, M.; Hakanen, J.; Calzada-Wack, J.; Tost, M.; Jennen, L.; Rathkolb, B.; Hans, W.; et al. Missing-in-metastasis MIM/MTSS1 promotes actin assembly at intercellular junctions and is required for integrity of kidney epithelia. J. Cell Sci. 2011, 124, 1245–1255. [Google Scholar] [CrossRef]
- Varjosalo, M.; Taipale, J. Hedgehog: Functions and mechanisms. Genes Dev. 2008, 22, 2454–2472. [Google Scholar] [CrossRef]
- Lee, S.H.; Kerff, F.; Chereau, D.; Ferron, F.; Klug, A.; Dominguez, R. Structural basis for the actin-binding function of missing-in-metastasis. Structure 2007, 15, 145–155. [Google Scholar] [CrossRef]
- Morimoto, S.; Nishimura, N.; Terai, T.; Manabe, S.; Yamamoto, Y.; Shinahara, W.; Miyake, H.; Tashiro, S.; Shimada, M.; Sasaki, T. Rab13 mediates the continuous endocytic recycling of occludin to the cell surface. J. Biol. Chem. 2005, 280, 2220–2228. [Google Scholar]
- Yamamura, R.; Nishimura, N.; Nakatsuji, H.; Arase, S.; Sasaki, T. The interaction of JRAB/MICAL-L2 with Rab8 and Rab13 coordinates the assembly of tight junctions and adherens junctions. Mol. Biol. Cell 2008, 19, 971–983. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Safari, F.; Suetsugu, S. The BAR Domain Superfamily Proteins from Subcellular Structures to Human Diseases. Membranes 2012, 2, 91-117. https://doi.org/10.3390/membranes2010091
Safari F, Suetsugu S. The BAR Domain Superfamily Proteins from Subcellular Structures to Human Diseases. Membranes. 2012; 2(1):91-117. https://doi.org/10.3390/membranes2010091
Chicago/Turabian StyleSafari, Fatemeh, and Shiro Suetsugu. 2012. "The BAR Domain Superfamily Proteins from Subcellular Structures to Human Diseases" Membranes 2, no. 1: 91-117. https://doi.org/10.3390/membranes2010091
APA StyleSafari, F., & Suetsugu, S. (2012). The BAR Domain Superfamily Proteins from Subcellular Structures to Human Diseases. Membranes, 2(1), 91-117. https://doi.org/10.3390/membranes2010091