Antioxidant Defenses in Plants with Attention to Prunus and Citrus spp.
Abstract
:1. Introduction
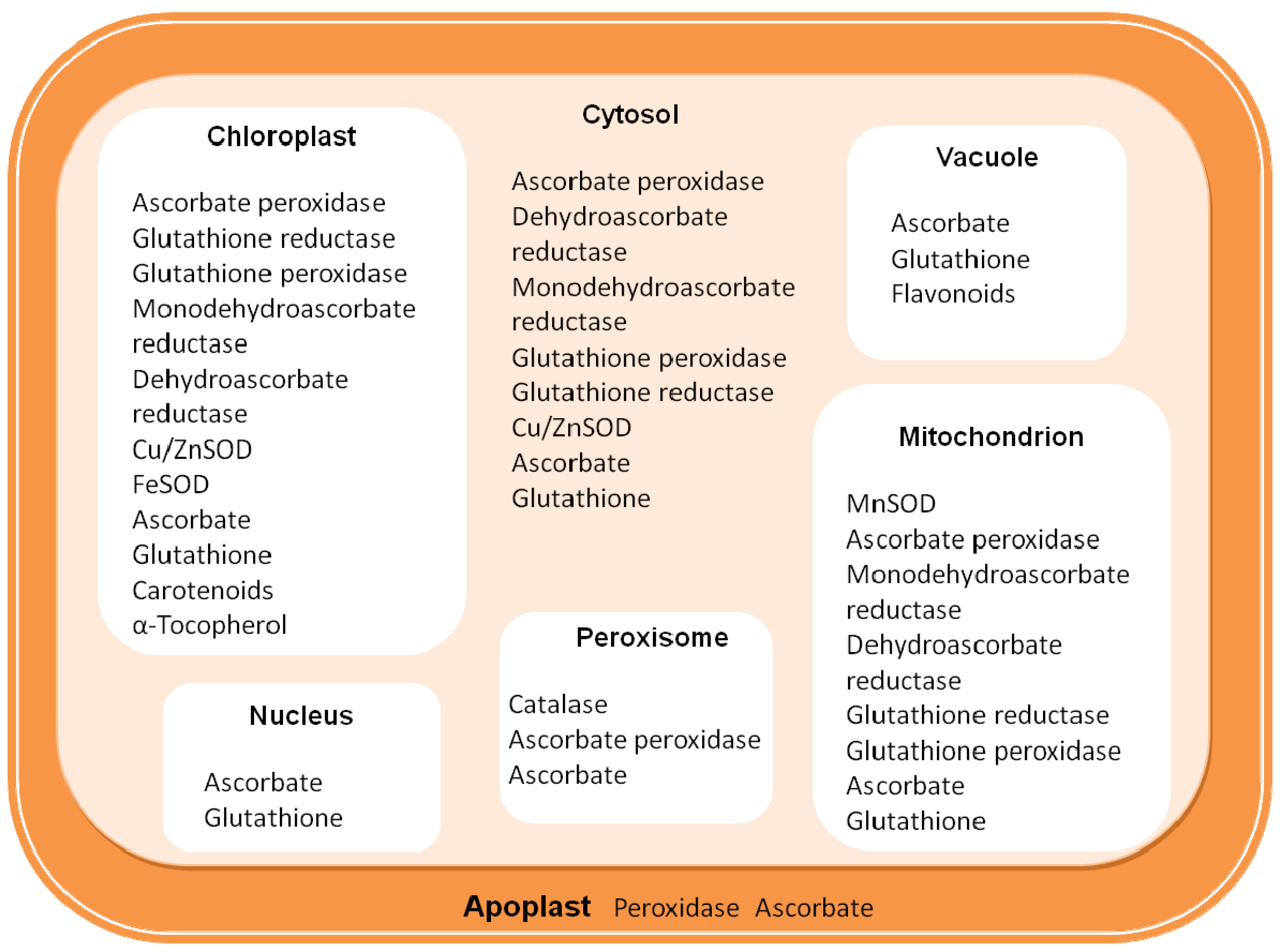
2. Defense against ROS
2.1. Enzymatic Component of ROS Scavenging Defenses
2.1.1. Superoxide Dismutase (SODs)
| SOD | ||
| O2•− + O2•− + 2H+ | → | O2 + H2O2 (K2 = 2.4 × 109 M−1 s−1) |
| CAT | ||
| H2O2 + H2O2 | → | 2H2O + O2 (K1 = 1.7 × 107 M−1 s−1) |
| PX | ||
| H2O2 + R(OH)2 | → | 2H2O + R(O)2 (K4 = 0.2–1 × 103 M−1 s−1) |
| Non enzymatic antioxidant molecules | Location | Primary ROS | |
|---|---|---|---|
| Ascorbate (vitamin C) | Chl, Cyt, Mit, Per, Apo | H2O2, O2•− | |
| Glutathione reduced (GSH) | Mit, Nuc, Per, Chl, Cyt, Apo Vac, | H2O2 | |
| β-Carotene | Chl, | 1O2 | |
| α-tocopherol (vitamin E) | Cell and plastid membrane | ROOH, 1O2 | |
| Zeaxanthin | Chl, | 1O2 | |
| Antioxidant enzymes | |||
| Enzyme | EC number | ||
| Superoxide dismutase (SOD) | 1.15.1.1 | Cyt, Apo, (Cu/ZnSOD);Chl, (Cu/ZnSOD; FeSOD); Mit, (MnSOD); Per, (Cu/ZnSOD) | O2•− |
| Ascorbate peroxidase (APX) | 1.11.1.11 | Chl, Cyt, Mit, Per, Apo | H2O2 |
| Catalase (CAT) | 1.11.1.6 | Per | H2O2 |
| Peroxidase (non-specific) | 1.11.1.7 | Cyt; CW | H2O2 |
| Glutathione peroxidase (GPX) | 1.11.1.19 | Cyt, Mit, | H2O2, ROOH |
| Glutathione reductase (GR) | 1. 6.4.2 | Mit, Cyt, Chl, Per | ROOH |
2.1.2. Catalase (CATs)
| Species | Class I | Class II | Class III |
|---|---|---|---|
| Gossypium hirsutum (cotton) | SU2 | SU1 | |
| Nicotiana plumbaginifolia | Cat1 | Cat2 | Cat3 |
| Ricinus communis L. (castor bean) | CAT2 | CAT1 | |
| Zea mays (maize) | CAT-2 | CAT-3 | CAT-1 |
| Arabidopsis thaliana | CAT2 | CAT1 | CAT3 |
| Lycopersicon esculentum (tomato) | TOMCAT1 | ||
| Solanum tuberosum (potato) | Cat2St | ||
| Prunus persica (peach) | Cat1 | Cat2 |
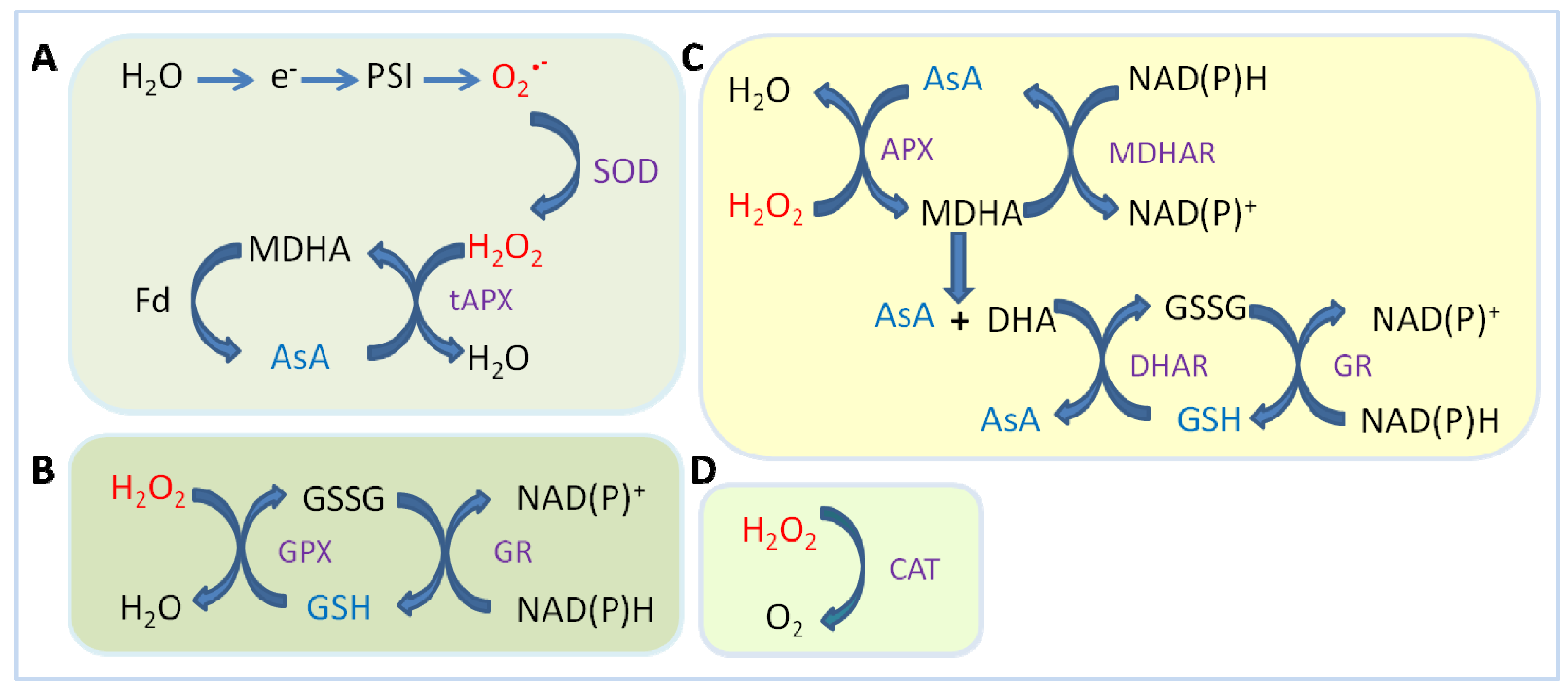
2.1.3. Ascorbate Peroxidase (APX)
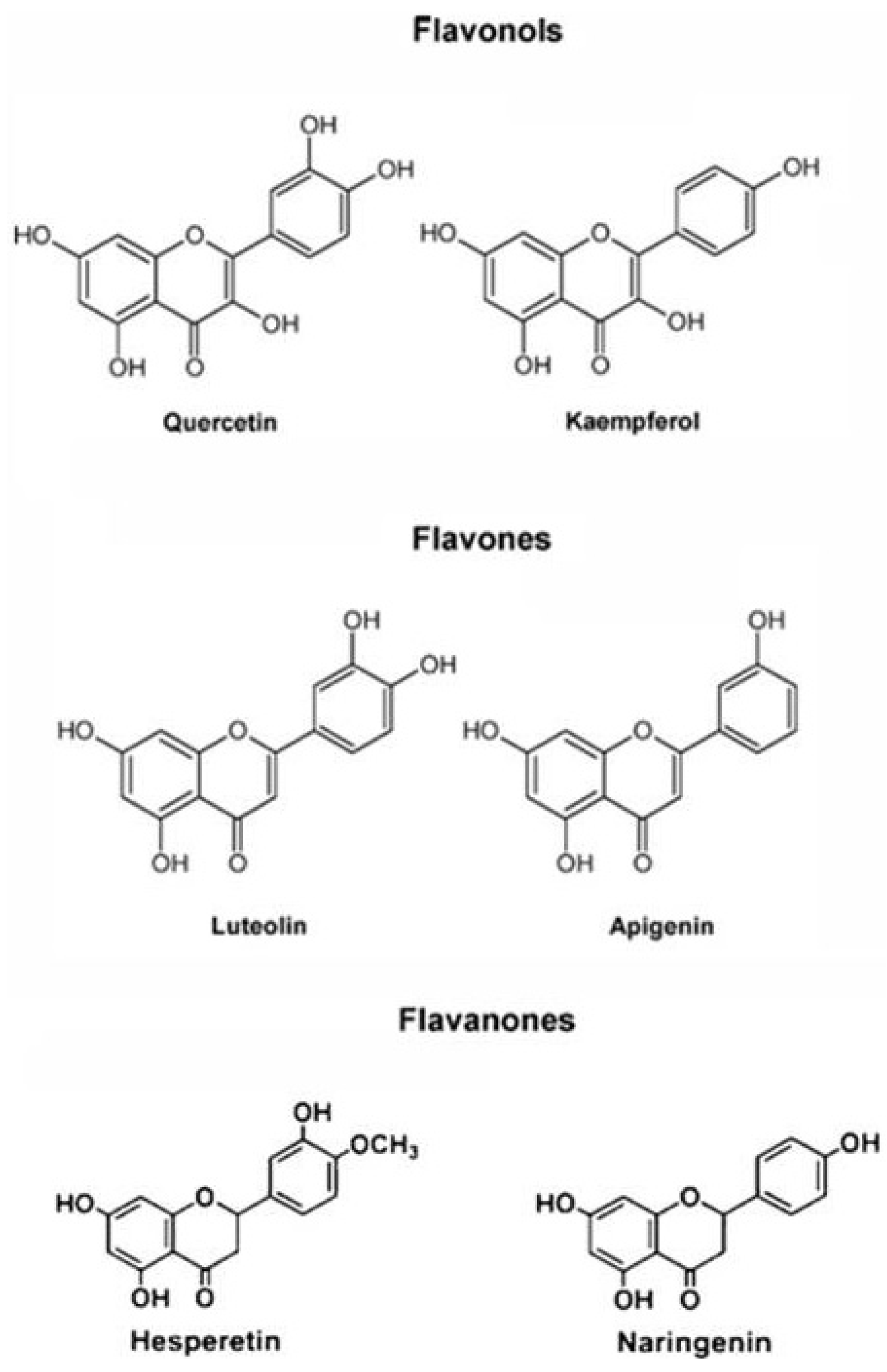
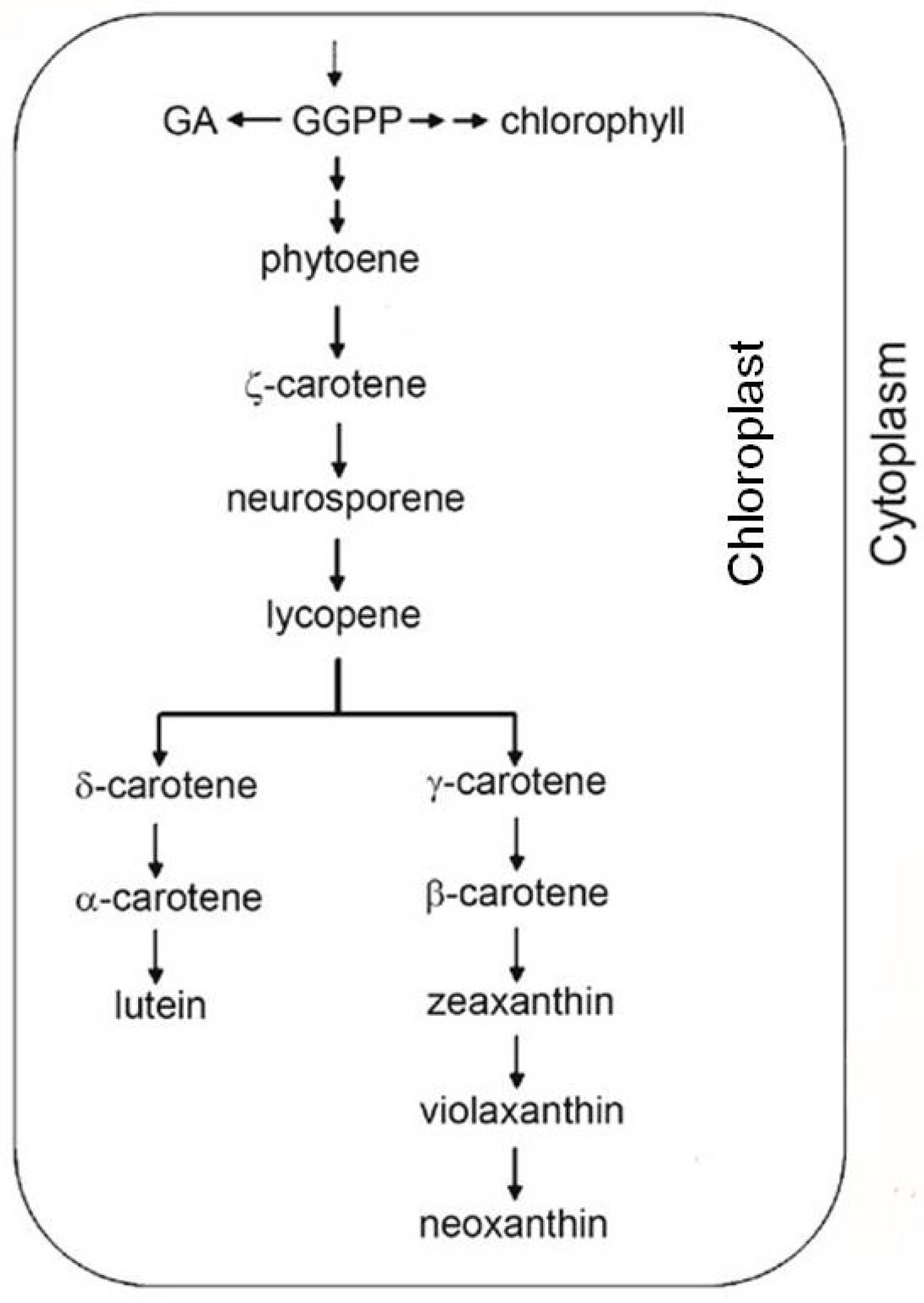
2.2. Antioxidant Molecules

3. Antioxidant System during Development, Maturation and Fruit Ripening with Particular Attention to Prunus and Citrus spp.
| Compounds | Means ± SE | |
|---|---|---|
| Abibi et al. [145] | Cantin et al. [142] | |
| Vitamin C | 4.0 ± 0.1 | 3.7 ± 1.0 |
| Total phenolics | 32.6 ± 0.7 | 36.4 ± 11.0 |
| Flavonoids | 12.5 ± 0.6 | 8.8 ± 0.4 |
| Anthocyanins | 3.2 ± 0.2 | 3.0 ± 0.3 |
| RAC | 464.2 ± 12.5 | 405.0 ± 4.9 |
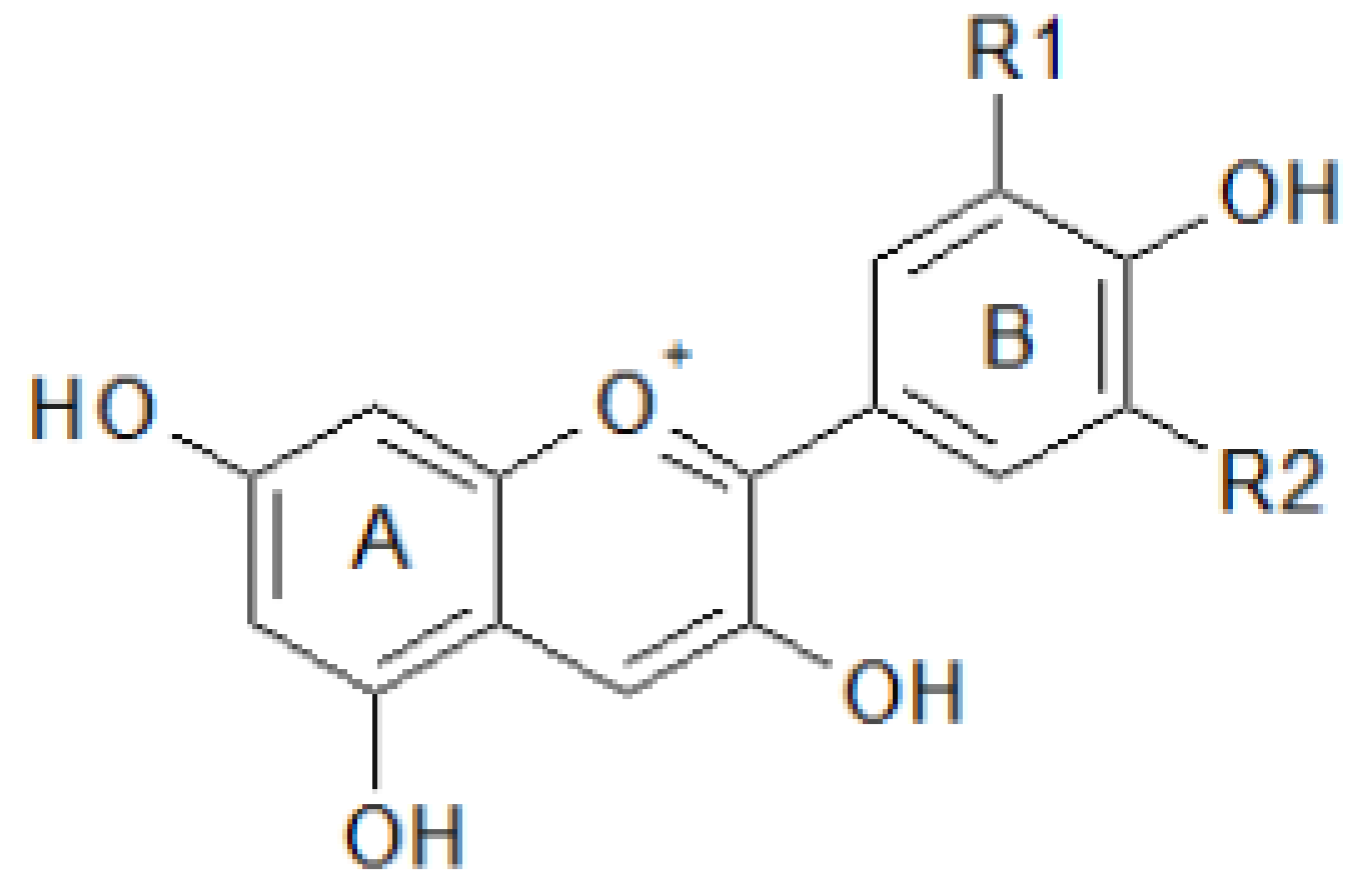
| Compounds | Content range | |
|---|---|---|
| Ascorbic acid vitamin C
mg/100g fresh weight | Nectarines white flesh | 5–14 |
| Nectarines yellow flesh | 5–7 | |
| Peaches white flesh | 6–8 | |
| Peaches yellow flesh | 4–13 | |
| Total phenolics
mg/100g fresh weight | Nectarines white flesh | 14–102 |
| Nectarines yellow flesh | 18–54 | |
| Peaches white flesh | 28–111 | |
| Peaches yellow flesh | 21–61 | |
| Carotenoids | Nectarines white flesh | 7–11 |
| µg/100g fresh weight | Nectarines yellow flesh | 80–157 |
| Peaches white flesh | 8–17 | |
| Peaches yellow flesh | 95–197 | |

4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Halliwell, B. Reactive species and antioxidants. Redox biology is fundamental theme of aerobic life. Plant Physiol. 2006, 141, 312–322. [Google Scholar] [CrossRef]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signaling in the response of plants to abiotic stress. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant Cell Environ. 2009, 33, 453–467. [Google Scholar] [PubMed]
- Dowling, D.K.; Simmons, L.W. Life-history evolution reactive oxygen species as universal constraints in life history. Proc. R. Soc. B 2009, 276, 1737–1745. [Google Scholar] [CrossRef]
- Grassmann, J.; Hippeli, S.; Elstner, E.F. Plant’s defence and its benefits for animals and medicine: Role of phenolics and terpenoids in avoiding oxygen stress. Plant Physiol. Biochem. 2002, 40, 471–478. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; van Breusegem, F. The reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Polle, A. Dissecting the superoxide dismutase-ascorbateglutathione-pathway in chloroplasts by metabolic modeling. Computer simulations as a step towards flux analysis. Plant Physiol. 2001, 126, 445–462. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Alsher, R.; Erturk, N.G.; Heath, L.S. Role of Superoxide dismutase in controlling oxidative stress in plants. J. Exp. Bot. 2002, 53, 1331–1341. [Google Scholar] [CrossRef]
- Scandalios, J.G. Oxidative stress: Molecular perception and transduction of signals triggering antioxidant gene defenses. Br. J. Med. Biol. Res. 2005, 38, 995–1014. [Google Scholar] [CrossRef]
- Kliebenstein, D.J.; Monde, R.A.; Last, R.L. Superoxide dismutase in Arabidopsis: An eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 1998, 118, 637–650. [Google Scholar] [CrossRef]
- Fink, R.C.; Scandalios, J.G. Molecular evolution and structure: Function relationships of the superoxide dismutase gene families in angiosperms and their relationship to other eukaryotic and prokaryotic superoxide dismutases. Arch. Biochem. Biophys. 2002, 399, 19–36. [Google Scholar] [CrossRef]
- Bowler, C.; van Camp, W.; van Montagu, M.; Inzé, D. Superoxide dismutase in plants. Crit. Rev. Plant Sci. 1994, 13, 199–218. [Google Scholar] [CrossRef]
- Van Camp, W.; Inzé, D.; van Montagu, M. The regulation and function of tobacco superoxide dismutases. Free Radic. Biol. Med. 1997, 23, 515–520. [Google Scholar] [CrossRef]
- Perl-Treves, R.; Galun, E. The tomato Cu/Zn superoxide dismutase genes are developmentally regulated and respond to light and stress. Plant Mol. Biol. 1991, 17, 745–760. [Google Scholar] [CrossRef]
- Sakamoto, A.; Ohsuga, H.; Tanaka, K. Nucleotide sequences of two cDNA clones encoding different Cu/Zn-superoxide dismutases expressed in developing rice seed (Oryza sativa L.). Plant Mol. Biol. 1992, 19, 323–327. [Google Scholar] [CrossRef]
- Kaminaka, H.; Morita, S.; Yokoi, H.; Masumura, T.; Tanaka, K. Molecular cloning and characterization of a cDNA for plastidic copper/zinc-superoxide dismutase in rice (Oryza sativa L.). Plant Cell Physiol. 1997, 38, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Kaminaka, H.; Morita, S.; Tokumoto, M.; Yokoyama, H.; Masumura, T.; Tanaka, K. Molecular cloning and characterization of a cDNA for an iron-superoxide dismutase in rice (Oryza sativa L.). Biosci. Biotechnol. Biochem. 1999, 63, 302–308. [Google Scholar] [CrossRef]
- Katyshev, A.I.; Rogozin, I.B.; Konstantinov, Y.M. Identification of New Superoxide Dismutase Transcripts in Plants by EST Analysis: Alternative Polyadenylation and Splicing Events in Computational Structural and Functional Genomics and Transcriptomics. In Proceedings of the Fifth International Conference on Bioinformatics of Genome Regulation and Structure BGRS, Novosibirsk, Russia, 16–22 July 2006; pp. 61–64.
- Molina-Rueda, J.J.; Tsai, C.J.; Kirby, E.G. The Populus superoxide dismutase gene family and its responses to drought stress in transgenic poplar overexpressing a pine cytosolic glutamine synthetase (GS1a). PLos One 2012, 8, e564211. [Google Scholar]
- Srivastava, V.; Srivastava, M.K.; Nilsson, R.; Rouhier, N.; Melzer, M.; Wingsle, G. Alternative splicing studies of the reactive oxygen species gene network in populus reveal two isoforms of high-isoelectric-point superoxide dismutase. Plant Physiol. 2009, 149, 1848–1859. [Google Scholar] [CrossRef]
- Feng, W.; Hongbin, W.; Bing, L.; Jinfa, W. Cloning and characterization of a novel splicing isoform of the iron-superoxide dismutase gene in rice (Oryza sativa L.). Plant Cell Rep. 2006, 24, 734–742. [Google Scholar] [CrossRef]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by down regulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef]
- Gill, S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Van Camp, W.; Willekens, H.; Bowler, C.; Van Montagu, M.; Inzé, D.; Reupold-Popp, P.; Sandermann, H., Jr.; Langebartels, C. Elevated levels of superoxide dismutase protect transgenic plants against ozone damage. Nat. Biotechnol. 1994, 12, 165–168. [Google Scholar] [CrossRef]
- Longa, M.A.; Del Rio, L.A.; Palma, J.M. Superoxide dismutases of chestnut leaves Castanea sativa: Characterization and study of their involvement in natural leaf senescence. Physiol. Plant. 1994, 92, 227–232. [Google Scholar] [CrossRef]
- Pastori, G.M.; Del Rio, L.A. Natural senescence of pea leaves an activated oxygen-mediated function for peroxisomes. Plant Physiol. 1997, 113, 411–418. [Google Scholar] [PubMed]
- Bagnoli, F.; Giannino, D.; Caparrini, S.; Camussi, A.; Mariotti, D.; Racchi, M.L. Molecular cloning, characterization and expression of a Mn-superoxide dismutase gene from peach (Prunus persica [L.] Batsch). Mol. Genet. Genomics 2002, 267, 321–328. [Google Scholar] [CrossRef]
- Swanson, S.; Gilroy, S. ROS in plant development. Physiol. Plant. 2010, 138, 384–392. [Google Scholar] [CrossRef]
- Bagnoli, F.; Capuana, M.; Racchi, M.L. Developmental changes of catalase and superoxide dismutase isoenzymes in zygotic and somatic embryos of horse chestnut. Aust. J. Plant Physiol. 1998, 25, 909–913. [Google Scholar] [CrossRef]
- Ma, L.; Xie, L.; Lin, G.; Jiang, S.; Chen, H.; Li, H.; Takac, T.; Samaj, J.; Xu, C. Histological changes and differences in activities of some antioxidant enzymes and hydrogen peroxide content during somatic embryogenesis of Musa AAA cv. Yueyoukang 1. Sci. Hortic. 2012, 144, 87–92. [Google Scholar] [CrossRef]
- Faize, M.; Faize, L.; Petri, C.; Barba-Espin, G.; Diaz-Vivancos, P.; Clemente-Moreno, M.J.; Koussa, T.; Hernandez, J.A. Cu/Zn superoxide dismutase and ascorbate peroxidase enhance in vitro shoot multiplication in transgenic plum. J. Plant Physiol. 2013, 170, 625–632. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Scandalios, J.G. Oxidative Stress and the Molecular Biology of Antioxidant Defenses; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1997. [Google Scholar]
- Willekens, H.; Langebartels, C.; Tiré, C.; van Montagu, M.; Inzé, D.; van Camp, W. Differential expression of catalase genes in Nicotiana plumbaginifolia (L.). Proc. Natl. Acad. Sci. USA 1994, 91, 10450–10454. [Google Scholar] [CrossRef]
- Scandalios, J.G. The Antioxidant Enzyme Genes Cat and Sod of Maize: Regulation, Functional Significance, and Molecular Biology. In Isozymes: Current Topics in Biological and Medical Research, Molecular and Cellular Biology; Rattazzi, M.C., Scandalios, J.G., Whitt, G.S., Eds.; Alan R. Liss: New York, NY, USA, 1987; pp. 19–44. [Google Scholar]
- Willekens, H.; Inzé, D.; van Montagu, M.; van Camp, W. Catalases in plants. Mol. Breed. 1995, 1, 207–228. [Google Scholar] [CrossRef]
- Frugoli, J.A.; Zhong, H.H.; Nuccio, M.L.; McCourt, P.; McPeek, M.A.; Thomas, T.L.; McClung, C. Catalase is encoded by a multigene family in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1996, 112, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Scandalios, J.G.; Guan, L.M.; Polidoros, A. Catalases in Plants: Gene Structure, Properties, Regulation, and Expression. In Oxidative Stress and the Molecular Biology of Antioxidant Defenses; Scandalios, J.G., Ed.; Cold Spring Harbor Laboratory Press: Plainview, NY, USA, 1997; pp. 343–406. [Google Scholar]
- Bagnoli, F.; Danti, S.; Magherini, V.; Cozza, R.; Innocenti, A.M.; Racchi, M.L. Molecular cloning, characterization and expression of two catalase genes from peach (Prunus persica). Funct. Plant Biol. 2004, 3, 349–357. [Google Scholar]
- Suzuki, M.; Ario, T.; Hattori, T.; Nakamura, K.; Asahi, T. Isolation and characterization of two tightly linked catalase genes from castor bean that are differentially regulated. Plant Mol. Biol. 1994, 25, 507–516. [Google Scholar] [CrossRef]
- Higo, K.; Higo, H. Cloning and characterization of the rice CatA catalase gene, a homologue of the maize Cat3 gene. Plant Mol. Biol. 1996, 30, 505–521. [Google Scholar] [CrossRef]
- Drory, A.; Woodson, W.R. Molecular cloning and nucleotide sequence of a cDNA encoding catalase from tomato. Plant Physiol. 1992, 100, 1605–1606. [Google Scholar] [CrossRef] [PubMed]
- Niebel, A.; Heungens, K.; Barthels, N.; Inzé, D.; van Montagu, M.; Gheysen, G. Characterization of a pathogen-induced potato catalase and its systemic expression upon nematode and bacterial infection. Mol. Plant Microbe Interact. 1995, 8, 371–378. [Google Scholar] [CrossRef]
- Ni, W.; Trelease, R.N. Post-translational regulation of catalase isozyme expression in cottonseeds. Plant Cell 1991, 3, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Willekens, H.; Villarroel, R.; van Montagu, M.; Inzé, D.; van Camp, W. Molecular identification of catalases from Nicotiana plumbaginifolia (L.). FEBS Lett. 1994, 352, 79–83. [Google Scholar] [CrossRef]
- Willekens, H.; Chamnongpol, S.; Davey, M.; Schraudner, M.; Langebartels, C.; van Montagu, M.; Inzé, D.; van Camp, W. Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J. 1997, 16, 4806–4816. [Google Scholar] [CrossRef]
- Suzuki, M.; Miyamoto, R.; Hattori, T.; Nakamura, K.; Asahi, T. Differential regulation of the expression in transgenic tobacco of the gene for b-glucuronidase under the control of the 5′-upstream regions of two catalase genes from castor bean. Plant Cell Physiol. 1995, 36, 273–279. [Google Scholar] [PubMed]
- Skadsen, R.W.; Schulze-Lefert, P.; Herbst, J.M. Molecular cloning, characterization and expression analysis of two catalase isozyme genes in barley. Plant Mol. Biol. 1995, 29, 1005–1014. [Google Scholar] [CrossRef]
- Mullen, R.T.; Gifford, D.J. Purification and characterization of catalase from loblolly pine (Pinus taeda L.) megagametophytes. Plant Physiol. 1993, 103, 477–483. [Google Scholar] [PubMed]
- Racchi, M.L.; Chiusi, A.P.; Giannini, R. Catalase isozymes as biochemical markers of different developmental stages in cypress (Cupressus sempervirens). Can. J. For. Res. 1996, 26, 1629–1633. [Google Scholar] [CrossRef]
- Racchi, M.L.; Bagnoli, F.; Balla, I.; Danti, S. Differential activity of catalase and superoxide dismutase in seedlings and in vitro micro-propagated oak (Quercus robur L.). Plant Cell Rep. 2001, 20, 169–174. [Google Scholar] [CrossRef]
- Dat, J.; Vandenabeele, S.; Vranová, E.; van Montagu, M.; Inzé, D.; van Breusegem, F. Dual action of the active oxygen species during plant stress responses. Cell. Mol. Life Sci. 2000, 57, 779–795. [Google Scholar] [CrossRef]
- Davletova, S.; Rizhsky, L.; Liang, H.; Shengqiang, D.; Oliver, D.; Coutu, J.; Shulaev, V.; Schlauch, K.; Mittler, R. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. The Plant Cell 2004, 17, 268–281. [Google Scholar] [PubMed]
- Noctor, G.; Foyer, C.H. Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef]
- Asada, K. Production and Scavenging of Active Oxygen in Chloroplasts. In Current Communications in Cell and Molecular Biology 5. Molecular Biology of Free Radical Research Scavenging Systems; Scandalios, J.G., Ed.; Cold Spring Harbour Laboratory Press: New York, NY, USA, 1992; pp. 173–192. [Google Scholar]
- Chew, O.; Whelan, J.; Millar, A.H. Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J. Biol. Chem. 2003, 278, 46869–46877. [Google Scholar] [CrossRef]
- Teixeira, F.K.; Menezes-Benavente, L.; Margis, R.; Margis-Pinheiro, M. Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: Inferences from the rice genome. J. Mol. Evol. 2004, 59, 761–770. [Google Scholar] [CrossRef]
- Teixeira, F.K.; Menezes-Benavente, L.; Galvão, V.C.; Margis, R.; Margis-Pinheiro, M. Rice ascorbate peroxidase gene family encodes functionally diverse isoforms localized in different subcellular compartments. Planta 2006, 224, 300–314. [Google Scholar] [CrossRef]
- Ishikawa, T.; Yoshimura, K.; Tamoi, M.; Takeda, T.; Shigeoka, S. Alternative mRNA splicing of 3′-terminal exons generates ascorbate peroxidase isoenzymes in spinach (Spinacia oleracea) chloroplasts. Biochem. J. 1997, 328, 795–800. [Google Scholar] [CrossRef] [PubMed]
- Caverzan, A.; Passaia, G.; Barcellos Rosa, S.; Werner Ribeiro, C.; Lazzarotto, F.; Margis-Pinheiro, M. Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 2012, 35, 1011–1019. [Google Scholar] [CrossRef]
- Sato, Y.; Masuta, Y.; Saito, K.; Murayama, S.; Ozawa, K. Enhanced chilling tolerance at the booting stage in rice by transgenic over-expression of the ascorbate peroxidase gene, OsAPXa. Plant Cell Rep. 2011, 30, 299–406. [Google Scholar]
- Sun, W.H.; Duan, M.; Shu, D.F.; Yang, S.; Meng, Q.W. Over-expression of tomato tAPX gene in tobacco improves tolerance to high or low temperature stress. Biol. Plant. 2010, 54, 614–620. [Google Scholar] [CrossRef]
- Sun, W.H.; Duan, M.; Shu, D.F.; Yang, S.; Meng, Q.W. Over-expression of StAPX in tobacco improves seed germination and increases early seedling tolerance to salinity and osmotic stresses. Plant Cell Rep. 2010, 29, 917–926. [Google Scholar] [CrossRef]
- Koussevitzky, S.; Suzuki, N.; Huntington, S.; Armijo, L.; Sha, W.; Cortes, D.; Shulaev, V.; Mittler, R. Ascorbate peroxidase 1 plays a key role in the response of Arabidopsis thaliana to stress combination. J. Biol. Chem. 2008, 283, 34197–34203. [Google Scholar] [CrossRef]
- Badawi, G.H.; Kawano, N.; Yamauchi, Y.; Shimada, E.; Sasaki, R.; Kubo, A.; Tanaka, K. Over-expression of ascorbate peroxidase in tobacco chloroplasts enhances the tolerance to salt stress and water deficit. Physiol. Plant. 2004, 121, 231–238. [Google Scholar] [CrossRef]
- Li, Y.-J.; Hai, R.-L.; Du, X.-H.; Jiang, X.-N.; Lu, H. Over-expression of a Populus peroxisomal ascorbate peroxidase (PpAPX) gene in tobacco plants enhances stress tolerance. Plant Breed. 2009, 128, 404–410. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Rizhsky, L.; Hegie, A.; Koussevitzky, S.; Mittler, R. Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiol. 2007, 144, 1777–1785. [Google Scholar] [CrossRef]
- Dietz, K.J. Peroxiredoxins in plants and cyanobacteria. Antioxid. Redox. Signal. 2011, 15, 1129–1159. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine, 4th ed.; Oxford University Press: Oxford, UK, 2007. [Google Scholar]
- Arrigoni, O.; De Tullio, M.C. Ascorbic acid, much more than just an antioxidant. Biochim. Biophys. Acta 2002, 1569, 1–9. [Google Scholar] [CrossRef]
- Noctor, G.; Gomez, L.; Vanacker, H.; Foyer, C.H. Interactions between biosynthesis, compartmentation and transport in the control of glutathione homeostasis and signaling. J. Exp. Bot. 2002, 53, 1283–1304. [Google Scholar] [CrossRef]
- Zechmann, B.; Stumpe, M.; Mauch, F. Immunocytochemical determination of the subcellular distribution of ascorbate in plants. Planta 2011, 233, 1–12. [Google Scholar] [CrossRef]
- Pignocchi, C.; Kiddle, G.; Hernández, I.; Foster, S.J.; Asensi, A.; Taybi, T.; Barnes, J.; Foyer, C.H. Ascorbate oxidase-dependent changes in the redox state of the apoplast modulate gene transcript accumulation leading to modified hormone signaling and orchestration of defence processes in tobacco. Plant Physiol. 2006, 141, 423–435. [Google Scholar] [CrossRef]
- Xiang, C.; Werner, B.L.; Christensen, E.M.; Oliver, D.J. The biological functions of glutathione revisited in Arabidopsis transgenic plants with altered glutathione levels. Plant Physiol. 2001, 126, 564–574. [Google Scholar] [CrossRef]
- May, M.J.; Vernoux, T.; Sánchez-Fernández, R.; van Montagu, M.; Inzé, D. Evidence for posttranscriptional activation of γ-glutamylcysteine synthetase during plant stress responses. Proc. Natl. Acad. Sci. USA 1998, 95, 12049–12054. [Google Scholar] [CrossRef]
- Xiang, C.; Oliver, D.J. Glutathione metabolic genes coordinately respond to heavy metals and jasmonic acid in Arabidopsis. Plant Cell 1998, 10, 1539–1550. [Google Scholar] [CrossRef] [PubMed]
- Hicks, L.M.; Cahoon, R.E.; Bonner, E.R.; Rivard, R.S.; Sheffield, J.; Jez, J.M. Thiol-based regulation of redox-active glutamate-cysteine ligase from Arabidopsis thaliana. Plant Cell 2007, 19, 2653–2661. [Google Scholar] [CrossRef] [PubMed]
- Pietrini, F.; Iannelli, M.A.; Pasqualini, S.; Massacci, A. Interaction of cadmium with glutathione and photosynthesis in developing leaves and chloroplasts of Phragmites australis (Cav.) Trin. ex Steudel. Plant Physiol. 2003, 133, 829–837. [Google Scholar] [CrossRef]
- Sun, Q.; Yec, Z.H.; Wang, X.R.; Wong, M.H. Cadmium hyperaccumulation leads to an increase of glutathione rather than phytochelatins in the cadmium hyperaccumulator Sedum alfredii. J. Plant Physiol. 2007, 164, 1489–1498. [Google Scholar] [CrossRef] [PubMed]
- Navrot, N.; Collin, V.; Gualberto, J.; Gelhaye, E.; Hirasawa, M.; Rey, P.; Knaff, D.B.; Issakidis, E.; Jacquot, J.-P.; Rouhier, N. Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses. Plant Physiol. 2006, 142, 1364–1379. [Google Scholar] [CrossRef]
- Harborne, J.B. Plant Flavonoids in Biology and Medicine; Cody, V., Middleton, E., Harborne, J.B., Eds.; Alan R. Liss: New York, NY, USA, 1986; pp. 15–24. [Google Scholar]
- Bors, W.; Heller, W.; Michel, C.; Saran, M. Flavonoids as antioxidants: Determination of radical-scavenging efficiencies. Methods Enzymol. 1990, 186, 343–355. [Google Scholar] [CrossRef]
- Pietta, P.G. Flavonoids as antioxidants. J. Nat. Prod. 2000, 63, 1035–1042. [Google Scholar] [CrossRef]
- Pourcel, L.; Routaboul, J.M.; Cheynier, V.; Lepiniec, L.; Debeaujon, I. Flavonoid oxidation in plants: From biochemical properties to physiological functions. Trend Plant Sci. 2007, 12, 29–36. [Google Scholar]
- Solovchenko, A. Photoprotection in Plants; Springer-Verlag: Berlin, Heidelberg, Germany, 2010; pp. 143–158. [Google Scholar]
- Hirschberg, J. Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 2001, 4, 210–218. [Google Scholar] [CrossRef]
- Lu, S.; Li, L. Carotenoid metabolism: Biosynthesis, regulation, and beyond. J. Integr. Plant Biol. 2008, 50, 778–785. [Google Scholar] [CrossRef]
- Stahl, W.; Sies, H. Antioxidant activity of carotenoids. Mol. Asp. Med. 2003, 24, 345–351. [Google Scholar] [CrossRef]
- Tang, G. Bioconversion of dietary Provitamin A carotenoids to Vitamin A in humans. Am. J. Clin. Nutr. 2010, 91, 1468S–1473S. [Google Scholar] [CrossRef]
- Paiva, S.A.; Russell, R.M. Beta-carotene and other carotenoids as antioxidants. J. Am. Coll. Nutr. 1999, 18, 426–433. [Google Scholar] [CrossRef]
- Giuliano, G.; Bartley, G.E.; Scolnik, P.A. Regulation of carotenoid biosynthesis during tomato development. Plant Cell 1993, 5, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Bramley, P.M. Regulation of carotenoid formation during tomato fruit ripening and development. J. Exp. Bot. 2002, 53, 2107–2113. [Google Scholar] [CrossRef]
- Munné-Bosch, S. Alpha-tocopherol: A multifaceted molecule in plants. Vitam. Horm. 2007, 76, 375–392. [Google Scholar] [CrossRef]
- Munné-Bosch, S. The role of alpha-tocopherol in plant stress tolerance. J. Plant Physiol. 2005, 162, 743–748. [Google Scholar] [CrossRef]
- Munné-Bosch, S.; Weiler, E.W.; Alegre, L.; Müller, M.; Düchting, P.; Falk, J. Alpha-tocopherol may influence cellular signaling by modulating jasmonic acid levels in plants. Planta 2007, 225, 681–691. [Google Scholar] [CrossRef]
- Hincha, D.K. Effects of alpha-tocopherol (vitamin E) on the stability and lipid dynamics of model membranes mimicking the lipid composition of plant chloroplast membranes. FEBS Lett. 2008, 582, 3687–3692. [Google Scholar] [CrossRef]
- Zhu, C.; Sanahuja, G.; Yuan, D.; Farre, G.; Arjo, G.; Berman, J.; Zorrilla Lopez, U.; Banakar, R.; Bai, C.; Perez-Massot, E.; et al. Biofortification of plants with altered antioxidant content and composition: Genetic engineering strategies. Plant Biotechnol. J. 2013, 11, 129–141. [Google Scholar] [CrossRef]
- Faltin, Z.; Holland, D.; Velcheva, M.; Tsapovetsky, M.; Roeckel-Drevet, P.; Handa, A.K.; Abu-Abied, M.; Friedman-Einat, M.; Eshdat, Y.; Perl, A. Glutathione peroxidase regulation of reactive oxygen species level is crucial for in vitro plant differentiation. Plant Cell Physiol. 2010, 51, 1151–1162. [Google Scholar] [CrossRef]
- Joo, J.H.; Bae, Y.S.; Lee, J.S. Role of auxin-induced reactive oxygen species in root gravitropism. Plant Physiol. 2001, 126, 1055–1060. [Google Scholar] [CrossRef]
- Muller, K.; Linkies, A.; Vreeburg, R.A.; Fry, S.C.; Krieger-Liszkay, A.; Leubner-Metzger, G. In vivo cell wall loosening by hydroxyl radicals during cress (Lepidium sativum L.) seed germination and elongation growth. Plant Physiol. 2009, 150, 1855–1865. [Google Scholar] [CrossRef]
- Pedreira, J.; Sanz, N.; Pena, M.J.; Sanchez, M.; Queijeiro, E.; Revilla, G.; Zarra, I. Role of apoplastic ascorbate and hydrogen peroxide in the control of cell growth in pine hypocotyls. Plant Cell Physiol. 2004, 45, 530–534. [Google Scholar] [CrossRef]
- Schopfer, P. Hydroxyl radical-induced cell-wall loosening in vitro and in vivo: Implications for the control of elongation growth. Plant J. 2001, 28, 679–688. [Google Scholar] [CrossRef]
- Giovannoni, J. Molecular biology of fruit maturation and ripening. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 57, 735–749. [Google Scholar]
- Giovannoni, J. Genetic regulation of fruit development and ripening. Plant Cell 2004, 16, S170–S180. [Google Scholar] [CrossRef]
- Carrari, F.; Baxter, C.; Usadel, B.; Urbanczyk-Wochniak, E.; Zanor, M.I.; Nunes-Nesi, A.; Nikiforova, V.; Centero, D.; Ratzka, A.; Pauly, M.; et al. Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol. 2006, 142, 1380–1396. [Google Scholar] [CrossRef]
- Faurobert, M.; Mihr, C.; Bertin, N.; Pawlowski, T.; Negroni, L.; Sommerer, N.; Causse, M. Major proteome variations associated with cherry tomato pericarp development and ripening. Plant Physiol. 2007, 143, 1327–1346. [Google Scholar] [CrossRef]
- Lara, M.V.; Borsani, J.; Budde, C.O.; Lauxmann, M.A.; Lombardo, V.A.; Murray, R.; Andreo, C.S.; Drincovich, M.F. Biochemical and proteomic analysis of ‘Dixiland’ peach fruit (Prunus persica) upon heat treatment. J. Exp. Bot. 2009, 60, 4315–4333. [Google Scholar] [CrossRef]
- Nilo, R.; Saffie, C.; Lilley, K.; Baeza-Yates, R.; Cambiazo, V.; Campos-Vargas, R.; González, M.; Meisel, L.A.; Retamales, J.; Silva, H.; Orellana, A. Proteomic analysis of peach fruit mesocarp softening and chilling injury using difference gel electrophoresis (DIGE). BMC Genomics 2010, 11, 43. [Google Scholar] [CrossRef]
- Lombardo, V.A.; Osorio, S.; Borsani, J.; Lauxmann, M.A.; Bustament, C.; Budde, C.; Andreo, C.S.; Lara, M.V.; Alisdair, R.; Fernie Drincovich, M.F. Metabolic profiling during peach fruit development and ripening reveals the metabolic networks that underpin each developmental stage. Plant Physiol. 2011, 157, 1696–1710. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, Z.; Jiang, L.; Jiang, J.; Luo, H.; Fu, L. Effect of post-harvest heat treatment on proteome change of peach fruit during ripening. J. Proteomics 2011, 74, 1135–1149. [Google Scholar] [CrossRef]
- Prinsi, B.; Negri, A.S.; Fedeli, C.; Morgutti, S.; Negrini, N.; Cocucci, M.; Espen, L. Peach fruit ripening: A proteomic comparative analysis of the mesocarp of two cultivars with different flesh firmness at two ripening stages. Phytochemistry 2011, 72, 1251–1262. [Google Scholar] [CrossRef]
- Hu, H.; Liu, Y.; Shi, G.L.; Wu, R.J.; Yang, A.Z.; Wang, Y.M.; Hua, B.G.; Wang, Y.N. Proteomic analysis of peach endocarp and mesocarp during early fruit development. Physiol. Plant. 2011, 142, 390–406. [Google Scholar] [CrossRef]
- Nilo, R.P.; Campos-Vargas, R.; Orellana, A. Assessment of Prunus persica fruit softening using a proteomics approach. J. Proteomics 2012, 75, 1618–1638. [Google Scholar] [CrossRef]
- Guarino, C.; Arena, S.; De Simone, L.; D’Ambrosio, C.; Santoro, S.; Rocco, M.; Scaloni, A.; Marra, M. Proteomic analysis of the major soluble components in Annurca apple flesh. Mol. Nutr. Food Res. 2007, 51, 255–262. [Google Scholar] [CrossRef]
- Sarry, J.E.; Sommerer, N.; Sauvage, F.X.; Bergoin, A.; Rossignol, M.; Albagnac, G.; Romieu, C. Grape berry biochemistry revisited upon proteomic analysis of the mesocarp. Proteomics 2004, 4, 201–215. [Google Scholar] [CrossRef]
- Giribaldi, M.; Perugini, I.; Sauvage, F.X.; Schubert, A. Optimization of protein extraction and solubilization for mature grape berry clusters. Proteomics 2007, 7, 3154–3170. [Google Scholar] [CrossRef]
- Deytieux, C.; Geny, L.; Lapaillerie, D.; Claverol, S.; Bonneu, M.; Donèche, B. Proteome analysis of grape skins during ripening. J. Proteomics 2007, 58, 1851–1862. [Google Scholar]
- Deluc, L.G.; Grimplet, J.; Wheatley, M.D.; Tillett, R.L.; Quilici, D.R.; Osborne, C.; Schooley, D.A.; Schlauch, K.A.; Cushman, J.C.; Cramer, G.R. Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics 2007, 8, 429. [Google Scholar] [CrossRef]
- Zhang, J.W.; Ma, H.Q.; Feng, J.D.; Zheng, L.; Wang, Z.; Chen, S.W. Grape berry plasma membrane proteome analysis and its differential expression during ripening. J. Exp. Bot. 2008, 59, 2979–2990. [Google Scholar] [CrossRef]
- Negri, A.S.; Prinsi, B.; Rossoni, M.; Failla, O.; Scienza, A.; Cocucci, M.; Espen, L. Proteome changes in the skin of the grape cultivar Barbera among different stages of ripening. BMC Genomics 2008, 8, 378. [Google Scholar]
- Lücker, J.; Laszczak, M.; Smith, D.; Lund, S.T. Generation of a predicted protein database from EST data and application to iTRAQ analyses in grape (Vitis vinifera cv. Cabernet Sauvignon) berries at ripening initiation. BMC Genomics 2009, 10, 50. [Google Scholar] [CrossRef]
- Zamboni, A.; Di Carli, M.; Guzzo, F.; Stocchero, M.; Zenoni, S.; Ferrarini, A.; Tononi, P.; Toffali, K.; Desiderio, A.; Lilley, K.S.; et al. Identification of putative stage-specific grapevine berry biomarkers and omics data integration into networks. Plant Physiol. 2010, 154, 1439–1459. [Google Scholar] [CrossRef]
- Martínez-Esteso, M.J.; Sellés-Marchart, S.; Lijavetzky, D.; Pedreño, M.A.; Bru-Martínez, R. A DIGE-based quantitative proteomic analysis of grape berry flesh development and ripening reveals key events in sugar and organic acid metabolism. J. Exp. Bot. 2011, 62, 2521–2569. [Google Scholar] [CrossRef]
- Bianco, L.; Lopez, L.; Scalone, A.G.; Di Carli, M.; Desiderio, A.; Benvenuto, E.; Perrotta, G. Strawberry proteome characterization and its regulation during fruit ripening and in different genotypes. J. Proteomics 2009, 72, 586–607. [Google Scholar] [CrossRef]
- Osorio, S.; Bombarely, A.; Giavalisco, P.; Usadel, B.; Stephens, C.; Araguez, I.; Medina-Escobar, N.; Botella, M.A.; Fernie, A.R.; Valpuesta, V. Demethylation of oligogalacturonides by FaPE1 in the fruits of the wild strawberry Fragaria vesca triggers metabolic and transcriptional changes associated with defence and development of the fruit. J. Exp. Bot. 2011, 62, 2855–2873. [Google Scholar] [CrossRef] [PubMed]
- Katz, E.; Fon, M.; Eigenheer, R.A.; Phinney, B.S.; Fass, J.N.; Lin, D.W.; Sakda, A.; Blumwald, E. A label-free differential quantitative mass spectrometry method for the characterization and identification of protein changes during citrus fruit development. Proteome Sci. 2010, 8, 68. [Google Scholar] [CrossRef]
- Palma, J.M.; Corpas, F.J.; Del Río, L.A. Proteomics as an approach to the understanding of the molecular physiology of fruit development and ripening. J. Proteomics 2011, 74, 1230–1243. [Google Scholar] [CrossRef]
- Qin, G.; Meng, X.; Wang, Q.; Tian, S. Oxidative damage of mitochondrial proteins contributes to fruit senescence: A redox proteomics analysis. J. Proteome Res. 2009, 8, 2449–2462. [Google Scholar] [CrossRef]
- Jimenez, A.; Creissen, G.; Kular, B.; Firmin, J.; Robinson, S.; Verhoeyen, M.; Mullineaux, P. Changes in oxidative processes and components of the antioxidant system during tomato fruit ripening. Planta 2002, 214, 751–758. [Google Scholar] [CrossRef]
- Trainotti, L.; Bonghi, C.; Ziliotto, F.; Zanin, D.; Rasori, A.; Casadoro, G.; Ramina, A.; Tonutti, P. The use of microarray µPEACH1.0 to investigate transcriptome changes during transition from preclimacteric to climacteric phase in peach fruit. Plant Sci. 2006, 170, 606–813. [Google Scholar] [CrossRef]
- Kazt, E.; Fon, M.; Lee, Y.J.; Phinney, B.S.; Sadka, A.; Blumwald, E. The citrus fruit proteome: insights into the citrus fruit metabolism. Planta 2007, 226, 989–1005. [Google Scholar] [CrossRef]
- Muccilli, V.; Licciardello, C.; Fontanini, D.; Russo, M.P.; Cunsolo, V.; Saletti, R.; Recupero, G.R.; Foti, S. Proteome analysis of Citrus sinensis L. (Osbeck) flesh at ripening time. J. Proteomics 2009, 73, 134–152. [Google Scholar] [CrossRef]
- Wang, H.; Cao, G.; Prior, R.L. Total antioxidant capacity of fruits. J. Agric. Food Chem. 1996, 44, 701–705. [Google Scholar] [CrossRef]
- Wolfe, K.L.; Kang, X.; He, X.; Dong, M.; Zhan, G.Q.; Liu, R.H. Cellular antioxidant activity of common fruits. J. Agric. Food Chem. 2008, 56, 8418–8426. [Google Scholar] [CrossRef]
- Fu, L.; Xu, B.T.; Xu, X.R.; Gan, R.Y.; Zhang, Y.; Xia, E.Q.; Li, H.B. Antioxidant capacities and total phenolic contents of 62 fruits. Food Chem. 2011, 129, 345–350. [Google Scholar] [CrossRef]
- Benzie, I.F.; Strain, J.J. The ferric reducing ability of plasma as a measure of “antioxidant power” the FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef]
- Cao, G.; Prior, R.L. Measurement of oxygen radical absorbance capacity in biological samples. Methods Enzymol. 1999, 299, 50–62. [Google Scholar] [CrossRef]
- Evelson, P.; Travacio, M.; Repetto, M. Evaluation of total reactive antioxidant potential of tissue homogenates and their cytosols. Arch. Biochem. Biophys. 2001, 388, 261–266. [Google Scholar] [CrossRef]
- Ou, B.; Hampsch-Woodill, M.; Prior, R.L. Development and validation of an improved oxygen radical absorbance capacity assay using fluorescein as the fluorescent probe. J. Agric. Food Chem. 2001, 49, 4619–4626. [Google Scholar] [CrossRef]
- Scalzo, J.; Politi, A.; Pellegrini, N.; Mezzetti, B.; Battino, M. Plant genotype affects total antioxidant capacity and phenolic contents in fruit. Nutrition 2005, 21, 207–213. [Google Scholar] [CrossRef]
- FAOSTAT. 2013. Available online: http://www.faostat.fao.org (accessed on 9 November 2013).
- Génard, M.; Lescourret, F.; Gómez, L.; Habib, R. Changes in fruit sugar concentrations in response to assimilate supply, metabolism and dilution: A modelling approach applied to peach fruit (Prunus persica). Tree Physiol. 2003, 23, 373–385. [Google Scholar] [CrossRef]
- Crisosto, C.H.; Crisosto, G.M. Relationship between ripe soluble solids concentration (RSSC) and consumer acceptance of high and low acid melting flesh peach and nectarine [Prunus persica (L.) Batsch] cultivars. Postharvest Biol. Technol. 2005, 38, 239–246. [Google Scholar] [CrossRef]
- Gil, M.I.; Tomas-Barberan, F.A.; Hess-Pierce, B.; Kader, A.A. Antioxidant capacities, phenolic compounds, carotenoids and vitamin C contents of nectarine, peach and plum cultivars from California. J. Agric. Food Chem. 2002, 50, 4976–4982. [Google Scholar] [CrossRef]
- Cantin, C.M.; Moreno, M.A.; Gogorcena, Y. Evaluation of the antioxidant capacity, phenolic compounds, and vitamin c content of different peach and nectarine [Prunus persica (L.) Batsch] Breeding Progenies. J. Agric. Food Chem. 2009, 57, 4586–4592. [Google Scholar] [CrossRef]
- Camejo, D.; Marti, M.C.; Roman, P.; Ortiz, A.; Jimenez, A. Antioxidant system and protein pattern in peach fruits at two maturation stages. J. Agric. Food Chem. 2010, 58, 11140–11147. [Google Scholar] [CrossRef]
- Legua, P.; Hernandez, F.; Huertas, M.; Diaz-Mula, M.; Valero, D.; Serrano, M. Quality, bioactive compounds, and antioxidant activity of new flat-type peach and nectarine cultivars: A comparative study. J. Food Sci. 2011, 76, C729–C735. [Google Scholar] [CrossRef]
- Abidi, W.; Jiménez, S.; Moreno, M.Á.; Gogorcena, Y. Evaluation of Antioxidant Compounds and Total Sugar Contentin a Nectarine [Prunus persica (L.) Batsch] Progeny. Int. J. Mol. Sci. 2011, 12, 6919–6935. [Google Scholar] [CrossRef]
- Montevecchi, G.; Vasile Simone, G.; Masino, F.; Bignami, C.; Antonelli, A. Physical and chemical characterization of Pescabivona, a Sicilian white flesh peach cultivar [Prunus persica (L.) Batsch]. Food Res. Int. 2012, 45, 123–131. [Google Scholar] [CrossRef]
- Tomàs-Barberàn, F.A.; Gil, M.I.; Cremin, P.; Waterhouse, A.L.; Hess-Pierce, B.; Kader, A.A. HPLC-DAD-ESIMS Analysis of phenolic compounds in nectarines, peaches, and plums. J. Agric. Food Chem. 2001, 49, 4748–4760. [Google Scholar] [CrossRef] [PubMed]
- Cevallos-Casals, B.A.; Byrne, D.; Okie, W.R.; Cisneros-Zevallos, L. Selecting new peach and plum genotypes rich in phenolic compounds and enhanced functional properties. Food Chem. 2006, 96, 273–280. [Google Scholar] [CrossRef]
- Huang, R.H.; Xia, R.X.; Hu, L.M.; Lu, Y.M.; Wang, M.Y. Antioxidant activity and oxygen-scavenging system in orange pulp during fruit ripening and maturation. Sci. Hortic. 2007, 113, 166–172. [Google Scholar] [CrossRef]
- Chien, P.J.; Sheu, F.; Lin, H.R. Coating citrus (Murcott tangor) fruit with low molecular weight chitosan increases postharvest quality and shelf life. Food Chem. 2007, 100, 1160–1164. [Google Scholar] [CrossRef]
- Huang, R.H.; Liu, J.H.; Lu, Y.M.; Xia, R.X. Effect of salicylic acid on the antioxidant system in the pulp of ‘Cara Cara’ navel orange (Citrus sinensis L. Osbeck) at different storage temperatures. Postharvest Biol. Technol. 2008, 47, 168–175. [Google Scholar] [CrossRef]
- Rapisarda, P.; Bianco, M.L.; Pannuzzo, P.; Timpanaro, N. Effect of cold storage on vitamin C, phenolics and antioxidant activity of five orange genotypes [Citrus sinensis (L.) Osbeck]. Postharvest Biol. Technol. 2008, 49, 348–354. [Google Scholar] [CrossRef]
- Alquezar, B.; Rodrigo, M.J.; Zacarías, L. Regulation of carotenoid biosynthesis during fruit maturation in the red-fleshed orange mutant Cara Cara. Phytochemistry 2008, 69, 1997–2007. [Google Scholar] [CrossRef]
- Proteggente, A.R.; Saija, A.; de Pasquale, A.; Rice-Evans, C.A. The compositional characterization and antioxidant activity of fresh juices from sicilian sweet Orange (Citrus sinensis L. Osbeck) varieties. Free Radic. Res. 2003, 37, 681–687. [Google Scholar] [CrossRef]
- Camarda, L.; di Stefano, V.; Fatta Del Bosco, S.; Schillaci, D. Antiproliferative activity of Citrus juices and HPLC evaluation of their flavonoid composition. Fitoterapia 2007, 78, 426–429. [Google Scholar] [CrossRef]
- Licciardello, C.; Russo, M.P.; Vale’, G.; Reforgiato, G.R. Identification of differentially expressed genes in the flesh of blood and common oranges. Tree Genet. Genomes 2008, 4, 315–331. [Google Scholar] [CrossRef]
- Maccarone, E.; Maccarrone, A.; Perrini, G.; Rapisarda, P. Anthocyanins of the Moro orange juice. Ann. Chim. (Rome) 1983, 73, 533–539. [Google Scholar]
- Maccarone, E.; Maccarrone, A.; Rapisarda, P. Acylated anthocyanins from oranges. Ann. Chim. (Rome) 1985, 75, 79–86. [Google Scholar]
- Rapisarda, P.; Bellomo, S.E.; Intrigliolo, F. Anthocyanins in Blood Oranges: Composition and Biological Activity. In Recent Research Developments in Agricultural and Food Chemistry; Pandalai, S.G., Ed.; Research Signpost: Trivandrum, India, 2001; pp. 217–230. [Google Scholar]
- Nagy, S. Vitamin C contents of Citrus fruit and their products: A review. J. Agric. Food Chem. 1980, 28, 8–18. [Google Scholar] [CrossRef]
- Vinson, J.A.; Bose, P. Comparative bioavailability to humans of ascorbic acid alone or in a citrus extract. Am. J. Clin. Nutr. 1988, 48, 601–604. [Google Scholar] [PubMed]
- Riso, P.; Visioli, F.; Gardana, C.; Grande, S.; Brusamolino, A.; Galvano, F.; Galvano, G.; Porrini, M. Effects of blood orange juice intake on antioxidant bioavailability and on different markers related to oxidative stress. J. Agric. Food Chem. 2005, 53, 941–947. [Google Scholar] [CrossRef]
- Rapisarda, P.; Intelisano, S. Sample preparation for vitamin C analysis of pigmented orange juice. Italian J. Food Sci. 1996, 8, 251–256. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Racchi, M.L. Antioxidant Defenses in Plants with Attention to Prunus and Citrus spp. Antioxidants 2013, 2, 340-369. https://doi.org/10.3390/antiox2040340
Racchi ML. Antioxidant Defenses in Plants with Attention to Prunus and Citrus spp. Antioxidants. 2013; 2(4):340-369. https://doi.org/10.3390/antiox2040340
Chicago/Turabian StyleRacchi, Milvia Luisa. 2013. "Antioxidant Defenses in Plants with Attention to Prunus and Citrus spp." Antioxidants 2, no. 4: 340-369. https://doi.org/10.3390/antiox2040340
APA StyleRacchi, M. L. (2013). Antioxidant Defenses in Plants with Attention to Prunus and Citrus spp. Antioxidants, 2(4), 340-369. https://doi.org/10.3390/antiox2040340




