Revised Taxonomy of Rhabdoviruses Infecting Fish and Marine Mammals
Abstract
:Simple Summary
Abstract
1. Introduction
2. Virus Classification and the Virus Species Concept
3. The Taxonomic Structure of the Family Rhabdoviridae
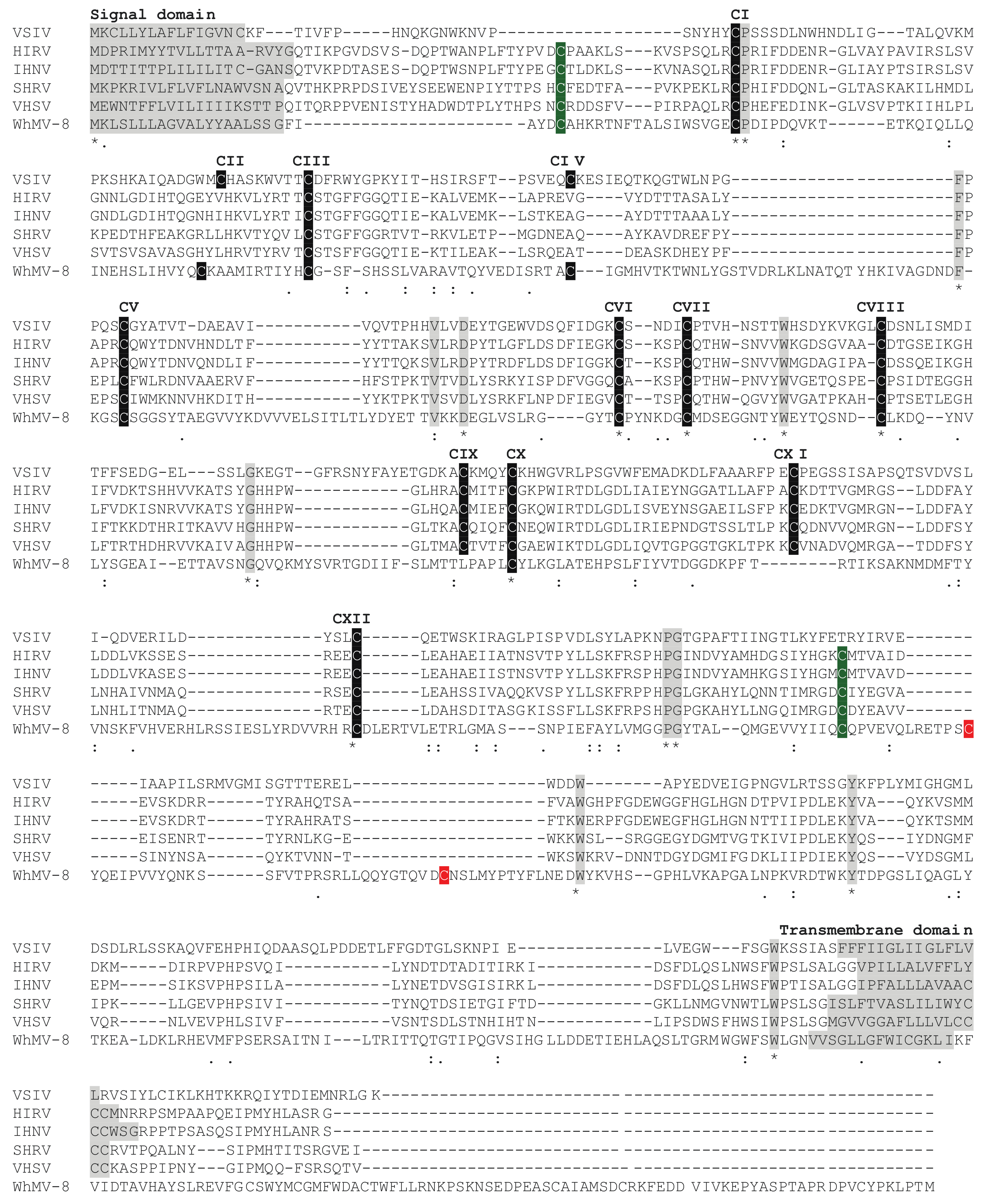
4. The Subfamily Alpharhabdovirinae
5. Subfamily Gammarhabdovirinae
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Walker, P.J.; Firth, C.; Widen, S.G.; Blasdell, K.R.; Guzman, H.; Wood, T.G.; Paradkar, P.N.; Holmes, E.C.; Tesh, R.B.; Vasilakis, N. Evolution of genome size and complexity in the Rhabdoviridae. PLoS Path. 2015, 11, e1004664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietzgen, R.G.; Kondo, H.; Goodin, M.M.; Kurath, G.; Vasilakis, N. The family Rhabdoviridae: Mono- and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins. Virus Res. 2017, 227, 158–170. [Google Scholar] [CrossRef] [Green Version]
- Whitfield, A.E.; Huot, O.B.; Martin, K.M.; Kondo, H.; Dietzgen, R.G. Plant rhabdoviruses-their origins and vector interactions. Curr. Opin. Virol. 2018, 33, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.J.; Winton, J.R. Emerging viral diseases of fish and shrimp. Vet. Res. 2010, 41, 51. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, B.; Beer, M.; Schutze, H.; Mettenleiter, T.C. Fish rhabdoviruses: Molecular epidemiology and evolution. Curr. Top. Microbiol. Immunol. 2005, 292, 81–117. [Google Scholar]
- Adams, M.J.; Lefkowitz, E.J.; King, A.M.; Harrach, B.; Harrison, R.L.; Knowles, N.J.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Mushegian, A.R.; et al. 50 years of the International Committee on Taxonomy of Viruses: Progress and prospects. Arch. Virol. 2017, 162, 1441–1446. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Krupovic, M.; Mushegian, A.; Kropinski, A.M.; Siddell, S.G.; Varsani, A.; Adams, M.J.; Davison, A.J.; Dutilh, B.E.; Harrach, B.; et al. The new scope of virus taxonomy: Partitioning the virosphere into 15 hierarchical ranks. Nat. Microbiol. 2020, 5, 668–674. [Google Scholar] [CrossRef]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Dempsey, D.M.; Dutilh, B.E.; Harrach, B.; Harrison, R.L.; Hendrickson, R.C.; Junglen, S.; et al. Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2019). Arch. Virol. 2019, 164, 2417–2429. [Google Scholar] [CrossRef] [Green Version]
- Zerbini, F.M.; Siddell, S.G.; Mushegian, A.; Walker, P.J.; Lefkowitz, E.J.; Adriaenssens, E.; Alfenas-Zerbini, P.; Duthil, B.; García, M.L.; Junglen, S.; et al. Differentiating between viruses and virus species by writing their names correctly. Arch. Virol. 2022, 167, 1231–1234. [Google Scholar] [CrossRef]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Alfenas-Zerbini, P.; Davison, A.J.; Dempsey, D.M.; Dutilh, B.E.; García, M.L.; et al. Changes to virus taxonomy and to the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2021). Arch. Virol. 2021, 166, 2633–2648. [Google Scholar] [CrossRef]
- Fenner, F. Classification and nomenclature of viruses. Second report of the International Committee on Taxonomy of Viruses. Intervirology 1976, 7, 1–115. [Google Scholar] [CrossRef]
- Walker, P.J.; Blasdell, K.R.; Calisher, C.H.; Dietzgen, R.G.; Kondo, H.; Kurath, G.; Longdon, B.; Stone, D.M.; Tesh, R.B.; Tordo, N.; et al. ICTV taxonomy profile: Rhabdoviridae. J. Gen. Virol. 2018, 99, 447–448. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.H.; Adkins, S.; Agwanda, B.R.; Al Kubrusli, R.; Alkhovsky, S.V.; Amarasinghe, G.K.; Avšič-Županc, T.; Ayllón, M.A.; Bahl, J.; Balkema-Buschmann, A.; et al. 2021 Taxonomic update of phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch. Virol. 2021, 166, 3513–3566. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.J.; Dietzgen, R.G.; Joubert, D.A.; Blasdell, K.R. Rhabdovirus accessory genes. Virus Res. 2011, 162, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Bourhy, H.; Cowley, J.A.; Larrous, F.; Holmes, E.C.; Walker, P.J. Phylogenetic relationships among rhabdoviruses inferred using the L polymerase gene. J. Gen. Virol. 2005, 86, 2849–2858. [Google Scholar] [CrossRef] [PubMed]
- Fijan, N.; Petrinec, Z.; Sulimanovic, D.; Zwillenberg, L.O. Isolation of the viral causative agent from the acute form of infectious dropsy of carp. Veterinarsky Arhiv 1971, 41, 125–138. [Google Scholar]
- Ashraf, U.; Lu, Y.; Lin, L.; Yuan, J.; Wang, M.; Liu, X. Spring viraemia of carp virus: Recent advances. J. Gen. Virol. 2016, 97, 1037–1051. [Google Scholar] [CrossRef]
- De Kinkelin, P.; Galinard, B. Isolation and identification of the causative agent of “red disease” on pike (Esox licius L. 1766). Nature 1972, 241, 465–467. [Google Scholar] [CrossRef]
- Dorson, M.; Torchy, C.; Chilmonczyk, S.; de Kinkelin, P.; Michel, C. A rhabdovirus pathogenic for perch, Perca fluviatilis L.:Isolation and preliminary study. J. Fish. Dis. 1984, 7, 241–245. [Google Scholar] [CrossRef]
- Bigarré, L.; Plassiart, G.; de Boisséson, C.; Pallandre, L.; Pozet, F.; Ledoré, Y.; Fontaine, P.; Lieffrig, F. Molecular investigations of outbreaks of Perch perhabdovirus infections in pike-perch. Dis. Aquat. Org. 2017, 127, 19–27. [Google Scholar] [CrossRef]
- Koski, P.; Hill, B.J.; Way, K.; Neuvonen, E.; Rintamaki, P. A rhabdovirus isolated from brown trout [Salmo trutta m. lacustris (L.)] with lesions in parenchymatous organs. Bull. Eur. Assn Fish. Pathol. 1992, 12, 177–180. [Google Scholar]
- Sano, T.; Nishimura, T.; Okamoto, N.; Fukuda, H. Studies on viral diseases of Japanese fishes. VII A rhabdovirus isolated from European eel, Anguilla anguilla. Bull. Jap. Soc. Sci. Fish. 1977, 43, 491–495. [Google Scholar] [CrossRef]
- Galinier, R.; van Beurden, S.; Amilhat, E.; Castric, J.; Schoehn, G.; Verneau, O.; Fazio, G.; Allienne, J.F.; Engelsma, M.; Sasal, P.; et al. Complete genomic sequence and taxonomic position of eel virus European X (EVEX), a rhabdovirus of European eel. Virus Res. 2012, 166, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sano, T. Viral diseases of cultured fishes in Japan. Fish. Pathol. 1976, 19, 221–226. [Google Scholar] [CrossRef]
- Nishimura, T.; Toba, M.; Ban, F.; Okamoto, N.; Sano, T. Eel rhabdovirus, EVA, EXEX and their infectivity to fishes. Fish. Path. 1981, 15, 173–184. [Google Scholar] [CrossRef]
- Pallandre, L.; Luo, D.; Feuvrier, C.; Lieffrig, F.; Pozet, F.; Dacheux, L.; Bigarré, L. Revisiting the classification of percid perhabdoviruses using new full-length genomes. Viruses 2020, 12, 649. [Google Scholar] [CrossRef] [PubMed]
- Osterhaus, A.D.; Broeders, H.W.; Teppema, J.S.; Kuiken, T.; House, J.A.; Vos, H.W.; Visser, I.K. Isolation of a virus with rhabdovirus morphology from a white-beaked dolphin (Lagenorhynchus albirostris). Arch. Virol. 1993, 133, 189–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegers, J.Y.; van de Bildt, M.W.; van Elk, C.E.; Schurch, A.C.; Tordo, N.; Kuiken, T.; Bodewes, R.; Osterhaus, A.D. Genetic relatedness of dolphin rhabdovirus with fish rhabdoviruses. Emerg. Inf. Dis. 2014, 20, 1081–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emelianchik, A.; Rodrigues, T.C.S.; Subramaniam, K.; Nielsen, O.; Burek-Huntington, K.A.; Rotstein, D.; Popov, V.L.; Stone, D.; Waltzek, T.B. Characterization of a novel rhabdovirus isolated from a stranded harbour porpoise (Phocoena phocoena). Virus Res. 2019, 273, e197742. [Google Scholar] [CrossRef] [PubMed]
- Axen, C.; Hakhverdyan, M.; Boutrup, T.S.; Blomkvist, E.; Ljunghager, F.; Alfjorden, A.; Hagstrom, A.; Olesen, N.J.; Juremalm, M.; Leijon, M.; et al. Emergence of a new rhabdovirus associated with mass mortalities in eelpout (Zoarces viviparous) in the Baltic Sea. J. Fish. Dis. 2017, 40, 219–229. [Google Scholar] [CrossRef]
- Tao, J.J.; Gui, J.F.; Zhang, Q.Y. Isolation and characterization of a rhabdovirus from co-infection of two viruses in mandarin fish. Aquaculture 2007, 262, 1–9. [Google Scholar] [CrossRef]
- Tao, J.J.; Zhou, G.Z.; Gui, J.F.; Zhang, Q.Y. Genomic sequence of mandarin fish rhabdovirus with an unusual small non-transcriptional ORF. Virus Res. 2008, 132, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Wang, Q.; Wang, Y.; Liu, C.; Liang, H.; Fang, X.; Wu, S. Genomic characterization and taxonomic position of a rhabdovirus from a hybrid snakehead. Arch. Virol. 2014, 159, 2469–2473. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wen, Y.; Hu, X.; Wang, W.; Liang, X.; Li, J.; Vakharia, V.; Lin, L. Breaking the host range: Mandarin fish is susceptible to a vesiculovirus derived from snakehead fish. J. Gen. Virol. 2015, 96, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Fan, Y.; Li, Z.; Zhao, J.; Zhou, Y.; Jiang, N.; Zeng, J.; Cain, K.; Zeng, L. Isolation, identification, and classification of a novel rhabdovirus from diseased Chinese rice-field eels (Monopterus albus). Arch. Virol. 2019, 164, 105–116. [Google Scholar] [CrossRef]
- Ma, D.; Deng, G.; Bai, J.; Li, S.; Yu, L.; Quan, Y.; Yang, X.; Jiang, X.; Zhu, Z.; Ye, X. A strain of Siniperca chuatsi rhabdovirus causes high mortality among cultured largemouth bass in South China. J. Aquat. Anim. Health 2013, 25, 197–204. [Google Scholar] [CrossRef]
- Lyu, S.J.; Yuan, X.M.; Zhang, H.Q.; Shi, W.D.; Hang, X.Y.; Liu, L.; Wu, Y.L. Isolation and characterization of a novel strain (YH01) of Micropterus salmoides rhabdovirus and expression of its glycoprotein by the baculovirus expression system. J. Zhejiang Uni. Sci. B 2019, 20, 728–739. [Google Scholar] [CrossRef]
- Zhang, Q.Y.; Tao, J.J.; Gui, L.; Zhou, G.Z.; Ruan, H.M.; Li, Z.Q.; Gui, J.F. Isolation and characterization of Scophthalmus maximus rhabdovirus. Dis. Aquat. Org. 2007, 74, 95–105. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.D.; Chen, X.; Tian, J.H.; Chen, L.J.; Li, K.; Wang, W.; Eden, J.S.; Shen, J.J.; Liu, L.; et al. The evolutionary history of vertebrate RNA viruses. Nature 2018, 556, 197–202. [Google Scholar] [CrossRef]
- Morzunov, S.P.; Winton, J.R.; Nichol, S.T. The complete genome structure and phylogenetic relationship of infectious hematopoietic necrosis virus. Virus Res. 1995, 38, 175–192. [Google Scholar] [CrossRef]
- Dixon, P.; Paley, R.; Alegria-Moran, R.; Oidtmann, B. Epidemiological characteristics of infectious hematopoietic necrosis virus (IHNV): A review. Vet. Res. 2016, 47, e63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurath, G. Fish novirhabdoviruses. In Rhabdoviruses: Molecular Taxonomy, Evolution, Genomics, Ecology, Cytopathology, and Control; Dietzgen, R.G., Kuzmin, I.V., Eds.; Caister Academic Press: Norfolk, UK, 2012; pp. 98–116. [Google Scholar]
- Jensen, M.H. Research on the virus of Egtved disease. Ann. N. Y. Acad. Sci. 1965, 126, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Skall, H.F.; Olesen, N.J.; Mellergaard, S. Viral haemorrhagic septicaemia virus in marine fish and its implications for fish farming—A review. J. Fish. Dis. 2005, 28, 509–529. [Google Scholar] [CrossRef] [PubMed]
- Einer-Jensen, K.; Ahrens, P.; Forsberg, R.; Lorenzen, N. Evolution of the fish rhabdovirus viral haemorrhagic septicaemia virus. J. Gen. Virol. 2004, 85, 1167–1179. [Google Scholar] [CrossRef] [PubMed]
- Snow, M.; Bain, N.; Black, J.; Taupin, V.; Cunningham, C.O.; King, J.A.; Skall, H.F.; Raynard, R.S. Genetic population structure of marine viral haemorrhagic septicaemia virus (VHSV). Dis. Aquat. Org. 2004, 61, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Emmenegger, E.J.; Moon, C.H.; Hershberger, P.K.; Kurath, G. Virulence of viral hemorrhagic septicemia virus (VHSV) genotypes Ia, IVa, IVb, and IVc in five fish species. Dis. Aquat. Org. 2013, 107, 99–111. [Google Scholar] [CrossRef] [Green Version]
- Oh, M.-J.; Choi, T.-J. A new rhabdovirus (HRV-like) isolated in Korea from cultured Japanese flounder (Paralichthys olivaceus). J. Fish. Pathol. 1998, 11, 129–136. [Google Scholar]
- Kim, W.S.; Oh, M.J. Hirame rhabdovirus (HIRRV) as the cause of a natural disease outbreak in cultured black seabream (Acanthopagrus schlegeli) in Korea. Arch. Virol. 2015, 160, 3063–3066. [Google Scholar] [CrossRef]
- Kim, D.H.; Oh, H.K.; Eou, J.I.; Seo, H.J.; Kim, S.K.; Oh, M.J.; Nam, S.W.; Choi, T.J. Complete nucleotide sequence of the hirame rhabdovirus, a pathogen of marine fish. Virus Res. 2005, 107, 1–9. [Google Scholar] [CrossRef]
- Seo, H.G.; Do, J.W.; Jung, S.H.; Han, H.J. Outbreak of hirame rhabdovirus infection in cultured spotted sea bass Lateolabrax maculatus on the western coast of Korea. J. Fish. Dis. 2016, 39, 1239–1246. [Google Scholar] [CrossRef] [Green Version]
- Kimura, T.; Yoshimizu, M.; Gorie, S. A new rhabdovirus isolated in Japan from cultured hirame (Japanese flounder) Paralichthys olivaceus and ayu Plecoglossus altivelis. Dis. Aquat. Org. 1986, 1, 209–217. [Google Scholar] [CrossRef]
- Borzym, E.; Matras, M.; Maj-Paluch, J.; Baud, M.; De Boisseson, C.; Talbi, C.; Olesen, N.J.; Bigarré, L. First isolation of hirame rhabdovirus from freshwater fish in Europe. J. Fish. Dis. 2014, 37, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Kasornchandra, J.; Engelking, H.M.; Lannan, C.N.; Rohovec, J.S.; Fryer, J.L. Characteristics of three rhabdoviruses from snakehead fish Ophicephalus striatus. Dis. Aquat. Org. 1992, 13, 89–94. [Google Scholar] [CrossRef]
- Johnson, M.C.; Maxwell, J.M.; Loh, P.C.; Leong, J.A. Molecular characterization of the glycoproteins from two warm water rhabdoviruses: Snakehead rhabdovirus (SHRV) and rhabdovirus of penaeid shrimp (RPS)/spring viremia of carp virus (SVCV). Virus Res. 1999, 64, 95–106. [Google Scholar] [CrossRef]
- Frerichs, G.N.; Millar, S.D.; Roberts, R.J. Ulcerative rhabdovirus in fish in South-East Asia. Nature 1986, 322, 216. [Google Scholar] [CrossRef] [PubMed]
- Olberding, K.P.; Frost, J.W. Electron microscopical observations of the structure of the virus of viral haemorrhagic septicaemia (VHS) of rainbow trout (Salmo gairdneri). J. Gen. Virol. 1975, 27, 305–312. [Google Scholar] [CrossRef]
- Zwillenberg, L.O.; Jensen, H.M.; Zwillenberg, H.H.L. Electron microscopy of the virus of viral haemorrhagie septicaemia of rainbow trout (Egtved virus). Arch. Gesamte Virusforsch. 1965, 17, 1–19. [Google Scholar] [CrossRef]
- Armend, D.F.; Chambers, V.C. Morphology of certain viruses of salmonid fishes. I. In vitro studies of sorne viruses causing hematopoietic necrosis. J. Fish. Res. Board Canada 1970, 27, 1285–1293. [Google Scholar] [CrossRef]
- Thoulouze, M.-I.; Bouguyon, E.; Carpentier, C.; Bremont, M. Essential role of the NV protein of novirhabdovirus for pathogenicity in rainbow trout. J. Virol. 2004, 78, 4098–4107. [Google Scholar] [CrossRef] [Green Version]
- Brémont, M. Reverse genetics on fish rhabdoviruses: Tools to study the pathogenesis of fish rhabdoviruses. Curr. Top. Microbiol, Immunol. 2005, 292, 119–141. [Google Scholar] [CrossRef]
- Kurath, G.; Leong, J.C. Characterization of infectious hematopoietic necrosis virus mRNA species reveals a nonvirion rhabdovirus protein. J. Virol. 1985, 53, 462–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schütze, H.; Mundt, E.; Mettenleiter, T.C. Complete genomic sequence of viral hemorrhagic septicemia virus, a fish rhabdovirus. Virus Genes 1999, 19, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.J.; Kongsuwan, K. Deduced structural model for animal rhabdovirus glycoproteins. J. Gen. Virol. 1999, 80, 1211–1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Einer-Jensen, K.; Krogh, T.N.; Roepstorff, P.; Lorenzen, N. Characterization of intramolecular disulfide bonds and secondary modifications of the glycoprotein from viral hemorrhagic septicemia virus, a fish rhabdovirus. J. Virol. 1998, 72, 10189–10196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roche, S.; Bressanelli, S.; Rey, F.A.; Gaudin, Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 2006, 313, 187–191. [Google Scholar] [CrossRef]
- Roche, S.; Rey, F.A.; Gaudin, Y.; Bressanelli, S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science 2007, 315, 843–848. [Google Scholar] [CrossRef] [Green Version]
- Li, C.X.; Shi, M.; Tian, J.H.; Lin, X.D.; Kang, Y.J.; Chen, L.J.; Qin, X.C.; Xu, J.; Holmes, E.C.; Zhang, Y.Z. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. eLife 2015, 4, e05378. [Google Scholar] [CrossRef]
- Wolf, Y.I.; Kazlauskas, D.; Iranzo, J.; Lucia-Sanz, A.; Kuhn, J.H.; Krupovic, M.; Dolja, V.V.; Koonin, E.V. Origins and evolution of the global RNA virome. mBio 2018, 9, e02329-18. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Eickbush, T.H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990, 9, 3353–3362. [Google Scholar] [CrossRef]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. TrimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Deviatkin, A.A.; Lukashev, A.N. Recombination in the rabies virus and other lyssaviruses. Inf. Genet. Evol. 2018, 60, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Wittmann, T.J.; Biek, R.; Hassanin, A.; Rouquet, P.; Reed, P.; Yaba, P.; Pourrut, X.; Real, L.A.; Gonzalez, J.-P.; Leroy, E.M. Isolates of Zaire ebolavirus from wild apes reveal genetic lineage and recombinants. Proc. Natl. Acad. Sci. USA 2007, 104, 17123–17127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, P.J.; Kim, L.M.; Ip, H.S.; Afonso, C.L. Evolutionary dynamics of Newcastle disease virus. Virology 2009, 391, 64–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, G.-Z.; Worobey, M. Homologous recombination in negative sense RNA viruses. Viruses 2011, 3, 1358–1373. [Google Scholar] [CrossRef]
- Chare, E.R.; Gould, E.A.; Holmes, E.C. Phylogenetic analysis reveals a low rate of homologous recombination in negative-sense RNA viruses. J. Gen. Virol. 2003, 84, 2691–2703. [Google Scholar] [CrossRef]
- Roche, S.; Albertini, A.A.; Lepault, J.; Bressanelli, S.; Gaudin, Y. Structures of vesicular stomatitis virus glycoprotein: Membrane fusion revisited. Cell Mol. Life Sci. 2008, 65, 1716–1728. [Google Scholar] [CrossRef] [Green Version]
- Holmes, E.C.; Duchene, S. Can sequence phylogenies safely infer the origin of the global virome? mBio 2019, 10, e00289-19. [Google Scholar] [CrossRef] [Green Version]
- Wolf, Y.I.; Kazlauskas, D.; Iranzo, J.; Lucia-Sanz, A.; Kuhn, J.H.; Krupovic, M.; Dolja, V.V.; Koonin, E.V. Reply to Holmes and Duchene, "Can sequence phylogenies safely infer the origin of the global virome?": Deep phylogenetic analysis of RNA viruses is highly challenging but not meaningless. mBio 2019, 10, e00542-19. [Google Scholar] [CrossRef] [Green Version]
- Abrescia, N.G.; Bamford, D.H.; Grimes, J.M.; Stuart, D.I. Structure unifies the viral universe. Ann. Rev. Biochem. 2012, 81, 795–822. [Google Scholar] [CrossRef]
- Krupovic, M.; Dolja, V.V.; Koonin, E.V. Origin of viruses: Primordial replicators recruiting capsids from hosts. Nat. Rev. Microbiol. 2019, 17, 449–458. [Google Scholar] [CrossRef]
- Walker, P.J.; Blasdell, K.R.; Joubert, D.A. Ephemeroviruses: Arthropod-borne rhabdoviruses of ruminants, with large and complex genomes. In Rhabdoviruses: Molecular Taxonomy, Evolution, Genomics, Ecology, Cytopathology and Control; Dietzgen, R.G., Kuzman, I.V., Eds.; Horizon Scientific Press: Norwich, UK, 2012. [Google Scholar]


| Subfamily | Genus | Species | Exemplar Virus | Abbrev. | Sample § | GenBank Accession | Intra-Species Divergence (%) # | Inter-Species Divergence (%) # | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L | N | G | L | N | G | |||||||
| Alpharhabdovirinae | Sprivivirus | Sprivivirus cyprinus | spring viremia of carp virus | SVCV | VR-1390 | U18101 | ≤2.7 | ≤3.4 | ≤8.7 | ≥11.8 | ≥8.2 | ≥25.4 |
| Sprivivirus esox | pike fry rhabdovirus | PFRV | F4 | FJ872827 | ≤6.7 | ≤5.6 | ≤16.7 | |||||
| Perhabdovirus | Perhabdovirus perca | perch rhabdovirus | PRV | Dorson | JX679246 | ≤4.5 | ≤2.7 | ≤3.3 | ≥13.1 | ≥15.9 | ≥16.7 | |
| Perhabdovirus trutta | lake trout rhabdovirus | LTRV | 903/87 | AF434991 | ≤5.2 | ≤6.1 | ≤11.6 | |||||
| Perhabdovirus anguilla | eel virus European X | EVEX | CV1153311 | FN557213 | ≤2.2 | ≤1.7 | ≤4.1 | |||||
| Perhabdovirus leman | Leman virus | LEMV | 18/193 | MN963996 | n.a. | n.a. | n.a. | |||||
| Cetarhavirus | Cetarhavirus lagenorhynchus | dolphin rhabdovirus | DRV | pxV1 | KF958252 | n.a. | n.a. | n.a. | 31.3 | 31.8 | 56.1 | |
| Cetarhavirus phocoena | harbour porpoise rhabdovirus | HPRV | WVL17017A | MN103537 | n.a. | n.a. | n.a. | |||||
| Siniperhavirus | Siniperhavirus zoarces | eelpout rhabdovirus | EPRV | FSK0523 | KR612230 | n.a. | n.a. | n.a. | ≥28.8 | ≥33.7 | ≥50.0 | |
| Siniperhavirus chuatsi | Siniperca chuatsi rhabdovirus | SCRV | n.a. | DQ399789 | ≤3.6 | ≤6.8 | ≤10.6 | |||||
| Scophrhavirus | Scophrhavirus maximus | Scophthalmus maximus rhabdovirus | SMRV | n.a. | HQ003891 | n.a. | n.a. | n.a. | 46.5 | 62.6 | 71.0 | |
| Scophrhavirus chanodichthys | Wuhan redfin culter dimarhabdovirus | WhRCDRV | DSYS6218 | MG600013 | n.a. | n.a. | n.a. | |||||
| Gammarhabdovirinae | Novirhabdovirus | Novirhabdovirus salmonid | infectious haematopoietic necrosis virus | IHNV | WRAC | L40883 | ≤2.4 | ≤12.6 | ≤10.2 | ≥15.1 | ≥35.8 | ≥22.9 |
| Novirhabdovirus piscine | viral haemorrhagic septicaemia virus | VHSV | Fil3 | Y18263 | ≤7.1 | ≤11.1 | ≤11.5 | |||||
| Novirhabdovirus hirame | hirame rhabdovirus | HIRRV | CA9703 | AF104985 | 0.4 | 0.8 | ≤1.2 | |||||
| Novirhabdovirus snakehead | snakehead rhabdovirus | SHRV | n.a. | AF147498 | n.a. | n.a. | n.a. | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walker, P.J.; Bigarré, L.; Kurath, G.; Dacheux, L.; Pallandre, L. Revised Taxonomy of Rhabdoviruses Infecting Fish and Marine Mammals. Animals 2022, 12, 1363. https://doi.org/10.3390/ani12111363
Walker PJ, Bigarré L, Kurath G, Dacheux L, Pallandre L. Revised Taxonomy of Rhabdoviruses Infecting Fish and Marine Mammals. Animals. 2022; 12(11):1363. https://doi.org/10.3390/ani12111363
Chicago/Turabian StyleWalker, Peter J., Laurent Bigarré, Gael Kurath, Laurent Dacheux, and Laurane Pallandre. 2022. "Revised Taxonomy of Rhabdoviruses Infecting Fish and Marine Mammals" Animals 12, no. 11: 1363. https://doi.org/10.3390/ani12111363






