Characterization of Antimicrobial Resistance Dissemination across Plasmid Communities Classified by Network Analysis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Number of Edges and Nodes between Each Plasmid
| Clustering program | Identity cutoff (%) | Edges | Connected nodes | Connected components | Nodes in LCC | LCC % in total | Number of communities in the LCC using multilevel method |
|---|---|---|---|---|---|---|---|
| BLASTP | 50 | 251,961 | 3529 | 57 | 3265 | 86.1% | 19 |
| BLASTP | 60 | 177,932 | 3444 | 70 | 2793 | 73.6% | 25 |
| BLASTP | 70 | 129,545 | 3358 | 90 | 2613 | 68.9% | 31 |
| BLASTP | 80 | 104,815 | 3229 | 127 | 2265 | 59.7% | 26 |
| BLASTP | 90 | 81,752 | 3060 | 162 | 1995 | 52.6% | 34 |
| BLASTP | 99 | 52,029 | 2633 | 236 | 1389 | 36.6% | 36 |
| BLASTP | 100 | 41,956 | 2496 | 259 | 1241 | 32.7% | 26 |
| UCLUST | 50 | 188,844 | 3524 | 71 | 3233 | 85.2% | ND |
| UCLUST | 60 | 147,899 | 3435 | 75 | 2762 | 72.8% | ND |
| UCLUST | 70 | 121,841 | 3349 | 98 | 2558 | 67.4% | ND |
| UCLUST | 80 | 101,902 | 3213 | 133 | 2241 | 59.1% | ND |
| UCLUST | 90 | 82,046 | 3057 | 174 | 1949 | 51.4% | ND |
| UCLUST | 99 | 52,578 | 2626 | 240 | 1369 | 36.1% | ND |
| UCLUST | 100 | 43,275 | 2501 | 263 | 1232 | 32.5% | ND |
2.2. Connected Components (CCs)
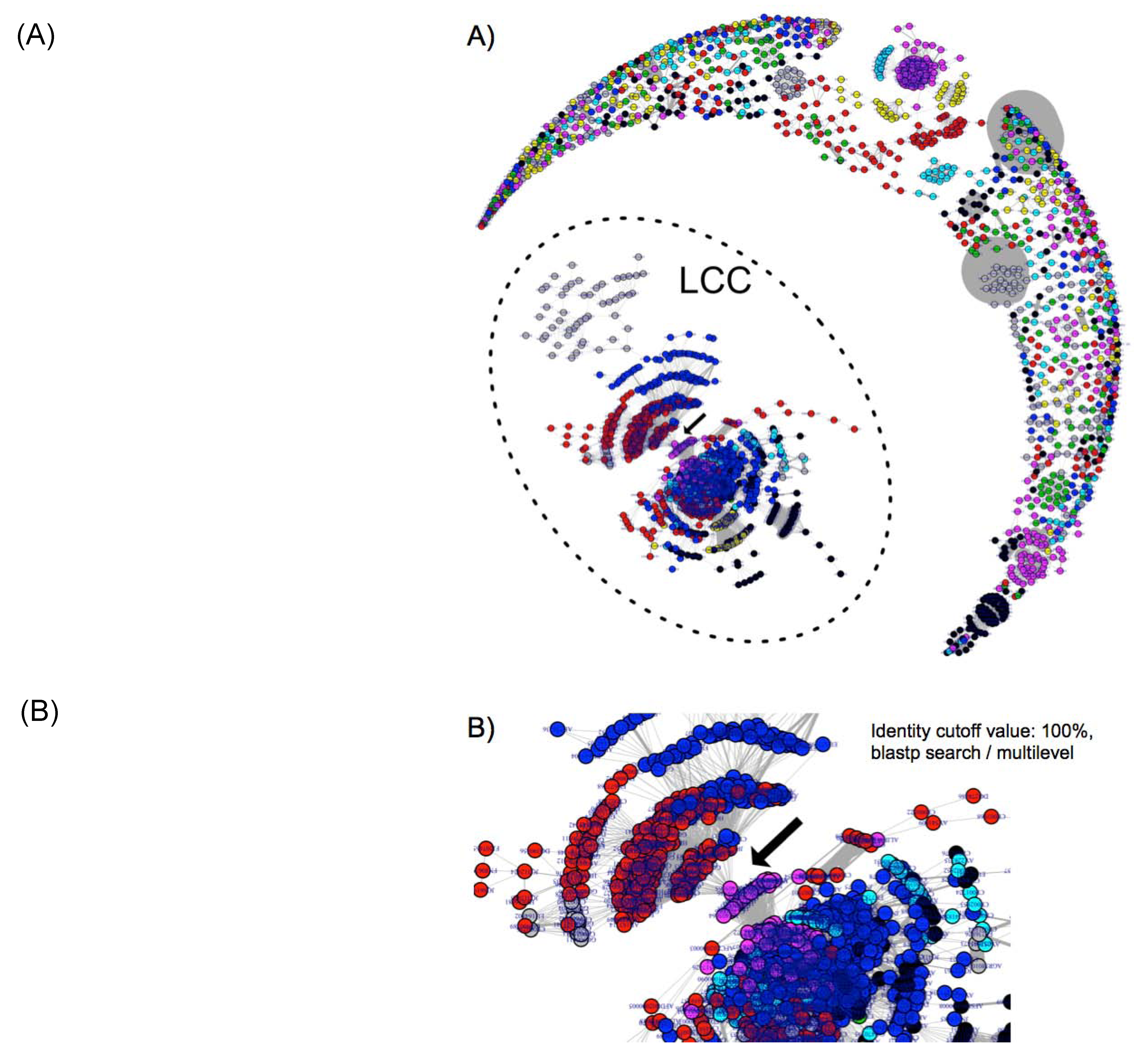
2.3. Community Network Analysis
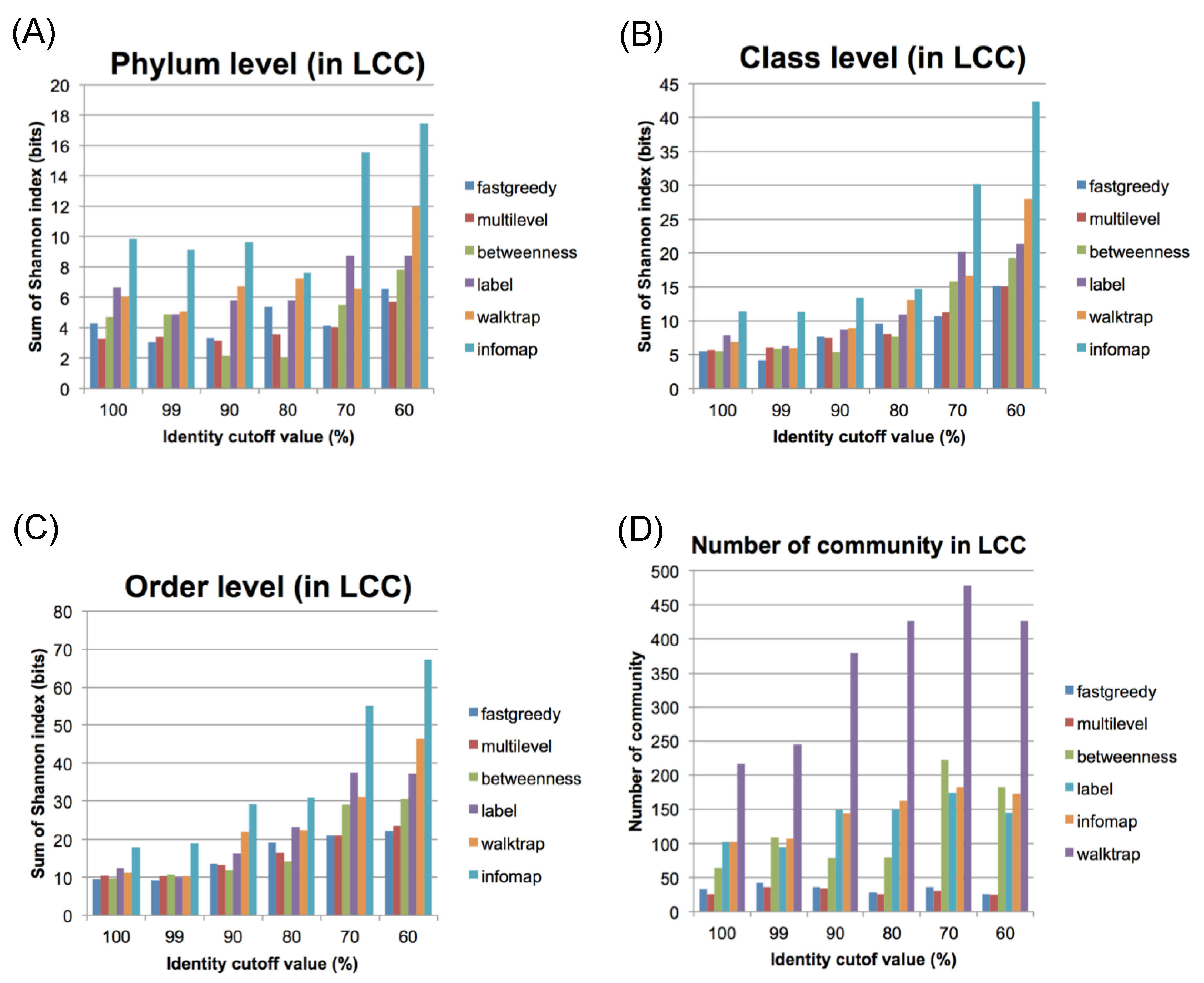
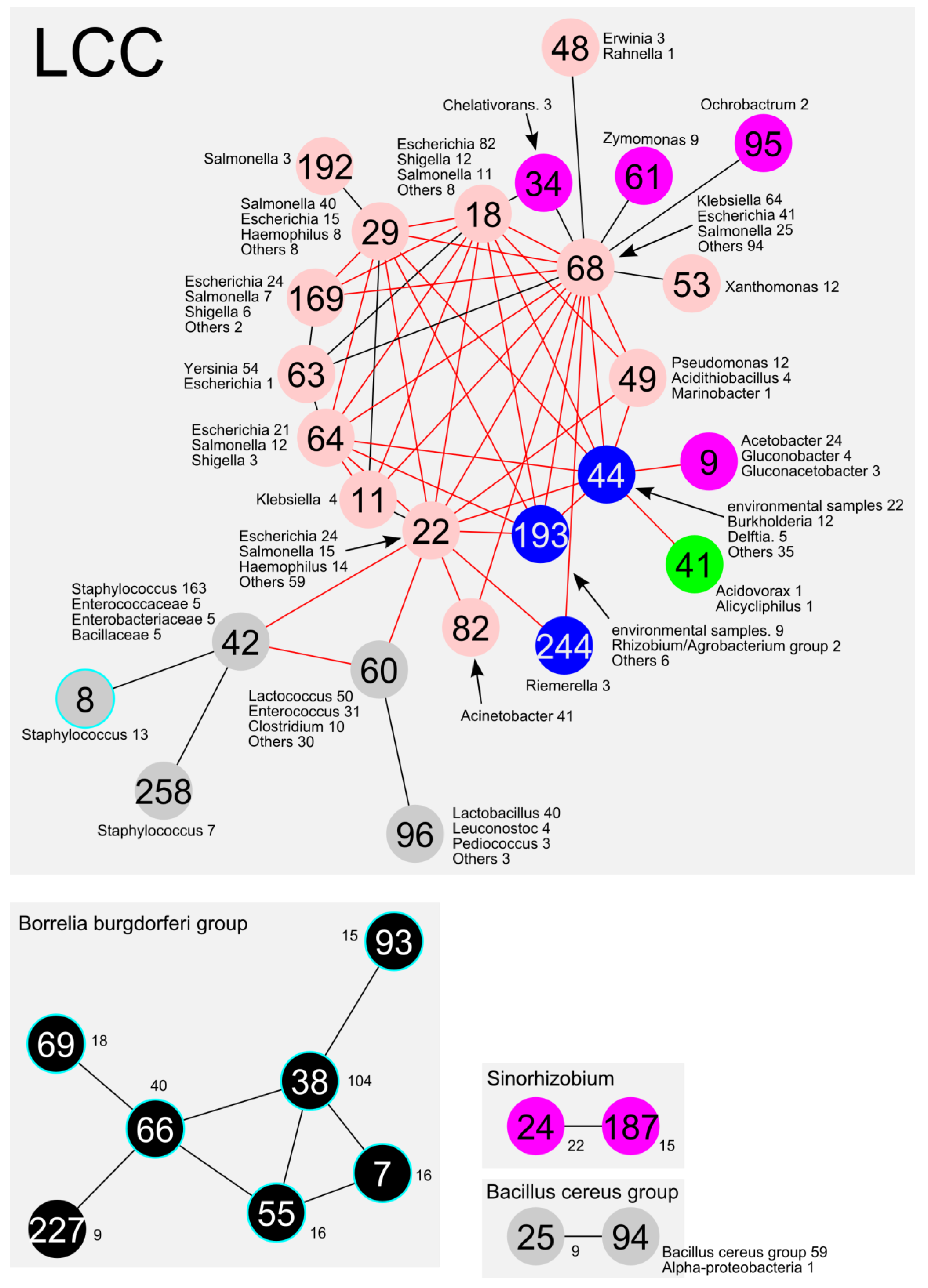
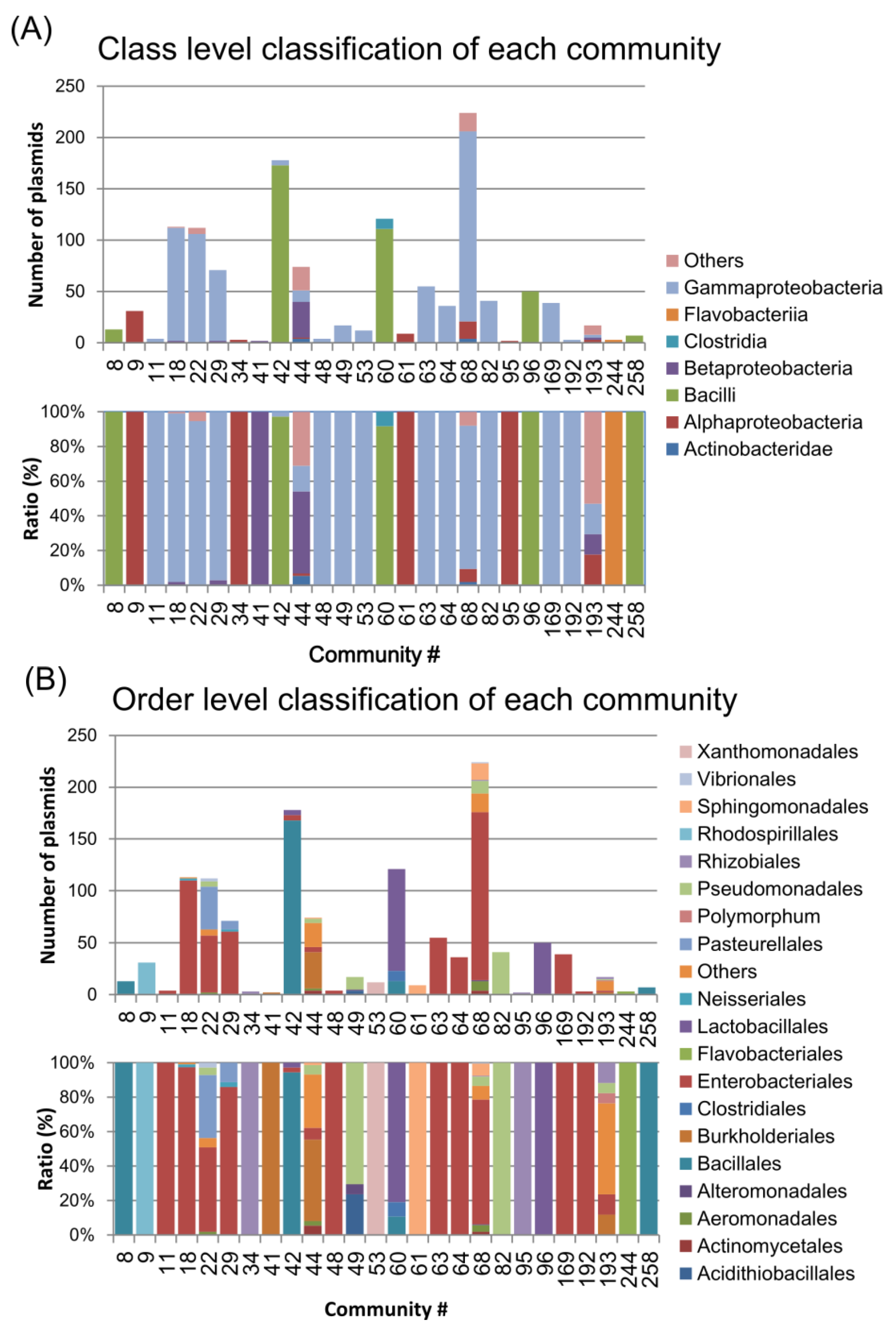
2.4. Connections between Communities
2.4.1. Genes Involved in the Connections between Communities
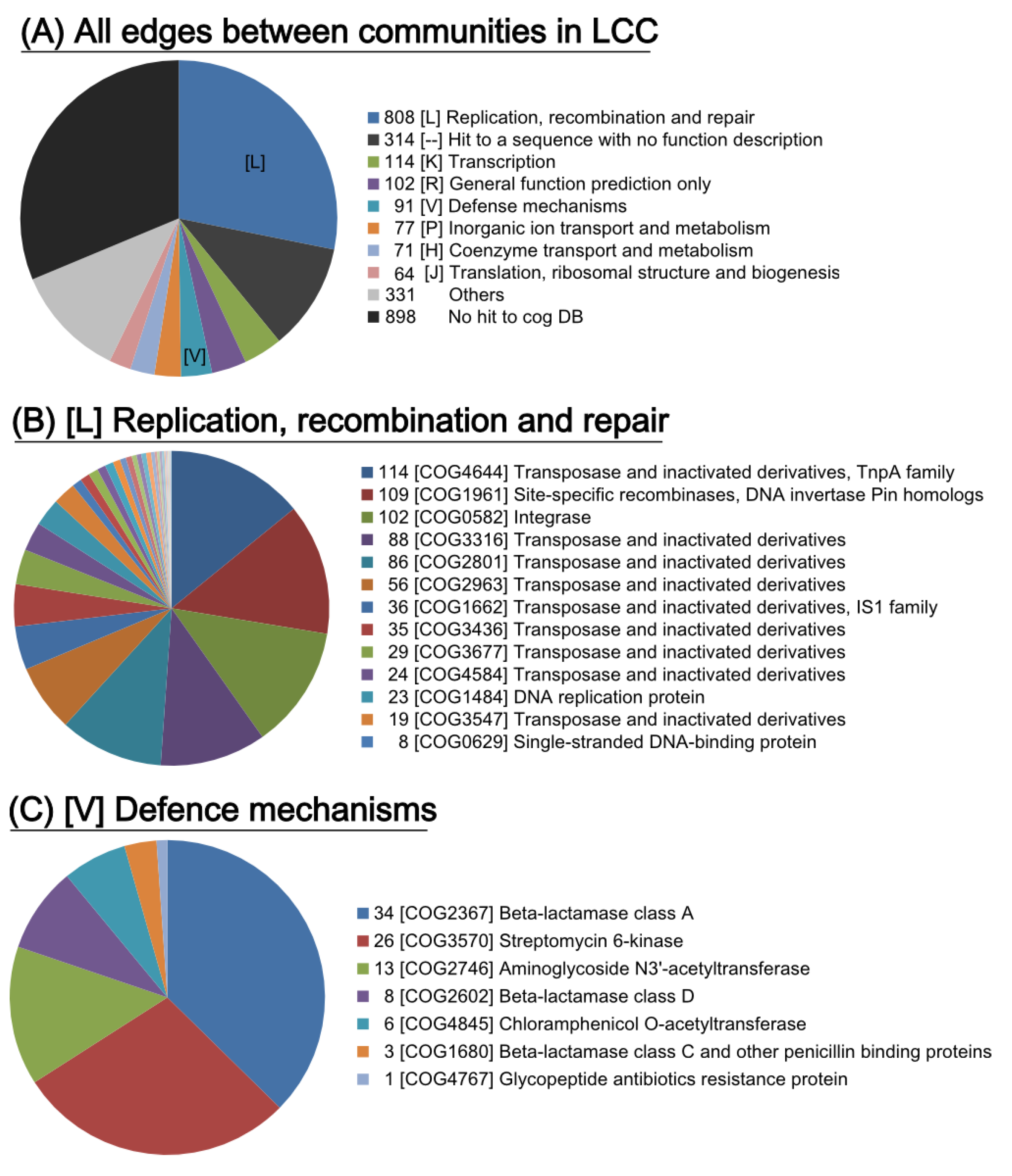
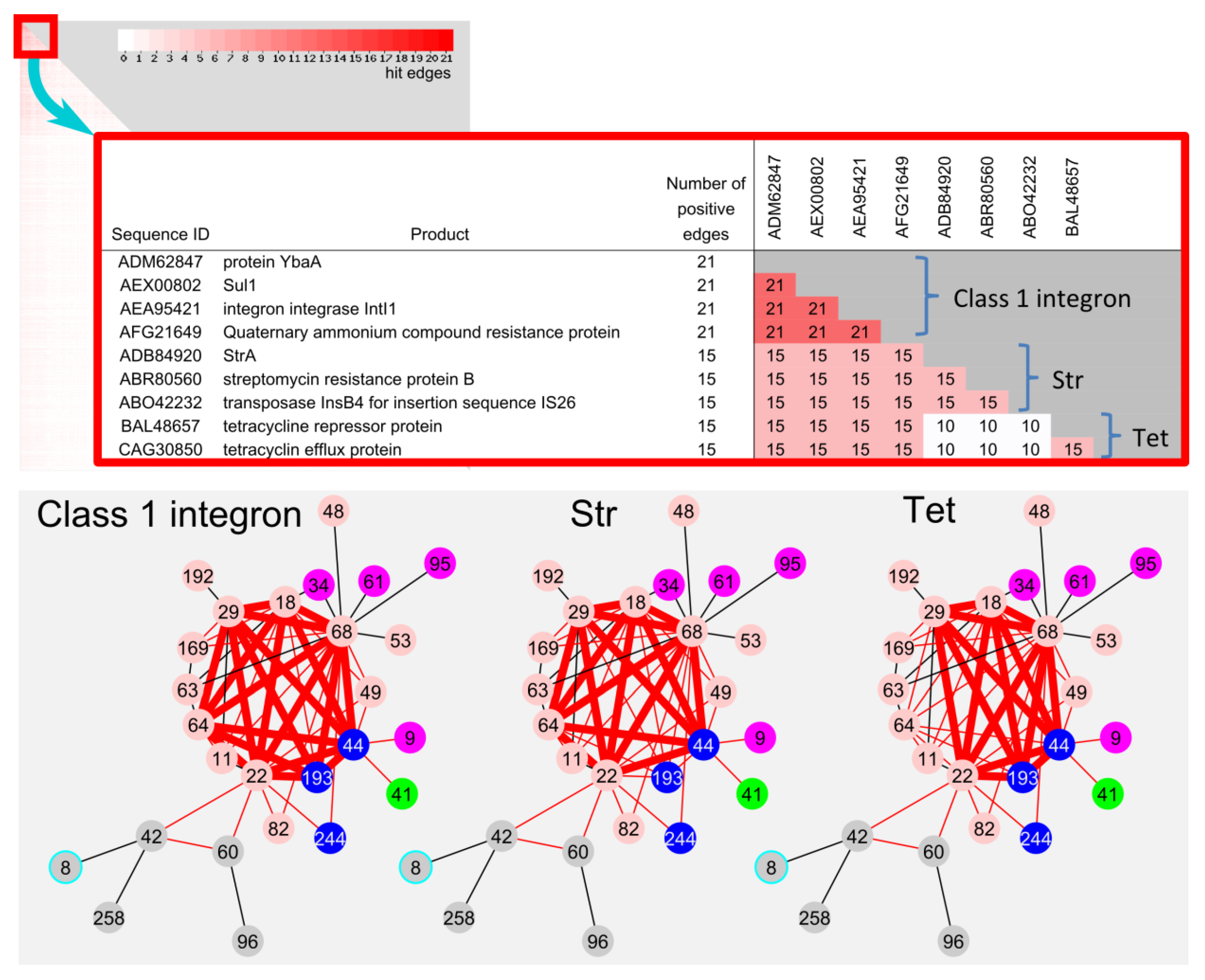
2.4.2. Frequently Identified Gene Combination among the Edges between Communities
| Order of gene groups | Community | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|
| co18 | co22 | co29 | co44 | co64 | co68 | co193 | |||
 | 1 | 1 | |||||||
 | 1 | 1 | |||||||
 | 1 | 1 | |||||||
 | 2 | 1 | 1 | 4 | |||||
 | 1 | 1 | |||||||
 | 1 | 1 | |||||||
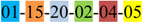 | 1 | 1 | |||||||
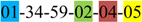 | 2 | 2 | |||||||
 | 1 | 1 | 2 | ||||||
 | 1 | 1 | |||||||
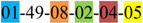 | 2 | 2 | |||||||
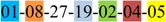 | 1 | 1 | |||||||
 | 1 | 1 | |||||||
 | 1 | 1 | |||||||
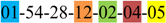 | 1 | 1 | |||||||
 | 1 | 1 | |||||||
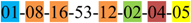 | 1 | 1 | |||||||
 | 1 | 1 | |||||||
 | 1 | 1 | |||||||
 | 1 | 1 | |||||||
 | 1 | 1 | |||||||
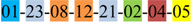 | 1 | 1 | |||||||
 | 1 | 1 | |||||||
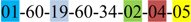 | 1 | 1 | |||||||
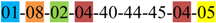 | 1 | 1 | |||||||
 | 1 | 1 | |||||||
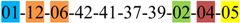 | 3 | 3 | |||||||
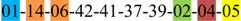 | 1 | 1 | |||||||
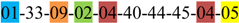 | 1 | 1 | |||||||
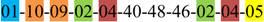 | 1 | 1 | |||||||
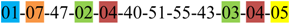 | 1 | 1 | |||||||
 | 1 | 1 | |||||||
| Total | 3 | 9 | 2 | 2 | 1 | 22 | 1 | 40 | |
| Gene group | Products | ||||||||
| 01 | integron integrase IntI1. | class 1 integron component | |||||||
| 02, 03 | quaternary ammonium compound-resistance protein QacEdelta1. | ||||||||
| 04 | sulphonamide resistant dihydropteroate synthase Sul1. | ||||||||
| 05 | protein YbaA. | ||||||||
| 06 | aminoglycoside 3'- N-acetyltransferase protein aacC. | aminoglycoside resistance | |||||||
| 07 | aminoglycoside 3'- N-acetyltransferase. | ||||||||
| 08 | aminoglycoside acetyltransferase. | ||||||||
| 09 | aminoglycoside adenylyltransferase. | ||||||||
| 10 | aminoglycoside N(6')-acetyltransferase. | ||||||||
| 11–14 | aminoglycoside resistance protein aadA. | ||||||||
| 15 | aminoglycoside-2'-adenylyltransferase. | ||||||||
| 16 | acetyltransferases aac(3)-Ia. | ||||||||
| 17 | putative aminoglycoside 6'- N-acetyltransferase. | ||||||||
| 18 | putative aminoglycoside adenyltransferase. | ||||||||
| 19 | class D β-lactamase OXA-10. | β-lactamase | |||||||
| 20 | class D β-lactamase OXA-21. | ||||||||
| 21 | class D β-lactamase OXA-2. | ||||||||
| 22 | class D β-lactamase OXA. | ||||||||
| 23 | metallo-β-lactamase GIM-1. | ||||||||
| 24 | metallo-β-lactamase IMP-6. | ||||||||
| 25 | metallo-β-lactamase VIM-1. | ||||||||
| 26 | chloramphenicol acetyltransferase catB2. | chloramphenicol resistance | |||||||
| 27 | chloramphenicol acetyltransferase catB8. | ||||||||
| 28, 29 | chloramphenicol acetyltransferase. | ||||||||
| 30 | chloramphenicol aminotransferase. | ||||||||
| 31 | chloramphenicol resistance protein CmlA1. | ||||||||
| 32 | dihydrofolate reductase DfrA5. | dihydrofolate reductase | |||||||
| 33-35 | dihydrofolate reductase. | ||||||||
| 36 | trimethoprim-resistant dihydrofolate reductase type I DhfrA1. | ||||||||
| 37–40 | transposase. | ||||||||
| 41 | molecular chaperone GroEL. | ||||||||
| 42 | molecular chaperone GroES. | ||||||||
| 43 | fluoroquinolone resistance protein QnrB2. | ||||||||
| 44 | quinolone-resistance determinant Qnr. | ||||||||
| 45–52 | Others | ||||||||
| 53-60 | hypothetical protein. | ||||||||
2.4.3. Connections between Plasmid Communities from Different Divisions
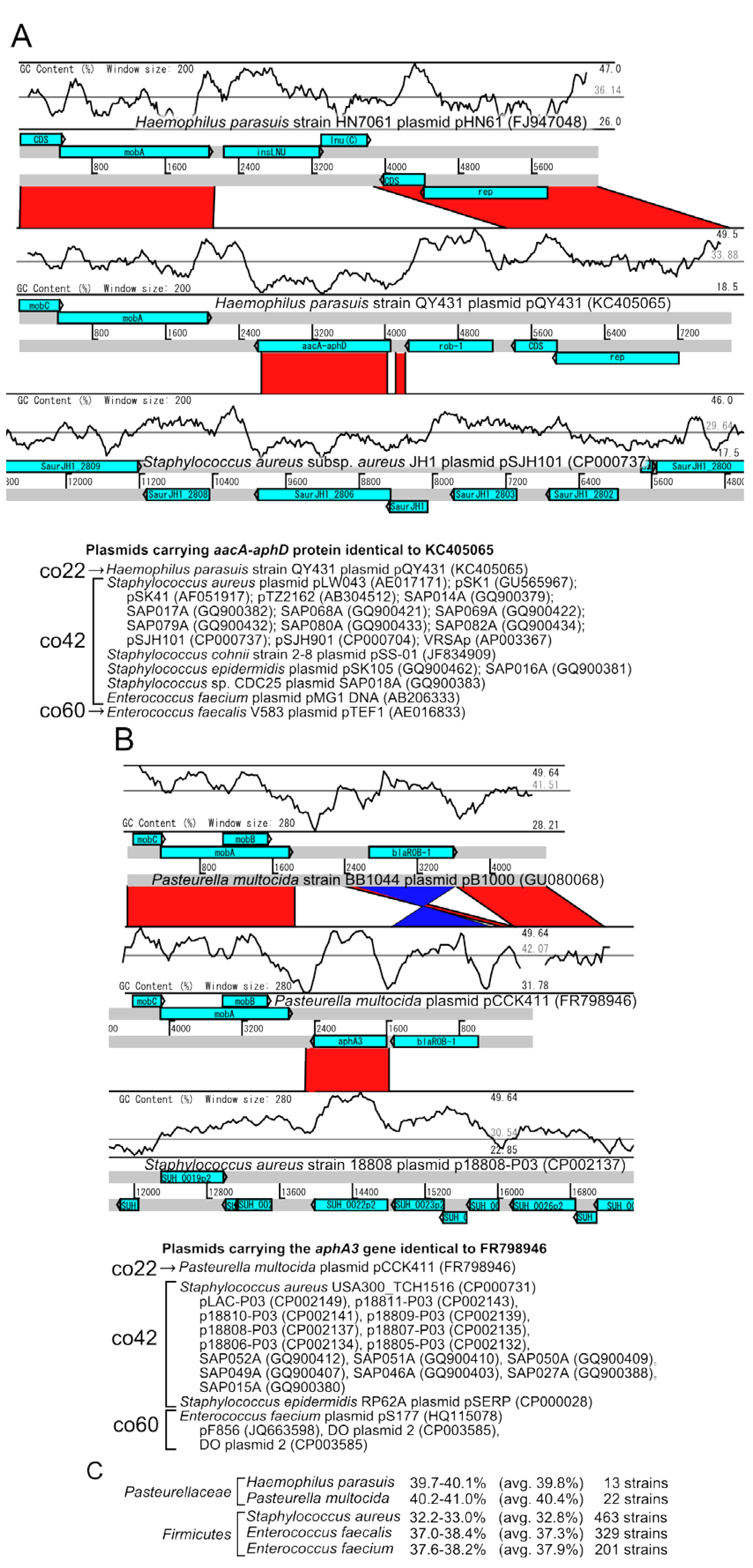
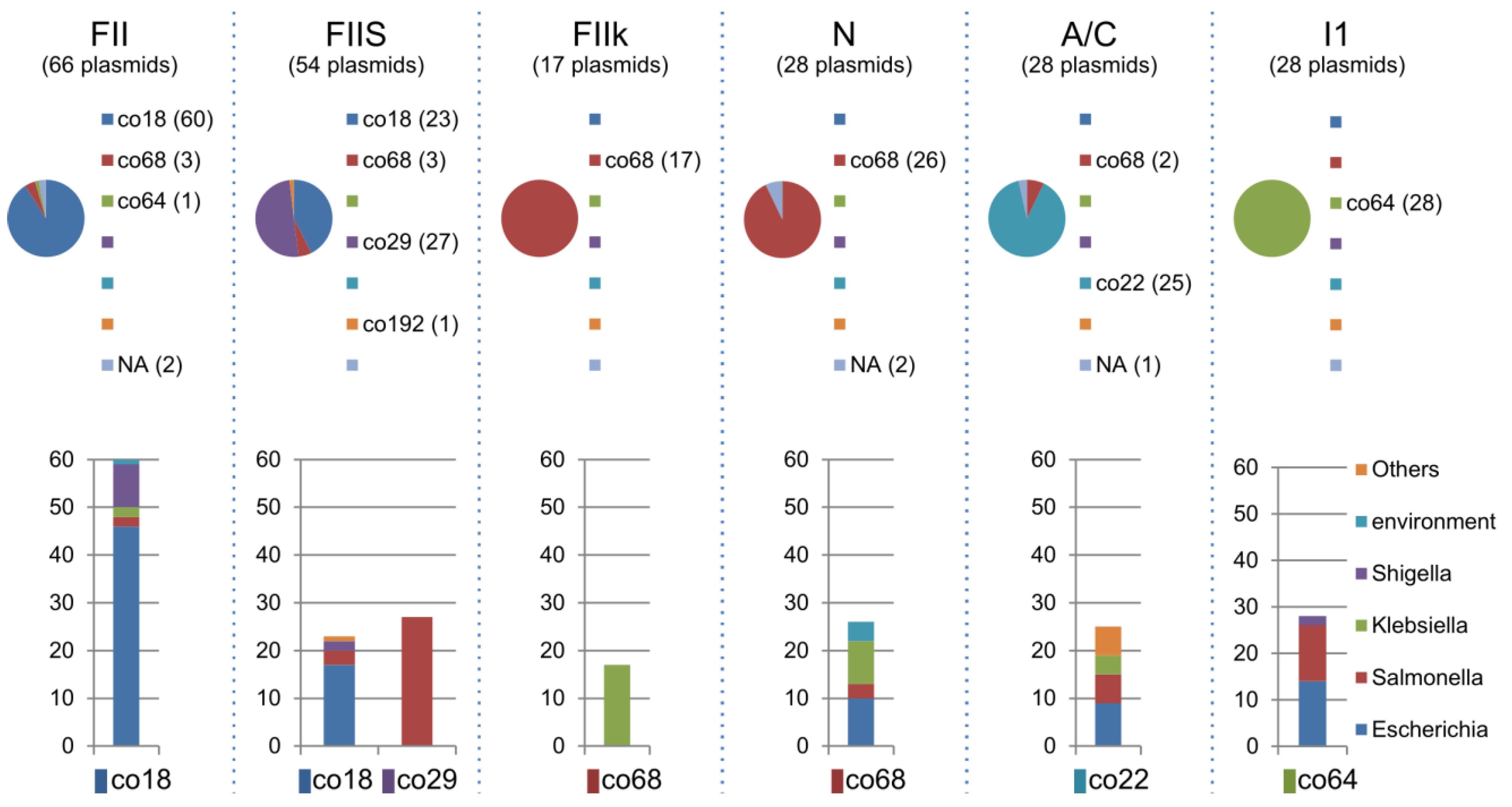
2.5. Distribution of Inc Types among Plasmid Communities
| Inc type | Community | |||||||
|---|---|---|---|---|---|---|---|---|
| co11 | co18 | co22 | co29 | co64 | co68 | co192 | co193 | |
| A/C | - | - | 25 | - | - | 2 | - | - |
| B/O | - | 3 | - | - | 1 | 1 | - | - |
| FIA | - | 5 | - | - | - | 3 | - | - |
| FIB | - | 5 | - | - | - | - | - | - |
| FIB-M | - | - | - | - | - | 1 | - | - |
| FII | - | 60 | - | - | 1 | 3 | - | - |
| FIIk | - | - | - | - | - | 17 | - | - |
| FIIS | - | 23 | - | 27 | - | 3 | 1 | - |
| HI1 | - | 1 | - | - | - | 1 | - | - |
| HI2 | - | - | - | - | - | 5 | - | - |
| HIB-M | 3 | - | - | - | - | - | - | - |
| I1 | - | - | - | - | 28 | - | - | - |
| K | - | 1 | - | - | 3 | - | - | - |
| L/M | - | - | - | - | - | 7 | - | - |
| N | - | - | - | - | - | 26 | - | - |
| P | - | - | - | - | - | - | - | 7 |
| R | - | - | 2 | - | - | 3 | - | - |
| T | - | 1 | - | - | - | 1 | - | - |
| U | - | - | - | - | - | 3 | - | - |
| W | - | - | 2 | - | - | 3 | - | - |
| X1 | - | 1 | - | 7 | - | - | - | - |
| X2 | - | - | - | - | - | 1 | - | - |
| Not Assigned | 1 | 13 | 83 | 37 | 3 | 144 | 2 | 10 |
| Total | 4 | 113 | 112 | 71 | 36 | 224 | 3 | 17 |
2.6. Distribution of AMR among Plasmid Communities
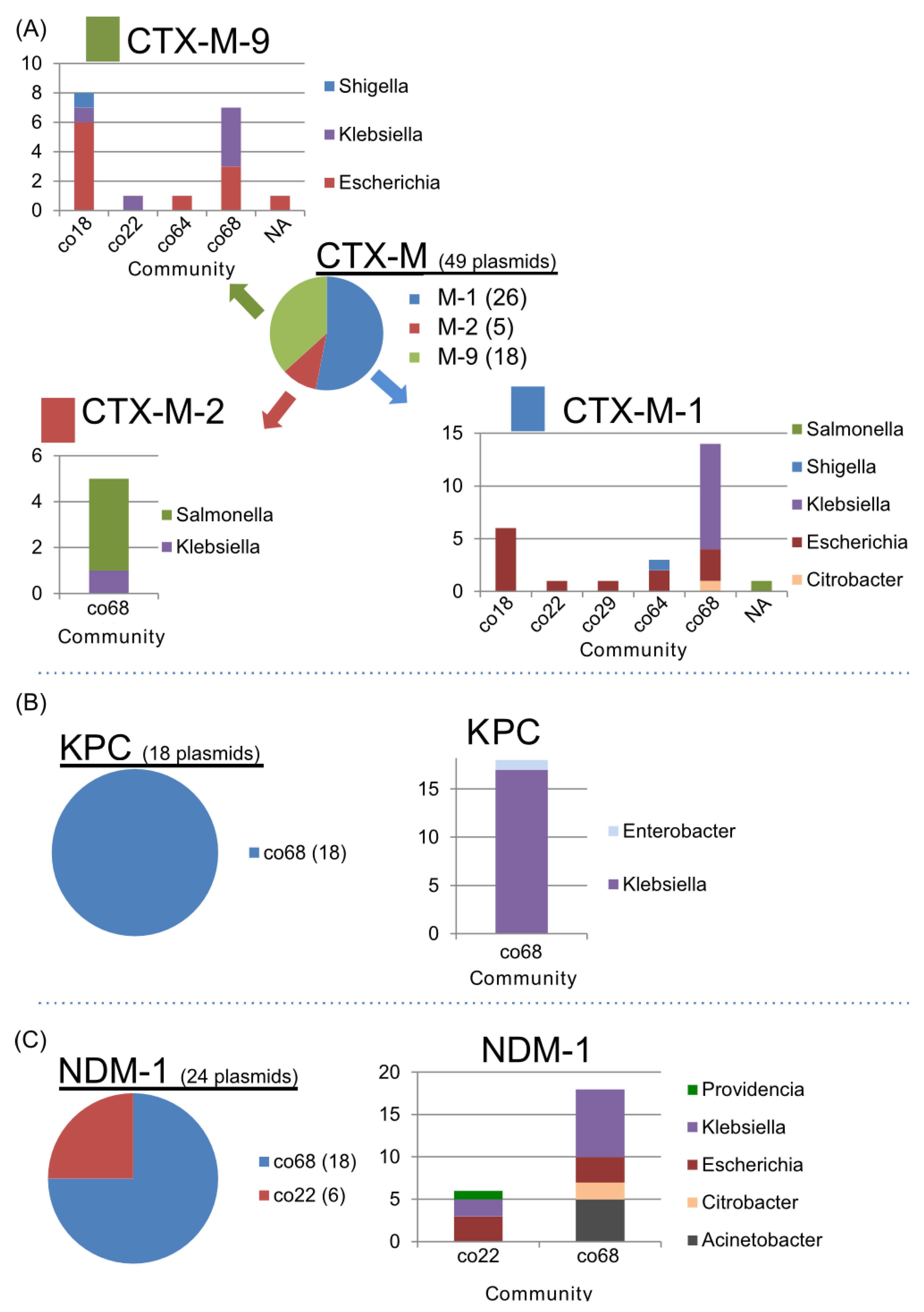
| Inc type | CTX-M | KPC | NDM-1 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| M-1 | M-2 | M-9 | 2 | 3 | 4 | 10 | co22 | co68 | |
| FII | 7 | 8 | 1 | ||||||
| FIIk | 4 | 5 | 2 | 1 | |||||
| N | 2 | 1 | 3 | 1 | 1 | 1 | |||
| A/C | 1 | 6 | |||||||
| L/M | 2 | 1 | 2 | ||||||
| I1 | 3 | ||||||||
| FIB-M | 1 | 1 | |||||||
| FIA | 1 | ||||||||
| FIIS | 1 | ||||||||
| HI1 | 1 | ||||||||
| HI2 | 1 | ||||||||
| K | 1 | ||||||||
| R | 1 | ||||||||
| Not Assigned | 5 | 4 | 4 | 4 | 1 | 1 | 12 | ||
| Total | 26 | 5 | 18 | 11 | 4 | 2 | 1 | 6 | 18 |
3. Experimental Section
3.1. Download and Selection of Complete Plasmids Sequences
3.2. Network Analysis
3.3. Inc Typing and Searching for Mobile Elements
3.4. Detection of CTX-M Genes
3.5. Detection of Other Antibiotic Resistance Genes
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Johnson, T.J.; Nolan, L.K. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 2009, 73, 750–774. [Google Scholar] [CrossRef]
- Norman, A.; Hansen, L.H.; Sorensen, S.J. Conjugative plasmids: Vessels of the communal gene pool. Philos. Trans. R Soc. Lond. B Biol. Sci. 2009, 364, 2275–2289. [Google Scholar] [CrossRef]
- Andam, C.P.; Fournier, G.P.; Gogarten, J.P. Multilevel populations and the evolution of antibiotic resistance through horizontal gene transfer. FEMS Microbiol. Rev. 2011, 35, 756–767. [Google Scholar] [CrossRef]
- Wiedenbeck, J.; Cohan, F.M. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 2011, 35, 957–976. [Google Scholar] [CrossRef]
- Skippington, E.; Ragan, M.A. Lateral genetic transfer and the construction of genetic exchange communities. FEMS Microbiol. Rev. 2011, 35, 707–735. [Google Scholar] [CrossRef]
- Harris, S.R.; Cartwright, E.J.; Torok, M.E.; Holden, M.T.; Brown, N.M.; Ogilvy-Stuart, A.L.; Ellington, M.J.; Quail, M.A.; Bentley, S.D.; Parkhill, J.; et al. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant staphylococcus aureus: A descriptive study. Lancet Infect. Dis. 2013, 13, 130–136. [Google Scholar] [CrossRef]
- He, M.; Miyajima, F.; Roberts, P.; Ellison, L.; Pickard, D.J.; Martin, M.J.; Connor, T.R.; Harris, S.R.; Fairley, D.; Bamford, K.B.; et al. Emergence and global spread of epidemic healthcare-associated clostridium difficile. Nat. Genet. 2013, 45, 109–113. [Google Scholar]
- Imperi, F.; Antunes, L.C.; Blom, J.; Villa, L.; Iacono, M.; Visca, P.; Carattoli, A. The genomics of acinetobacter baumannii: Insights into genome plasticity, antimicrobial resistance and pathogenicity. IUBMB Life 2011, 63, 1068–1074. [Google Scholar] [CrossRef]
- Walker, T.M.; Ip, C.L.; Harrell, R.H.; Evans, J.T.; Kapatai, G.; Dedicoat, M.J.; Eyre, D.W.; Wilson, D.J.; Hawkey, P.M.; Crook, D.W.; et al. Whole-genome sequencing to delineate mycobacterium tuberculosis outbreaks: A retrospective observational study. Lancet Infect. Dis. 2013, 13, 137–146. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Canton, R.; Akova, M.; Carmeli, Y.; Giske, C.G.; Glupczynski, Y.; Gniadkowski, M.; Livermore, D.M.; Miriagou, V.; Naas, T.; Rossolini, G.M.; et al. Rapid evolution and spread of carbapenemases among enterobacteriaceae in europe. Clin. Microbiol. Infect. 2012, 18, 413–431. [Google Scholar] [CrossRef]
- Halary, S.; Leigh, J.W.; Cheaib, B.; Lopez, P.; Bapteste, E. Network analyses structure genetic diversity in independent genetic worlds. Proc. Natl. Acad. Sci. USA 2010, 107, 127–132. [Google Scholar]
- Tamminen, M.; Virta, M.; Fani, R.; Fondi, M. Large-scale analysis of plasmid relationships through gene-sharing networks. Mol. Biol. Evol. 2012, 29, 1225–1240. [Google Scholar] [CrossRef]
- Fondi, M.; Bacci, G.; Brilli, M.; Papaleo, M.C.; Mengoni, A.; Vaneechoutte, M.; Dijkshoorn, L.; Fani, R. Exploring the evolutionary dynamics of plasmids: The acinetobacter pan-plasmidome. BMC Evol. Biol. 2010, 10, 59. [Google Scholar] [CrossRef]
- Brown Kav, A.; Sasson, G.; Jami, E.; Doron-Faigenboim, A.; Benhar, I.; Mizrahi, I. Insights into the bovine rumen plasmidome. Proc. Natl. Acad. Sci. USA 2012, 109, 5452–5457. [Google Scholar]
- Brolund, A.; Franzen, O.; Melefors, O.; Tegmark-Wisell, K.; Sandegren, L. Plasmidome-analysis of esbl-producing escherichia coli using conventional typing and high-throughput sequencing. PLoS One 2013, 8, e65793. [Google Scholar]
- Song, X.; Sun, J.; Mikalsen, T.; Roberts, A.P.; Sundsfjord, A. Characterisation of the plasmidome within enterococcus faecalis isolated from marginal periodontitis patients in norway. PLoS One 2013, 8, e62248. [Google Scholar]
- Lima-Mendez, G.; van Helden, J.; Toussaint, A.; Leplae, R. Reticulate representation of evolutionary and functional relationships between phage genomes. Mol. Biol. Evol. 2008, 25, 762–777. [Google Scholar] [CrossRef]
- Dwight Kuo, P.; Banzhaf, W.; Leier, A. Network topology and the evolution of dynamics in an artificial genetic regulatory network model created by whole genome duplication and divergence. Biosystems. 2006, 85, 177–200. [Google Scholar] [CrossRef]
- Fondi, M.; Fani, R. The horizontal flow of the plasmid resistome: Clues from inter-generic similarity networks. Environ. Microbiol. 2010, 12, 3228–3242. [Google Scholar] [CrossRef]
- The Igraph Library. Available online: http://igraph.sourceforge.net (accessed on 20 June 2013).
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Gillings, M.; Boucher, Y.; Labbate, M.; Holmes, A.; Krishnan, S.; Holley, M.; Stokes, H.W. The evolution of class 1 integrons and the rise of antibiotic resistance. J. Bacteriol. 2008, 190, 5095–5100. [Google Scholar] [CrossRef]
- Niven, D.F.; O’Reilly, T. Significance of v-factor dependency in the taxonomy of haemophilus species and related organisms. Int. J. Syst. Bacteriol. 1990, 40, 1–4. [Google Scholar] [CrossRef]
- Redfield, R.J.; Findlay, W.A.; Bosse, J.; Kroll, J.S.; Cameron, A.D.; Nash, J.H. Evolution of competence and DNA uptake specificity in the pasteurellaceae. BMC Evol. Biol. 2006, 6, 82. [Google Scholar] [CrossRef]
- Carattoli, A.; Miriagou, V.; Bertini, A.; Loli, A.; Colinon, C.; Villa, L.; Whichard, J.M.; Rossolini, G.M. Replicon typing of plasmids encoding resistance to newer beta-lactams. Emerg. Infect. Dis. 2006, 12, 1145–1148. [Google Scholar] [CrossRef]
- Bonnet, R. Growing group of extended-spectrum beta-lactamases: The ctx-m enzymes. Antimicrob. Agents Chemother. 2004, 48, 1–14. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar]
- Edgar, R.C. Search and clustering orders of magnitude faster than blast. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. Mega5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Liu, B.; Pop, M. Ardb—antibiotic resistance genes database. Nucleic Acids Res. 2009, 37, D443–D447. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; de Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yamashita, A.; Sekizuka, T.; Kuroda, M. Characterization of Antimicrobial Resistance Dissemination across Plasmid Communities Classified by Network Analysis. Pathogens 2014, 3, 356-376. https://doi.org/10.3390/pathogens3020356
Yamashita A, Sekizuka T, Kuroda M. Characterization of Antimicrobial Resistance Dissemination across Plasmid Communities Classified by Network Analysis. Pathogens. 2014; 3(2):356-376. https://doi.org/10.3390/pathogens3020356
Chicago/Turabian StyleYamashita, Akifumi, Tsuyoshi Sekizuka, and Makoto Kuroda. 2014. "Characterization of Antimicrobial Resistance Dissemination across Plasmid Communities Classified by Network Analysis" Pathogens 3, no. 2: 356-376. https://doi.org/10.3390/pathogens3020356
APA StyleYamashita, A., Sekizuka, T., & Kuroda, M. (2014). Characterization of Antimicrobial Resistance Dissemination across Plasmid Communities Classified by Network Analysis. Pathogens, 3(2), 356-376. https://doi.org/10.3390/pathogens3020356




